Genotype by Prenatal Environment Interaction for Postnatal Growth of Nelore Beef Cattle Raised under Tropical Grazing Conditions
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Overview
2.2. Traits
2.3. Genotypes
2.4. Definition of the Prenatal Environment
2.5. Data Quality Control
2.6. Multiple-Trait Reaction Norm Model
2.7. Single-Step Genomic-Wide Association Study (ssGWAS)
2.8. Reaction Norms
3. Results
3.1. Effect of Gestational Environment on the Phenotypic Scale
3.2. Covariance Components and Genetic Parameters
3.3. Heritability Estimates
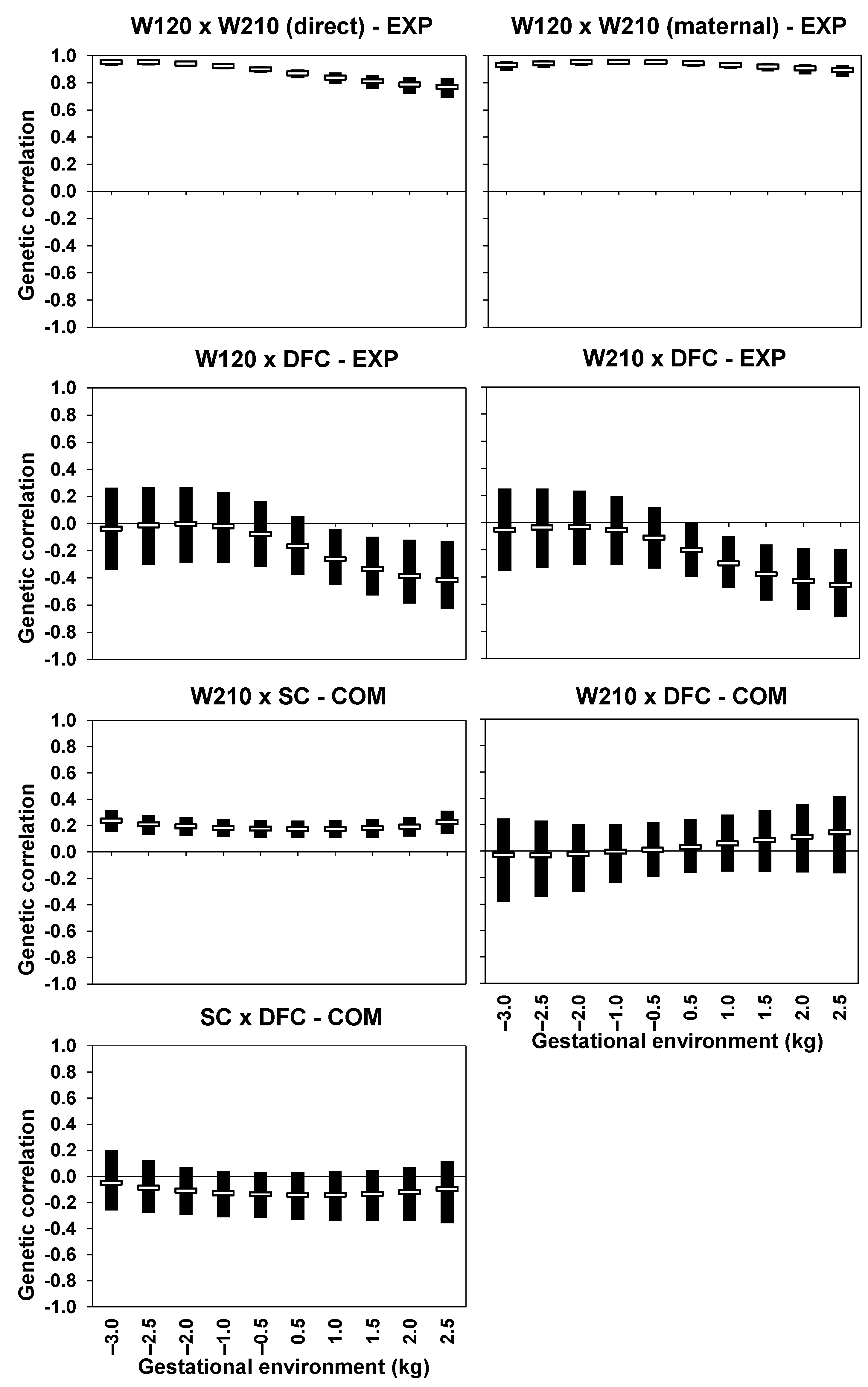
3.4. Intra-Trait Genetic Correlations
3.5. Inter-Trait Genetic Correlations
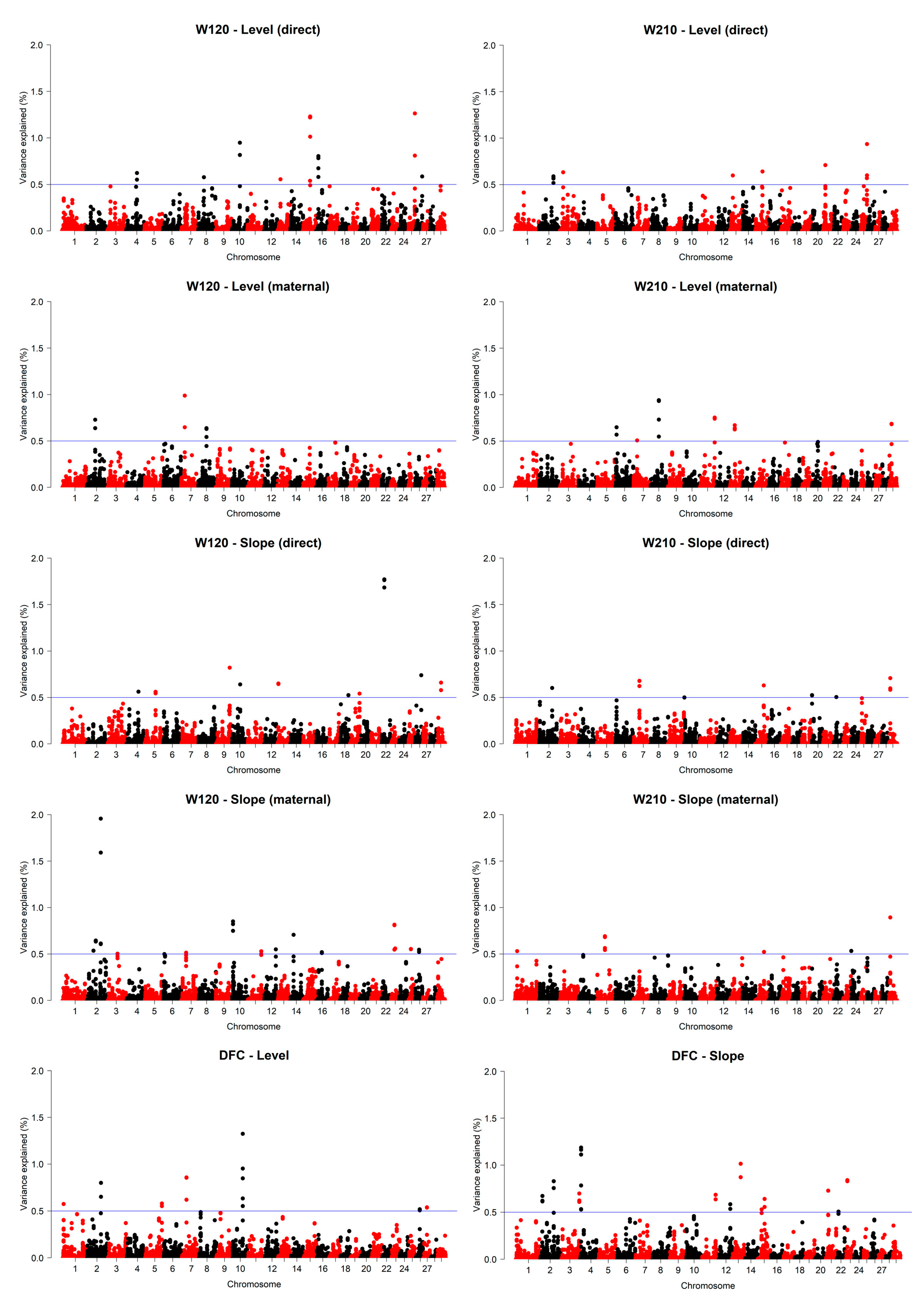
3.6. ssGWAS
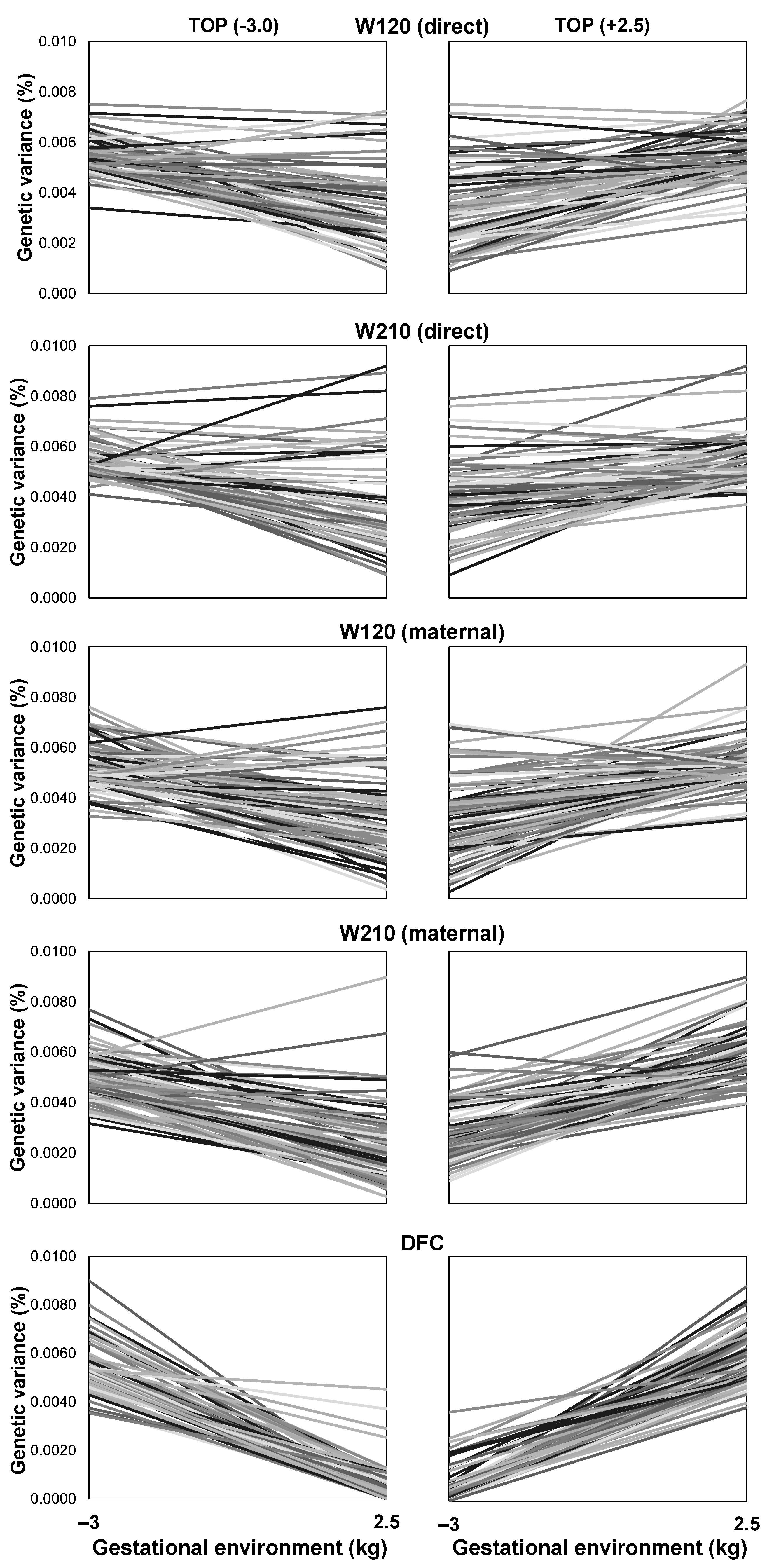
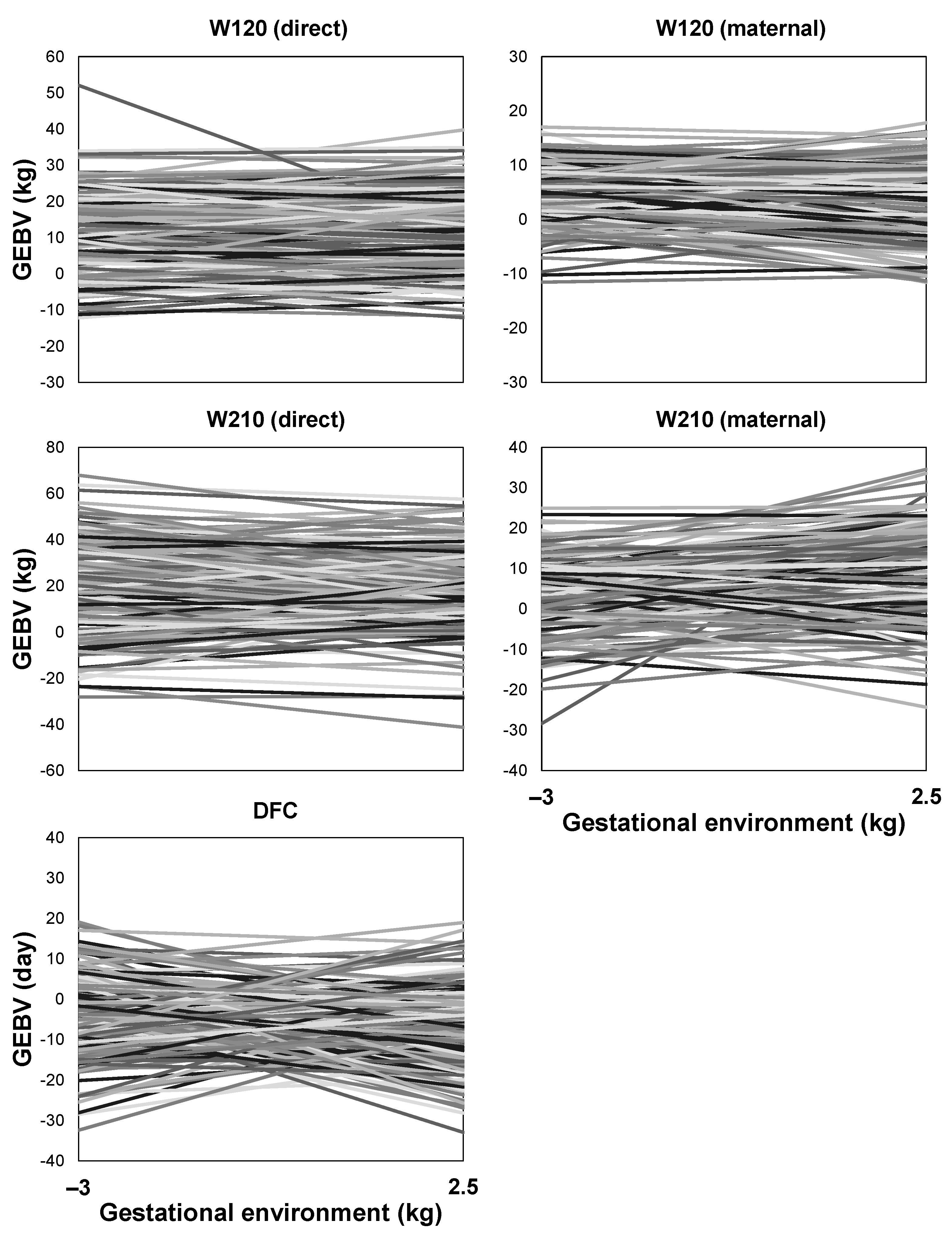
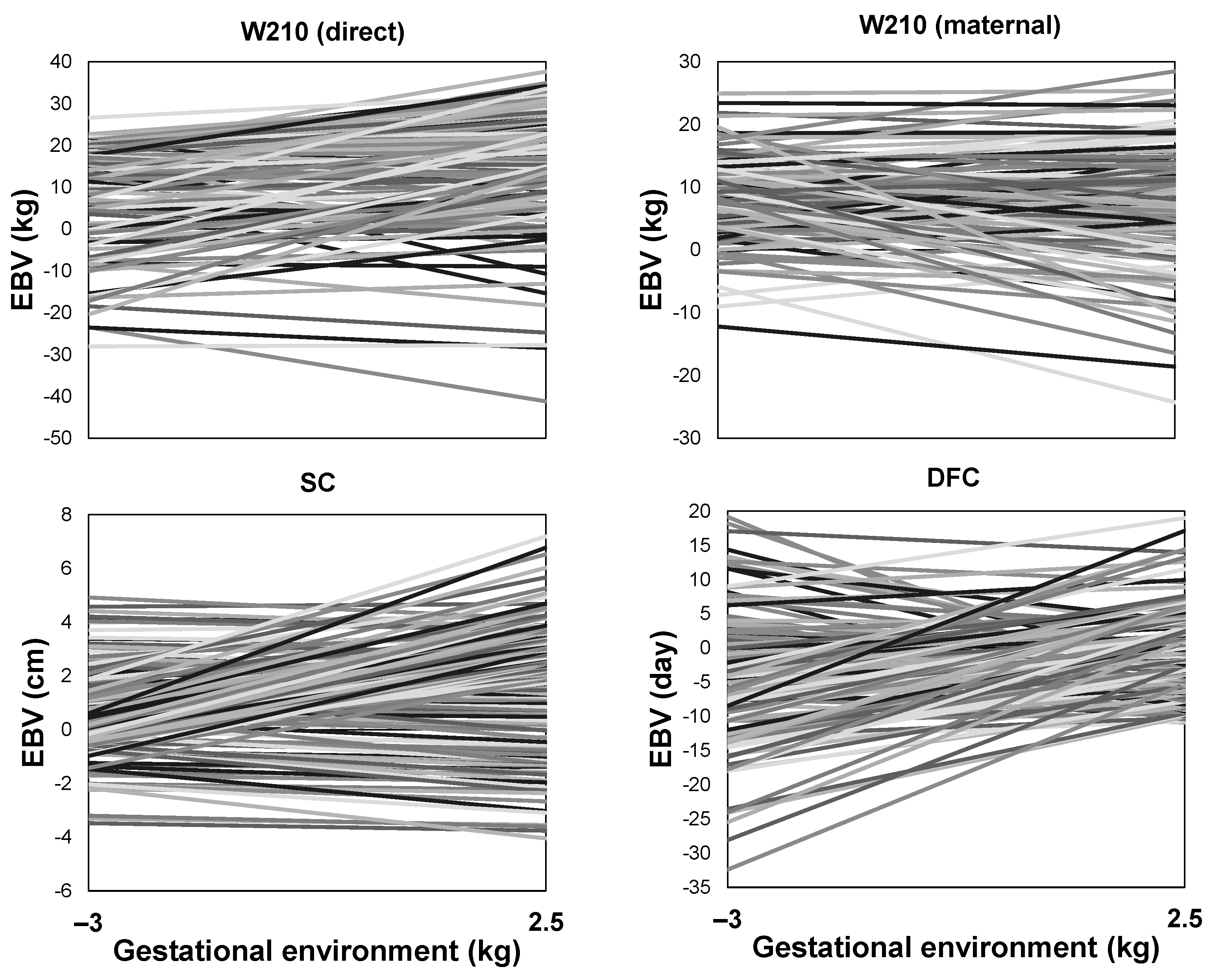
3.7. Reaction Norms
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement:
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Greenwood, P.L.; Cafe, L.M. Prenatal and Pre-Weaning Growth and Nutrition of Cattle: Long-Term Consequences for Beef Production. Animal 2007, 1, 1283–1296. [Google Scholar] [CrossRef] [PubMed]
- Funston, R.N.; Larson, D.M.; Vonnahme, K.A. Effects of Maternal Nutrition on Conceptus Growth and Offspring Performance: Implications for Beef Cattle Production. J. Anim. Sci. 2010, 88, E205–E215. [Google Scholar] [CrossRef]
- Tao, S.; Monteiro, A.P.A.; Thompson, I.M.; Hayen, M.J.; Dahl, G.E. Effect of Late-Gestation Maternal Heat Stress on Growth and Immune Function of Dairy Calves. J. Dairy Sci. 2012, 95, 7128–7136. [Google Scholar] [CrossRef]
- Greenwood, P.L.; Cafe, L.M.; Hearnshaw, H.; Hennessy, D.W. Consequences of Nutrition and Growth Retardation Early in Life for Growth and Composition of Cattle and Eating Quality of Beef. Recent Adv. Anim. Nutr. Aust. 2005, 15, 183–196. [Google Scholar]
- López Valiente, S.; Rodriguez, A.M.; Long, N.M.; Lacau-Mengido, I.M.; Maresca, S. The Degree of Maternal Nutrient Restriction during Late Gestation Influences the Growth and Endocrine Profiles of Offspring from Beef Cows. Anim. Prod. Sci. 2022, 62, 163–172. [Google Scholar] [CrossRef]
- Costa, T.C.; Du, M.; Nascimento, K.B.; Galvão, M.C.; Meneses, J.A.M.; Schultz, E.B.; Gionbelli, M.P.; Duarte, M.d.S. Skeletal Muscle Development in Postnatal Beef Cattle Resulting from Maternal Protein Restriction during Mid-Gestation. Animals 2021, 11, 860. [Google Scholar] [CrossRef] [PubMed]
- Micke, G.C.; Sullivan, T.M.; Soares Magalhaes, R.J.; Rolls, P.J.; Norman, S.T.; Perry, V.E.A. Heifer Nutrition during Early- and Mid-Pregnancy Alters Fetal Growth Trajectory and Birth Weight. Anim. Reprod. Sci. 2010, 117, 1–10. [Google Scholar] [CrossRef]
- Reynolds, L.P.; Caton, J.S.; Redmer, D.A.; Grazul-Bilska, A.T.; Vonnahme, K.A.; Borowicz, P.P.; Luther, J.S.; Wallace, J.M.; Wu, G.; Spencer, T.E. Evidence for Altered Placental Blood Flow and Vascularity in Compromised Pregnancies. J. Physiol. 2006, 572, 51–58. [Google Scholar] [CrossRef]
- Vonnahme, K.A.; Lemley, C.O. Programming the Offspring through Altered Uteroplacental Hemodynamics: How Maternal Environment Impacts Uterine and Umbilical Blood Flow in Cattle, Sheep and Pigs. Reprod. Fertil. Dev. 2012, 24, 97–104. [Google Scholar] [CrossRef]
- Sullivan, T.M.; Micke, G.C.; Greer, R.M.; Irving-Rodgers, H.F.; Rodgers, R.J.; Perry, V.E.A. Dietary Manipulation of Bos Indicus Heifers during Gestation Affects the Reproductive Development of Their Heifer Calves. Reprod. Fertil. Dev. 2009, 21, 773–784. [Google Scholar] [CrossRef]
- Evans, A.C.O.; Mossa, F.; Walsh, S.W.; Scheetz, D.; Jimenez-Krassel, F.; Ireland, J.L.H.; Smith, G.W.; Ireland, J.J. Effects of Maternal Environment during Gestation on Ovarian Folliculogenesis and Consequences for Fertility in Bovine Offspring. Reprod. Domest. Anim. 2012, 47, 31–37. [Google Scholar] [CrossRef] [PubMed]
- Monteiro, A.P.A.; Tao, S.; Thompson, I.M.; Dahl, G.E. Effect of Heat Stress during Late Gestation on Immune Function and Growth Performance of Calves: Isolation of Altered Colostral and Calf Factors. J. Dairy Sci. 2014, 97, 6426–6439. [Google Scholar] [CrossRef] [PubMed]
- Monteiro, A.P.A.; Guo, J.R.; Weng, X.S.; Ahmed, B.M.; Hayen, M.J.; Dahl, G.E.; Bernard, J.K.; Tao, S. Effect of Maternal Heat Stress during the Dry Period on Growth and Metabolism of Calves. J. Dairy Sci. 2016, 99, 3896–3907. [Google Scholar] [CrossRef] [PubMed]
- Skibiel, A.L.; Dado-Senn, B.; Fabris, T.F.; Dahl, G.E.; Laporta, J. In Utero Exposure to Thermal Stress Has Longterm Effects on Mammary Gland Microstructure and Function in Dairy Cattle. PLoS ONE 2018, 13, e0206046. [Google Scholar] [CrossRef] [PubMed]
- Laporta, J.; Ferreira, F.C.; Ouellet, V.; Dado-Senn, B.; Almeida, A.K.; De Vries, A.; Dahl, G.E. Late-Gestation Heat Stress Impairs Daughter and Granddaughter Lifetime Performance. J. Dairy Sci. 2020, 103, 7555–7568. [Google Scholar] [CrossRef]
- Duarte, M.S.; Gionbelli, M.P.; Paulino, P.V.R.; Serão, N.V.L.; Martins, T.S.; Tótaro, P.I.S.; Neves, C.A.; Valadares Filho, S.C.; Dodson, M.V.; Zhu, M.; et al. Effects of Maternal Nutrition on Development of Gastrointestinal Tract of Bovine Fetus at Different Stages of Gestation. Livest. Sci. 2013, 153, 60–65. [Google Scholar] [CrossRef]
- Alford, A.R.; Cafe, L.M.; Greenwood, P.L.; Griffith, G.R. Economic Effects of Nutritional Constraints Early in Life of Cattle. Anim. Prod. Sci. 2009, 49, 479–492. [Google Scholar] [CrossRef]
- Burrow, H.M. Importance of Adaptation and Genotype × Environment Interactions in Tropical Beef Breeding Systems. Animal 2012, 6, 729–740. [Google Scholar] [CrossRef]
- Holland, M.D.; Odde, K.G. Factors affecting calf birth weight: A review. Theriogenology 1992, 38, 769–798. [Google Scholar] [CrossRef]
- Chadio, S.; Kotsampasi, B. The Role of Early Life Nutrition in Programming of Reproductive Function. J. Dev. Orig. Health Dis. 2014, 5, 2–15. [Google Scholar] [CrossRef]
- Johnston, D.J.; Bunter, K.L. Days to Calving in Angus Cattle: Genetic and Environmental Effects, and Covariances with Other Traits. Livest. Prod. Sci. 1996, 45, 13–22. [Google Scholar] [CrossRef]
- Sargolzaei, M.; Chesnais, J.P.; Schenkel, F.S. A New Approach for Efficient Genotype Imputation Using Information from Relatives. BMC Genom. 2014, 15, 478. [Google Scholar] [CrossRef] [PubMed]
- de Paula Freitas, A.; Júnior, M.L.S.; Schenkel, F.S.; Mercadante, M.E.Z.; dos Santos Goncalves Cyrillo, J.N.; de Paz, C.C.P. Different Selection Practices Affect the Environmental Sensitivity of Beef Cattle. PLoS ONE 2021, 16, e0248186. [Google Scholar] [CrossRef] [PubMed]
- Aguilar, I.; Misztal, I.; Johnson, D.L.; Legarra, A.; Tsuruta, S.; Lawlor, T.J. Hot Topic: A Unified Approach to Utilize Phenotypic, Full Pedigree, and Genomic Information for Genetic Evaluation of Holstein Final Score. J. Dairy Sci. 2010, 93, 743–752. [Google Scholar] [CrossRef] [PubMed]
- Misztal, I.; Tsuruta, S.; Strabel, T.; Auvray, B.; Druet, T.; Lee, D.H. BLUPF90 and Related Programs (BGF90). In Proceedings of the 7th World Congress on Genetics Applied to Livestock Production, Montpellier, France, 19–23 August 2002; Volume 28. No. 07. [Google Scholar]
- Geweke, J. [Statistics, Probability and Chaos]: Comment: Inference and Prediction in the Presence of Uncertainty and Determinism. Stat. Sci. 1992, 7, 94–101. [Google Scholar] [CrossRef]
- Wang, H.; Misztal, I.; Aguilar, I.; Legarra, A.; Muir, W.M. Genome-Wide Association Mapping Including Phenotypes from Relatives without Genotypes. Genet. Res. 2012, 94, 73–83. [Google Scholar] [CrossRef] [PubMed]
- Aguilar, I.; Misztal, I.; Tsuruta, S.; Legarra, A.; Wang, H. PREGSF90-POSTGSF90: Computational Tools for the Implementation of Single-Step Genomic Selection and Genome-Wide Association with Ungenotyped Individuals in BLUPF90 Programs. In Proceedings of the 10th World Congress of Genetics Applied to Livestock Production, Vancouver, BC, Canada, 17–22 August 2014. [Google Scholar]
- Rosen, B.D.; Bickhart, D.M.; Schnabel, R.D.; Koren, S.; Elsik, C.G.; Tseng, E.; Rowan, T.N.; Low, W.Y.; Zimin, A.; Couldrey, C.; et al. De Novo Assembly of the Cattle Reference Genome with Single-Molecule Sequencing. Gigascience 2020, 9, giaa021. [Google Scholar] [CrossRef]
- Hu, Z.L.; Park, C.A.; Reecy, J.M. Building a Livestock Genetic and Genomic Information Knowledgebase through Integrative Developments of Animal QTLdb and CorrDB. Nucleic Acids Res. 2019, 47, D701–D710. [Google Scholar] [CrossRef] [PubMed]
- Roberts, J.; Funston, R.N.; Grings, E.E.; Petersen, M.K. TRIeNNIal RepRODUCTION SympOSIUm: Beef Heifer Development and Lifetime Productivity in Rangeland-Based Production Systems. J. Anim. Sci. 2016, 94, 2705–2715. [Google Scholar] [CrossRef]
- Robinson, D.L.; Café, L.M.; Greenwood, P.L. Meat Science and Muscle Biology Symposium: Developmental Programming in Cattle: Consequences for Growth, Efficiency, Carcass, Muscle, and Beef Quality Characteristics. J. Anim. Sci. 2013, 91, 1428–1442. [Google Scholar] [CrossRef]
- Sullivan, T.M.; Micke, G.C.; Greer, R.M.; Perry, V.E.A. Dietary Manipulation of Bos Indicus × Heifers during Gestation Affects the Prepubertal Reproductive Development of Their Bull Calves. Anim. Reprod. Sci. 2010, 118, 131–139. [Google Scholar] [CrossRef] [PubMed]
- Mossa, F.; Carter, F.; Walsh, S.W.; Kenny, D.A.; Smith, G.W.; Ireland, J.L.H.; Hildebrandt, T.B.; Lonergan, P.; Ireland, J.J.; Evans, A.C.O. Maternal Undernutrition in Cows Impairs Ovarian and Cardiovascular Systems in Their Offspring. Biol. Reprod. 2013, 88, 92. [Google Scholar] [CrossRef] [PubMed]
- Weller, M.M.D.C.A.; Fortes, M.R.S.; Marcondes, M.I.; Rotta, P.P.; Gionbeli, T.R.S.; Valadares Filho, S.C.; Campos, M.M.; Silva, F.F.; Silva, W.; Moore, S.; et al. Effect of Maternal Nutrition and Days of Gestation on Pituitary Gland and Gonadal Gene Expression in Cattle. J. Dairy Sci. 2016, 99, 3056–3071. [Google Scholar] [CrossRef]
- Harvey, K.M.; Cooke, R.F.; Marques, R.d.S. Supplementing Trace Minerals to Beef Cows during Gestation to Enhance Productive and Health Responses of the Offspring. Animals 2021, 11, 1159. [Google Scholar] [CrossRef]
- Chiaia, H.L.J.; de Lemos, M.V.A.; Venturini, G.C.; Aboujaoude, C.; Berton, M.P.; Feitosa, F.B.; Carvalheiro, R.; Albuquerque, L.G.; de Oliveira, H.N.; Baldi, F. Genotype × Environment Interaction for Age at First Calving, Scrotal Circumference, and Yearling Weight in Nellore Cattle Using Reaction Norms in Multitrait Random Regression Models. J. Anim. Sci. 2015, 93, 1503–1510. [Google Scholar] [CrossRef]
- Santana, M.L.; Eler, J.P.; Bignardi, A.B.; Menéndez-Buxadera, A.; Cardoso, F.F.; Ferraz, J.B.S. Multi-Trait Linear Reaction Norm Model to Describe the Pattern of Phenotypic Expression of Some Economic Traits in Beef Cattle across a Range of Environments. J. Appl. Genet. 2015, 56, 219–229. [Google Scholar] [CrossRef]
- Hay, E.H.; Roberts, A. Genomic Evaluation of Genotype by Prenatal Nutritional Environment Interaction for Maternal Traits in a Composite Beef Cattle Breed. Livest. Sci. 2019, 229, 118–125. [Google Scholar] [CrossRef]
- Alves, M.S.; Bignardi, A.B.; Zuim, D.M.; Silva, J.A.d.; Cardoso, M.G.R.; Piccoli, M.L.; Roso, V.M.; Carvalheiro, R.; Faro, L.E.; Pereira, R.J.; et al. Thermal Stress during Late Gestation Impairs Postnatal Growth and Provides Background for Genotype-Environment Interaction in Hereford-Braford and Angus-Brangus Cattle. Livest. Sci. 2022, 263, 105027. [Google Scholar] [CrossRef]
- Via, S.; Lande, R. Genotype-Environment Interaction and the Evolution of Phenotypic Plasticity. Evolution 1985, 39, 505. [Google Scholar] [CrossRef]
- Hay, E.H.; Roberts, A. Genotype × Prenatal and Post-Weaning Nutritional Environment Interaction in a Composite Beef Cattle Breed Using Reaction Norms and a Multi-Trait Model. J. Anim. Sci. 2018, 96, 444–453. [Google Scholar] [CrossRef]
- Boligon, A.A.; Mercadante, M.E.Z.; Forni, S.; Lôbo, R.B.; Albuquerque, L.G. Covariance Functions for Body Weight from Birth to Maturity in Nellore Cows. J. Anim. Sci. 2010, 88, 849–859. [Google Scholar] [CrossRef]
- Ribeiro, S.; Eler, J.P.; Pedrosa, V.B.; Rosa, G.J.M.; Ferraz, J.B.S.; Balieiro, J.C.C. Genotype×environment Interaction for Weaning Weight in Nellore Cattle Using Reaction Norm Analysis. Livest. Sci. 2015, 176, 40–46. [Google Scholar] [CrossRef]
- Carvalho Filho, I.; Silva, D.A.; Teixeira, C.S.; Silva, T.L.; Mota, L.F.M.; Albuquerque, L.G.; Carvalheiro, R. Heteroscedastic Reaction Norm Models Improve the Assessment of Genotype by Environment Interaction for Growth, Reproductive, and Visual Score Traits in Nellore Cattle. Animals 2022, 12, 2613. [Google Scholar] [CrossRef]
- Wang, X.; Lan, X.; Radunz, A.E.; Khatib, H. Maternal Nutrition during Pregnancy Is Associated with Differential Expression of Imprinted Genes and DNA Methyltranfereases in Muscle of Beef Cattle Offspring. J. Anim. Sci. 2015, 93, 35–40. [Google Scholar] [CrossRef]
- Polizel, G.H.G.; Espigolan, R.; Fantinato-Neto, P.; de Francisco Strefezzi, R.; Rangel, R.B.; de Carli, C.; Fernandes, A.C.; Dias, E.F.F.; Cracco, R.C.; de Almeida Santana, M.H. Different Prenatal Supplementation Strategies and Its Impacts on Reproductive and Nutrigenetics Assessments of Bulls in Finishing Phase. Vet. Res. Commun. 2022, 47, 457–471. [Google Scholar] [CrossRef]
- Polizel, G.H.G.; Fantinato-Neto, P.; Rangel, R.B.; Grigoletto, L.; Bussiman, F.d.O.; Cracco, R.C.; Garcia, N.P.; Ruy, I.M.; Ferraz, J.B.S.; Santana, M.H.d.A. Evaluation of Reproductive Traits and the Effect of Nutrigenetics on Bulls Submitted to Fetal Programming. Livest. Sci. 2021, 247, 104487. [Google Scholar] [CrossRef]
- Cardoso, F.F.; Tempelman, R.J. Linear Reaction Norm Models for Genetic Merit Prediction of Angus Cattle under Genotype by Environment Interaction. J. Anim. Sci. 2012, 90, 2130–2141. [Google Scholar] [CrossRef] [PubMed]
- Barcelos, S.d.S.; Nascimento, K.B.; Silva, T.E.d.; Mezzomo, R.; Alves, K.S.; de Souza Duarte, M.; Gionbelli, M.P. The Effects of Prenatal Diet on Calf Performance and Perspectives for Fetal Programming Studies: A Meta-Analytical Investigation. Animals 2022, 12, 2145. [Google Scholar] [CrossRef]
- Lima, A.O.; Malheiros, J.M.; Afonso, J.; Petrini, J.; Coutinho, L.L.; da Silva Diniz, W.J.; Bressani, F.A.; Tizioto, P.C.; de Oliveira, P.S.N.; Ribeiro, J.A.S.; et al. Prune Homolog 2 with BCH Domain (PRUNE2) Gene Expression Is Associated with Feed Efficiency-Related Traits in Nelore Steers. Mamm. Genome 2022, 33, 629–641. [Google Scholar] [CrossRef]
- Buss, C.E.; Afonso, J.; de Oliveira, P.S.N.; Petrini, J.; Tizioto, P.C.; Cesar, A.S.M.; Gustani-Buss, E.C.; Cardoso, T.F.; Rovadoski, G.A.; da Silva Diniz, W.J.; et al. Bivariate GWAS Reveals Pleiotropic Regions among Feed Efficiency and Beef Quality-Related Traits in Nelore Cattle. Mamm. Genome 2023, 34, 90–103. [Google Scholar] [CrossRef]
- So, E.Y.; Ouchi, T. BRAT1 Deficiency Causes Increased Glucose Metabolism and Mitochondrial Malfunction. BMC Cancer 2014, 14, 548. [Google Scholar] [CrossRef]
- Fortes, M.R.S.; Enculescu, C.; Neto, L.R.P.; Lehnert, S.A.; McCulloch, R.; Hayes, B. Candidate Mutations Used to Aid the Prediction of Genetic Merit for Female Reproductive Traits in Tropical Beef Cattle. Rev. Bras. Zootec. 2018, 47, 226. [Google Scholar] [CrossRef]
- Olivieri, B.F.; Mercadante, M.E.Z.; Cyrillo, J.N.D.S.G.; Branco, R.H.; Bonilha, S.F.M.; De Albuquerque, L.G.; De Oliveira Silva, R.M.; Baldi, F. Genomic Regions Associated with Feed Efficiency Indicator Traits in an Experimental Nellore Cattle Population. PLoS ONE 2016, 11, e0164390. [Google Scholar] [CrossRef] [PubMed]
- Littlejohn, B.P.; Price, D.M.; Neuendorff, D.A.; Carroll, J.A.; Vann, R.C.; Riggs, P.K.; Riley, D.G.; Long, C.R.; Welsh, T.H.; Rande, R.D. Prenatal Transportation Stress Alters Genome-Wide DNA Methylation in Suckling Brahman Bull Calves. J. Anim. Sci. 2018, 96, 5075–5099. [Google Scholar] [CrossRef] [PubMed]
- Li, R.W.; Li, C.; Gasbarre, L.C. The Vitamin D Receptor and Inducible Nitric Oxide Synthase Associated Pathways in Acquired Resistance to Cooperia Oncophora Infection in Cattle. Vet. Res. 2011, 42, 48. [Google Scholar] [CrossRef] [PubMed]
- Hoelker, M.; Salilew-Wondim, D.; Drillich, M.; Christine, G.B.; Ghanem, N.; Goetze, L.; Tesfaye, D.; Schellander, K.; Heuwieser, W. Transcriptional Response of the Bovine Endometrium and Embryo to Endometrial Polymorphonuclear Neutrophil Infiltration as an Indicator of Subclinical Inflammation of the Uterine Environment. Reprod. Fertil. Dev. 2012, 24, 778–793. [Google Scholar] [CrossRef]
- Liu, D.; Chen, Z.; Zhao, W.; Guo, L.; Sun, H.; Zhu, K.; Liu, G.; Shen, X.; Zhao, X.; Wang, Q.; et al. Genome-Wide Selection Signatures Detection in Shanghai Holstein Cattle Population Identified Genes Related to Adaption, Health and Reproduction Traits. BMC Genom. 2021, 22, 747. [Google Scholar] [CrossRef]
- Bédard, J.; Brûlé, S.; Price, C.A.; Silversides, D.W.; Lussier, J.G. Serine Protease Inhibitor-E2 (SERPINE2) Is Differentially Expressed in Granulosa Cells of Dominant Follicle in Cattle. Mol. Reprod. Dev. 2003, 64, 152–165. [Google Scholar] [CrossRef]
- Ramirez-Diaz, J.; Cenadelli, S.; Bornaghi, V.; Bongioni, G.; Montedoro, S.M.; Achilli, A.; Capelli, C.; Rincon, J.C.; Milanesi, M.; Passamonti, M.M.; et al. Identification of Genomic Regions Associated with Total and Progressive Sperm Motility in Italian Holstein Bulls. J. Dairy Sci. 2023, 106, 407–420. [Google Scholar] [CrossRef]
- Kelly, M.J.; Tume, R.K.; Fortes, M.; Thompson, J.M. Whole-Genome Association Study of Fatty Acid Composition in a Diverse Range of Beef Cattle Breeds. J. Anim. Sci. 2014, 92, 1895–1901. [Google Scholar] [CrossRef]
- Edea, Z.; Jung, K.S.; Shin, S.S.; Yoo, S.W.; Choi, J.W.; Kim, K.S. Signatures of Positive Selection Underlying Beef Production Traits in Korean Cattle Breeds. J. Anim. Sci. Technol. 2020, 62, 293–305. [Google Scholar] [CrossRef] [PubMed]



| Item | Experimental Data | ||||
|---|---|---|---|---|---|
| BW (kg) | W120 (kg) | W210 (kg) | SC (cm) | DFC (days) | |
| Animals in the pedigree, n | 10,350 | 9614 | 9573 | – | 2969 |
| Sires in the pedigree, n | 384 | 384 | 384 | – | 341 |
| Dams in the pedigree, n | 2540 | 2470 | 2469 | – | 1452 |
| Phenotypic records, n | 9816 | 9078 | 9003 | – | 2222 |
| Sires with progeny record, n | 370 | 370 | 370 | – | 306 |
| Dams with progeny record, n | 2519 | 2446 | 2447 | – | 1289 |
| Genotyped sires with progeny record, n | 116 | 116 | 116 | – | 70 |
| Genotyped dams with progeny record, n | 370 | 365 | 366 | – | 158 |
| Progeny records from genotyped sire, n | 3328 | 3077 | 3058 | – | 572 |
| Progeny records from genotyped dams, n | 1299 | 1199 | 1194 | – | 199 |
| Genotyped animals with phenotypic records, n | 1516 | 1503 | 1502 | – | 258 |
| Average of the trait | 28.92 | 122.91 | 188.99 | – | 347.80 |
| Standard deviation of the trait | 5.43 | 19.94 | 31.02 | – | 35.81 |
| Company data | |||||
| Animals in the pedigree, n | 356,730 | – | 216,707 | 120,619 | 25,072 |
| Sires in the pedigree, n | 3060 | – | 2476 | 2022 | 1318 |
| Dams in the pedigree, n | 133,863 | – | 99,553 | 67,291 | 35,950 |
| Phenotypic records, n | 287,705 | – | 146,020 | 52,259 | 22,405 |
| Sires with progeny record, n | 2342 | – | 1817 | 1418 | 765 |
| Dams with progeny record, n | 95,820 | – | 65,379 | 33,588 | 17,391 |
| Average of the trait | 31.87 | – | 188.65 | 27.62 | 340.47 |
| Standard deviation of the trait | 3.96 | – | 27.53 | 3.28 | 27.22 |
| Trait | Parameter | Experimental Data | Company Data | ||||
|---|---|---|---|---|---|---|---|
| Mean | SD | HPD95% | Mean | SD | HPD95% | ||
| W120 | 107.633 | 12.570 | 83.951 to 133.148 | – | – | – | |
| 87.426 | 13.384 | 62.855 to 115.186 | – | – | – | ||
| 53.373 | 5.747 | 42.060 to 64.750 | – | – | – | ||
| 0.166 | 0.019 | 0.129 to 0.202 | – | – | – | ||
| 110.854 | 4.826 | 101.550 to 120.200 | – | – | – | ||
| 0.177 | 0.094 | −0.016 to 0.353 | – | – | – | ||
| 0.215 | 0.129 | −0.051 to 0.460 | – | – | – | ||
| 0.164 | 0.045 | 0.083 to 0.260 | – | – | – | ||
| 0.170 | 0.044 | 0.099 to 0.267 | – | – | – | ||
| W210 | 286.698 | 30.428 | 230.108 to 349.021 | 98.198 | 5.570 | 87.560 to 109.307 | |
| 210.659 | 30.545 | 153.761 to 273.906 | 38.787 | 3.560 | 32.170 to 46.338 | ||
| 120.340 | 12.948 | 95.290 to 146.20 | 57.233 | 1.877 | 53.550 to 60.990 | ||
| 0.163 | 0.018 | 0.127 to 0.200 | 0.150 | 0.005 | 0.140 to 0.160 | ||
| 232.043 | 11.000 | 210.500 to 253.750 | 189.93 | 2.575 | 184.900 to 194.800 | ||
| −0.063 | 0.087 | −0.235 to 0.103 | 0.607 | 0.045 | 0.526 to 0.708 | ||
| 0.198 | 0.128 | −0.064 to 0.441 | 0.392 | 0.085 | 0.219 to 0.546 | ||
| 0.190 | 0.045 | 0.108 to 0.287 | 0.112 | 0.021 | 0.073 to 0.157 | ||
| 0.159 | 0.038 | 0.095 to 0.243 | 0.120 | 0.026 | 0.072 to 0.174 | ||
| SC | – | – | – | 38.837 | 1.485 | 35.978 to 41.789 | |
| – | – | – | 3.612 | 0.911 | 3.434 to 3.790 | ||
| – | – | – | 0.313 | 0.057 | 0.201 to 0.419 | ||
| – | – | – | 0.029 | 0.006 | 0.019 to 0.042 | ||
| DFC | 62.612 | 15.708 | 35.954 to 96.952 | 16.832 | 3.742 | 10.873 to 25.451 | |
| 222.314 | 9.133 | 204.400 to 240.700 | 239.606 | 3.124 | 233.100 to 245.400 | ||
| −0.073 | 0.188 | −0.422 to 0.311 | 0.550 | 0.130 | 0.286 to 0.754 | ||
| 0.651 | 0.230 | 0.309 to 1.195 | 0.484 | 0.210 | 0.181 to 0.990 | ||
| Experimental Data (EXP) | ||||
|---|---|---|---|---|
| W120 (direct effects: below diagonal, maternal above) | ||||
| GE | −2.5 | −1.0 | +1.0 | +2.5 |
| −2.5 | 1 | 0.901 (0.03) | 0.622 (0.08) | 0.343 (0.11) |
| −1.0 | 0.909 (0.02) | 1 | 0.899 (0.03) | 0.713 (0.06) |
| +1.0 | 0.639 (0.08) | 0.900 (0.03) | 1 | 0.947 (0.01) |
| +2.5 | 0.357 (0.12) | 0.710 (0.07) | 0.946 (0.01) | 1 |
| W210 (direct effects: below diagonal, maternal above) | ||||
| −2.5 | 1 | 0.908 (0.02) | 0.645 (0.07) | 0.371 (0.10) |
| −1.0 | 0.929 (0.01) | 1 | 0.904 (0.02) | 0.722 (0.06) |
| +1.0 | 0.646 (0.07) | 0.882 (0.03) | 1 | 0.948 (0.01) |
| +2.5 | 0.284 (0.11) | 0.617 (0.07) | 0.914 (0.02) | 1 |
| DFC (above) | ||||
| −2.5 | 1 | 0.885 (0.04) | 0.241 (0.17) | −0.294 (0.16) |
| −1.0 | – | 1 | 0.657 (0.10) | 0.172 (0.17) |
| +1.0 | – | – | 1 | 0.849 (0.05) |
| +2.5 | – | – | – | 1 |
| Company data (COM) | ||||
| W210 (direct effects: below diagonal, maternal above) | ||||
| −2.5 | 1 | 0.957 (0.01) | 0.869 (0.03) | 0.640 (0.06) |
| −1.0 | 0.959 (0.01) | 1 | 0.975 (0.01) | 0.834 (0.03) |
| +1.0 | 0.885 (0.03) | 0.889 (0.02) | 1 | 0.936 (0.01) |
| +2.5 | 0.722 (0.06) | 0.885 (0.03) | 0.961 (0.01) | 1 |
| SC (below), DFC (above) | ||||
| −2.5 | 1 | 0.818 (0.07) | 0.502 (0.18) | 0.102 (0.25) |
| −1.0 | 0.991 (0.00) | 1 | 0.904 (0.05) | 0.640 (0.14) |
| +1.0 | 0.969 (0.01) | 0.994 (0.00) | 1 | 0.904 (0.04) |
| +2.5 | 0.892 (0.02) | 0.945 (0.01) | 0.976 (0.01) | 1 |
| Trait | BTA | Position (bp) | Var (%) | Candidate Genes | QTL |
|---|---|---|---|---|---|
| W120 (direct) | 25 | 40,619,157–41,019,157 | 1.26 | GNA12, AMZ1, BRAT1, bta-mir-11980, IQCE, TTYH3, LFNG, bta-mir-12029, GRIFIN, CHST12, bta-mir-12019, EIF3B, SNX8, NUDT1, MRM2, MAD1L1 | Residual feed intake, conception rate, milking speed, average daily gain |
| 15 | 45,597,833–45,997,833 | 1.23 | RBMXL2, NLRP14, ZNF214, ZNF215, OR2D3, OR2D2, OR10A4, OR10A5, U6, OR10A5L, OR10A5G, OR6A2, OR6B18, OR6B17, OR2D4 | Milk-fat yield, body weight gain | |
| 10 | 52,283,853–52,683,853 | 0.95 | ALDH1A2, U6, POLR2M, MYZAP | Carcass weight, milk butyric acid content, milking speed, marbling score, m. paratuberculosis susceptibility, shear force, milk kappa-casein percentage, omega-6 to omega-3 fatty acid ratio, age at puberty | |
| W120 (maternal) | 7 | 21,907,628–22,307,628 | 0.99 | IRF1, SLC22A5, SLC22A4, bta-mir-2457, PDLIM4, P4HA2, bta-mir-12040 | Milking speed, m. paratuberculosis susceptibility, calving ease (maternal), daughter pregnancy rate, stillbirth (maternal), udder attachment, net merit, length of productive life, somatic cell score, stillbirth, udder depth |
| 2 | 54,729,312–55,129,312 | 0.73 | - | milk palmitoleic acid content, inseminations per conception, bovine respiratory disease susceptibility | |
| 8 | 52,728,463–53,128,463 | 0.64 | PRUNE2, FOXB2 | Residual feed intake, twinning, milk unglycosylated kappa-casein percentage, milk kappa-casein percentage, bovine tuberculosis susceptibility, milk protein yield | |
| W210 (direct) | 25 | 40,6191,57–41,019,157 | 0.94 | GNA12, AMZ1, BRAT1, bta-mir-11980, IQCE, TTYH3, LFNG, bta-mir-12029, GRIFIN, CHST12, bta-mir-12019, EIF3B, SNX8, NUDT1, MRM2, MAD1L1 | Residual feed intake, conception rate, milking speed, average daily gain |
| 21 | 14,794,265–15,194,265 | 0.71 | SLCO3A1 | Bovine tuberculosis susceptibility, age at first calving, kidney, pelvic, heart fat percentage, milk tridecylic acid content, somatic cell score | |
| 15 | 45,597,833–45,997,833 | 0.64 | RBMXL2, NLRP14, ZNF214, ZNF215, OR2D3, OR2D2, OR10A4, OR10A5, U6, OR10A5L, OR10A5G, OR6A2, OR6B18, OR6B17, OR2D4 | Milk-fat yield, body weight gain | |
| W210 (maternal) | 8 | 52,728,463–53,128,463 | 0.94 | PRUNE2, FOXB2 | Residual feed intake, twinning, milk unglycosylated kappa-casein percentage, milk kappa-casein percentage, bovine tuberculosis susceptibility, milk protein yield |
| 11 | 94,730,926–95,130,926 | 0.75 | DENND1A, LHX2 | cheese protein recovery, number of embryos, milk beta-lactoglobulin percentage, anti-müllerian hormone level, non-return rate | |
| 29 | 15,282,330–15,682,330 | 0.68 | - | milk kappa-casein percentage, milk glycosylated kappa-casein percentage, milk unglycosylated kappa-casein percentage, somatic cell score | |
| DFC | 10 | 66,575,785–66,975,785 | 1.32 | CDKN3, CNIH1 | Milking speed, bovine tuberculosis susceptibility, milk protein yield, body depth, PTA type, udder attachment, udder height, rump width, somatic cell score, stature, strength, udder depth |
| 7 | 29,486,606–29,886,606 | 0.86 | - | Clinical mastitis, body weight (birth), milk protein yield, body capacity, daughter pregnancy rate, stillbirth (maternal), udder attachment, length of productive life, somatic cell score, stillbirth, udder depth | |
| 2 | 89,254,217–89,654,217 | 0.80 | AOX4, AOX2, BZW1, CLK1, PPIL3, NIF3L1, ORC2, FAM126B, U6 | Conception rate, intramuscular fat, milk-fat yield |
| Trait | BTA | Position (bp) | Var (%) | Genes | QTL |
|---|---|---|---|---|---|
| W120 (direct) | 22 | 17,317,952–17,717,952 | 1.77 | SRGAP3, RAD18 | Milk tridecylic acid content, body weight (yearling), lean-meat yield, white spotting |
| 9 | 89,328,896–89,728,896 | 0.82 | 7SK, MYCT1, VIP | muscle magnesium content, muscle phosphorus content, teat placement—front, teat placement—rear, teat length, udder cleft | |
| 26 | 40,536,474–40,936,474 | 0.74 | PLPP4, WDR11 | milk-fat yield, stature, milk c14 index, milk myristoleic acid content, milk yield, milk protein yield | |
| W120 (maternal) | 2 | 89,254,217–89,654,217 | 1.96 | AOX4, AOX2, BZW1, CLK1, PPIL3, NIF3L1, ORC2, FAM126B, U6 | Conception rate, intramuscular fat, milk-fat yield |
| 10 | 5,768,117–6,168,117 | 0.85 | - | body weight (yearling), body weight gain, udder depth, conception rate | |
| 23 | 22,140,708–22,540,708 | 0.81 | MMUT, CENPQ, GLYATL3, C23H6orf141, U6, RHAG, CRISP2, CRISP3, 7SK, CRISP1 | milk protein percentage, milk glycosylated kappa-casein percentage, milk iron content, length of productive life, daughter pregnancy rate, stillbirth (maternal), calving ease, somatic cell score | |
| W210 (direct) | 29 | 6,736,030–7,136,030 | 0.71 | GRM5 | Shear force |
| 7 | 36,882,466–37,282,466 | 0.68 | - | Milk alpha-s1-casein percentage, milking speed, intramuscular fat | |
| 15 | 54,575,598–54,975,598 | 0.63 | RPS3, SNORD15, KLHL35, GDPD5, SERPINH1, MAP6, MOGAT2 | Milk-fat yield, first service conception, inseminations per conception, 305-day milk yield, milk rennet coagulation time, bovine respiratory disease susceptibility, conception rate | |
| W210 (maternal) | 29 | 6,736,030–7,136,030 | 0.89 | GRM5 | Shear force |
| 5 | 50,640,891–51,040,891 | 0.69 | PPM1H, MON2 | Milk-fat yield, milk yield, inhibin level, insulin-like growth factor 1 level, intramuscular fat | |
| 24 | 1,244,237–1,644,237 | 0.53 | - | Body weight (yearling) | |
| DFC | 11 | 76,492,925–76,892,925 | 1.09 | - | Body weight (yearling), milk alpha-lactalbumin percentage, lean-meat yield |
| 2 | 111,967,207–112,367,207 | 1.04 | WDFY1, U6, MRPL44, SERPINE2 | Metabolic body weight, fecal larva count, first service conception, heat tolerance, fertilization rate, conception rate, milk protein percentage, milk-fat yield | |
| 1 | 149,053,882–149,453,882 | 0.84 | HLCS, RIPPLY3, U6, PIGP, TTC3 | Conception rate, somatic cell score, teat length, length of productive life |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Santana, M.L.; Bignardi, A.B.; Pereira, R.J.; Oliveira Junior, G.A.; Freitas, A.P.; Carvalheiro, R.; Eler, J.P.; Ferraz, J.B.S.; Cyrillo, J.N.S.G.; Mercadante, M.E.Z. Genotype by Prenatal Environment Interaction for Postnatal Growth of Nelore Beef Cattle Raised under Tropical Grazing Conditions. Animals 2023, 13, 2321. https://doi.org/10.3390/ani13142321
Santana ML, Bignardi AB, Pereira RJ, Oliveira Junior GA, Freitas AP, Carvalheiro R, Eler JP, Ferraz JBS, Cyrillo JNSG, Mercadante MEZ. Genotype by Prenatal Environment Interaction for Postnatal Growth of Nelore Beef Cattle Raised under Tropical Grazing Conditions. Animals. 2023; 13(14):2321. https://doi.org/10.3390/ani13142321
Chicago/Turabian StyleSantana, Mário L., Annaiza B. Bignardi, Rodrigo J. Pereira, Gerson A. Oliveira Junior, Anielly P. Freitas, Roberto Carvalheiro, Joanir P. Eler, José B. S. Ferraz, Joslaine N. S. G. Cyrillo, and Maria E. Z. Mercadante. 2023. "Genotype by Prenatal Environment Interaction for Postnatal Growth of Nelore Beef Cattle Raised under Tropical Grazing Conditions" Animals 13, no. 14: 2321. https://doi.org/10.3390/ani13142321
APA StyleSantana, M. L., Bignardi, A. B., Pereira, R. J., Oliveira Junior, G. A., Freitas, A. P., Carvalheiro, R., Eler, J. P., Ferraz, J. B. S., Cyrillo, J. N. S. G., & Mercadante, M. E. Z. (2023). Genotype by Prenatal Environment Interaction for Postnatal Growth of Nelore Beef Cattle Raised under Tropical Grazing Conditions. Animals, 13(14), 2321. https://doi.org/10.3390/ani13142321






