Identification and Characterization of the Very-Low-Density Lipoprotein Receptor Gene from Branchiostoma belcheri: Insights into the Origin and Evolution of the Low-Density Lipoprotein Receptor Gene Family
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Amphioxus Cultivation
2.2. Cloning the Full-Length cDNA of the AmphiVLDLR Gene
2.3. Data Collection, Sequence Alignment, Phylogenetic Analysis, Selective Pressure Analysis, and Gene Annotation
2.4. Real-Time Quantitative PCR Analysis of AmphiVLDLR
3. Results
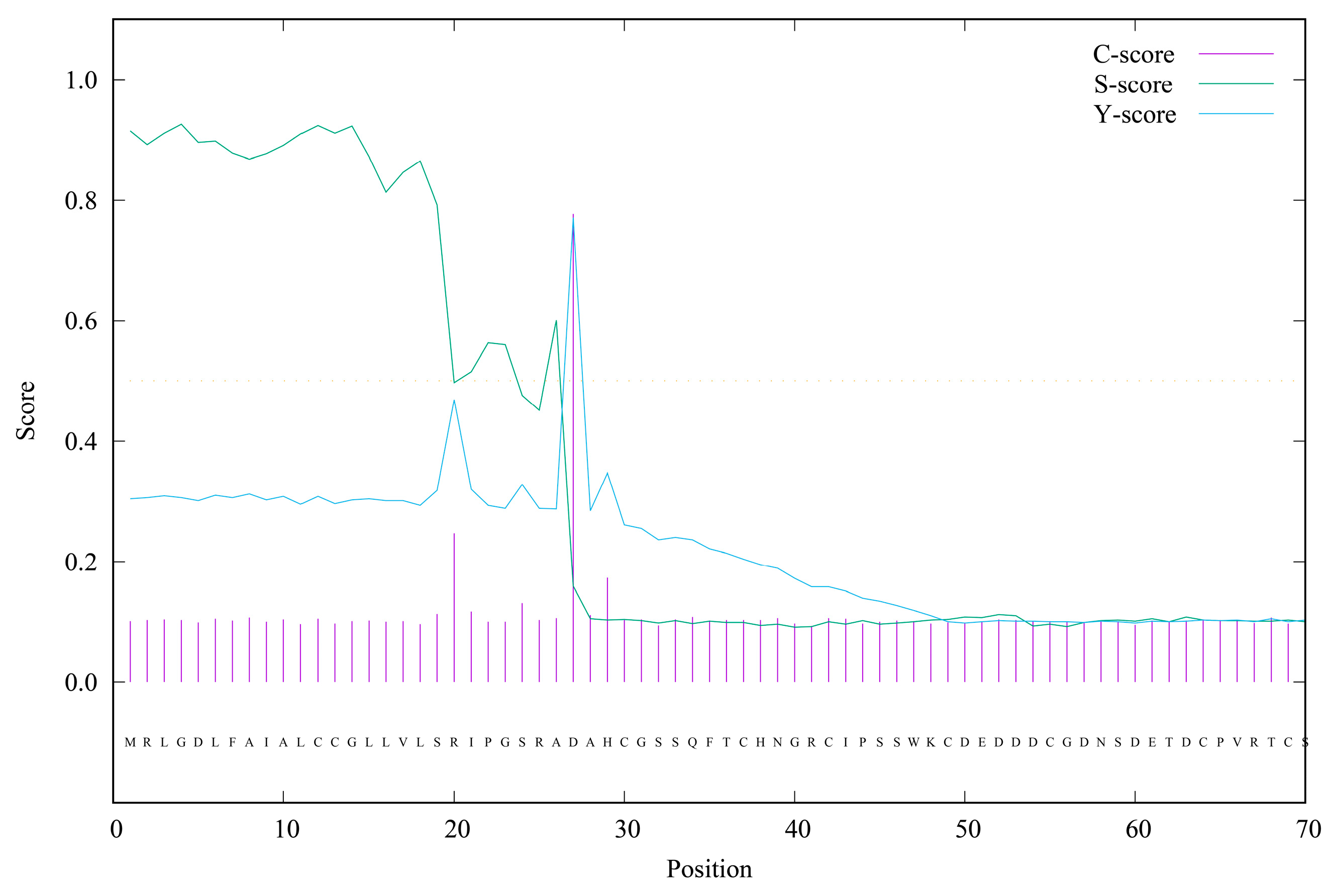
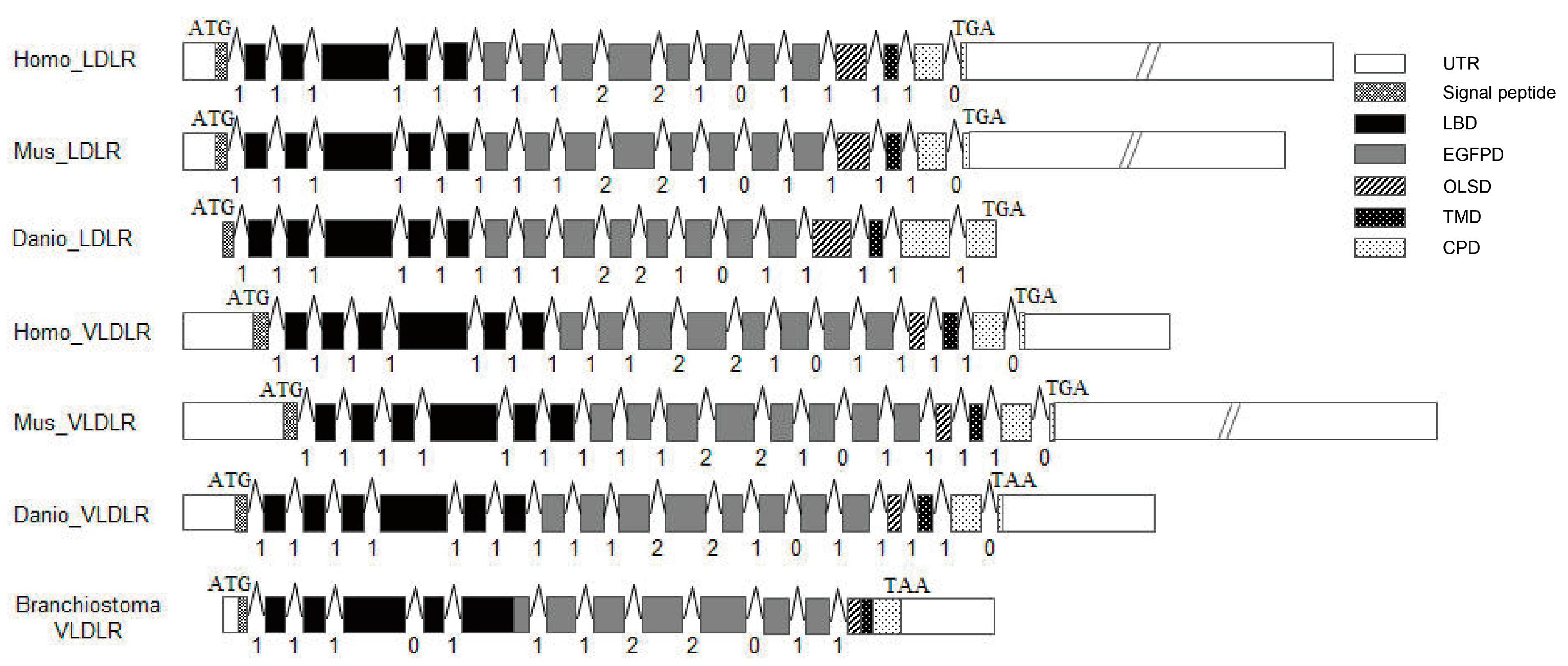
3.1. Cloning and Characterization of the AmphiVLDLR Gene
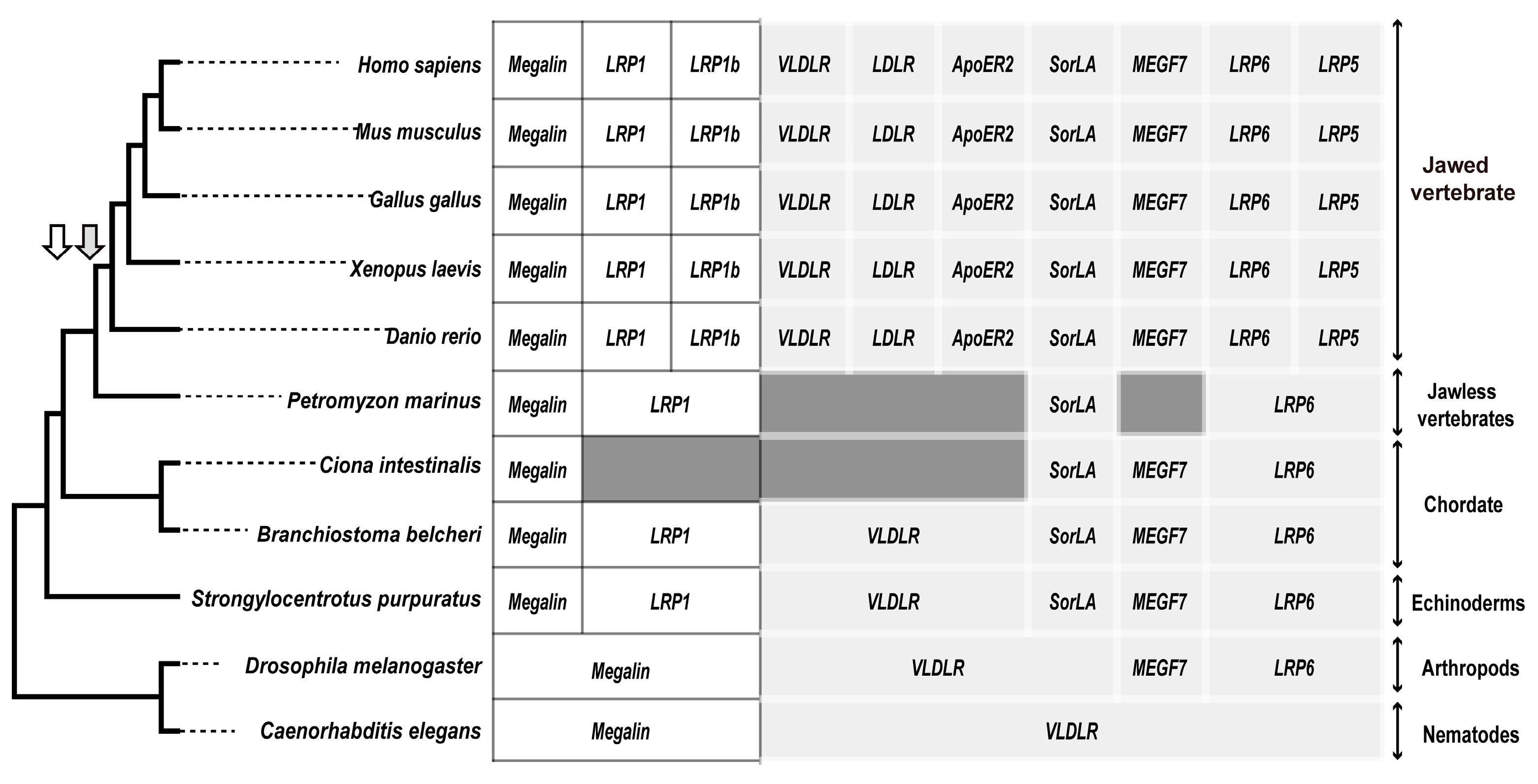
3.2. The Presence of LDLR Family Genes in Different Animals
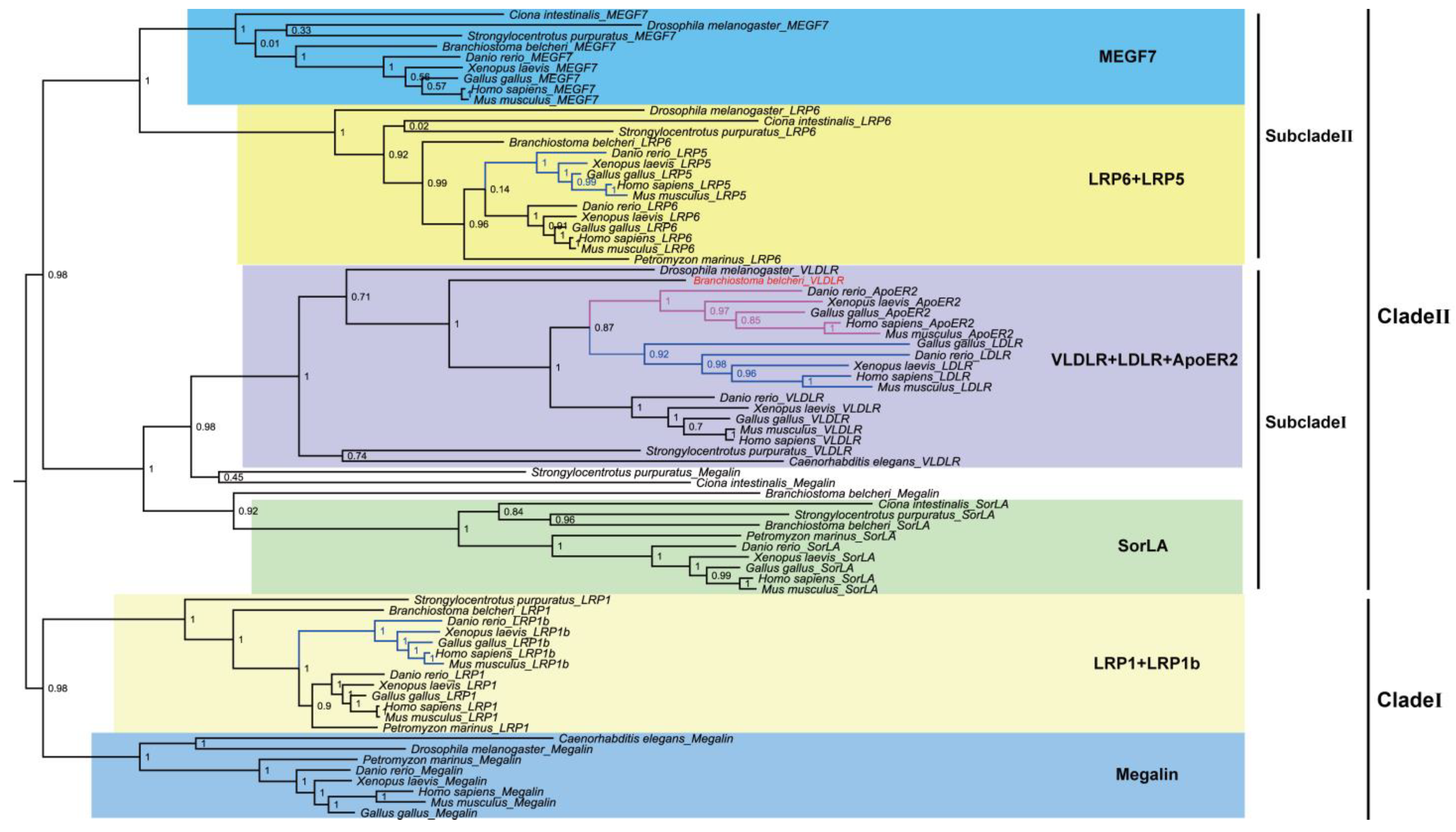
3.3. Alignments and Phylogenetic Analysis of AmphiVLDLR
3.4. AmphiVLDLR Expression in Different Tissues
4. Discussion
4.1. AmphiVLDLR Is a New Member of the VLDLR Family
4.2. Evolution of LDLR Family Genes
4.3. VLDLR Tissue Expression Pattern
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Takahashi, S.; Kawarabayasi, Y.; Nakai, T.; Sakai, J.; Yamamoto, T. Rabbit very low density lipoprotein receptor: A low density lipoprotein receptor-like protein with distinct ligand specificity. Proc. Natl. Acad. Sci. USA 1992, 89, 9252–9256. [Google Scholar] [CrossRef]
- Ashrafi, K.; Chang, F.Y.; Watts, J.L.; Fraser, A.G.; Kamath, R.S.; Ahringer, J.; Ruvkun, G. Genome-wide RNAi analysis of Caenorhabditis elegans fat regulatory genes. Nature 2003, 421, 268–272. [Google Scholar] [CrossRef] [PubMed]
- Agulleiro, M.J.; Andre, M.; Morais, S.; Cerda, J.; Babin, P.J. High transcript level of fatty acid-binding protein 11 but not of very low-density lipoprotein receptor is correlated to ovarian follicle atresia in a teleost fish (Solea senegalensis). Biol. Reprod. 2007, 77, 504–516. [Google Scholar] [CrossRef] [PubMed]
- Imai, H.; Oomiya, Y.; Kikkawa, S.; Shoji, W.; Hibi, M.; Terashima, T.; Katsuyama, Y. Dynamic changes in the gene expression of zebrafish Reelin receptors during embryogenesis and hatching period. Dev. Growth Differ. 2012, 54, 253–263. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Li, S.J.; Yu, W.H.; Xin, Q.W.; Li, C.; Feng, Y.P.; Peng, X.L.; Gong, Y.Z. Cloning and expression profiling of the VLDLR gene associated with egg performance in duck (Anas platyrhynchos). Genet. Sel. Evol. 2011, 43, 29. [Google Scholar] [CrossRef]
- Bujo, H.; Hermann, M.; Kaderli, M.O.; Jacobsen, L.; Sugawara, S.; Nimpf, J.; Yamamoto, T.; Schneider, W.J. Chicken oocyte growth is mediated by an eight ligand binding repeat member of the LDL receptor family. EMBO J. 1994, 13, 5165–5175. [Google Scholar] [CrossRef]
- Tiebel, O.; Oka, K.; Robinson, K.; Sullivan, M.; Martinez, J.; Nakamuta, M.; Ishimura-Oka, K.; Chan, L. Mouse very low-density lipoprotein receptor (VLDLR): Gene structure, tissue-specific expression and dietary and developmental regulation. Atherosclerosis 1999, 145, 239–251. [Google Scholar] [CrossRef]
- Sakai, J.; Hoshino, A.; Takahashi, S.; Miura, Y.; Ishii, H.; Suzuki, H.; Kawarabayasi, Y.; Yamamoto, T. Structure, chromosome location, and expression of the human very low density lipoprotein receptor gene. J. Biol. Chem. 1994, 269, 2173–2182. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, S.; Suzuki, J.; Kohno, M.; Oida, K.; Tamai, T.; Miyabo, S.; Yamamoto, T.; Nakai, T. Enhancement of the binding of triglyceride-rich lipoproteins to the very low density lipoprotein receptor by apolipoprotein E and lipoprotein lipase. J. Biol. Chem. 1995, 270, 15747–15754. [Google Scholar] [CrossRef]
- Webb, J.C.; Patel, D.D.; Jones, M.D.; Knight, B.L.; Soutar, A.K. Characterization and tissue-specific expression of the human ‘very low density lipoprotein (VLDL) receptor’ mRNA. Hum. Mol. Genet. 1994, 3, 531–537. [Google Scholar] [CrossRef]
- Willnow, T.E. The low-density lipoprotein receptor gene family: Multiple roles in lipid metabolism. J. Mol. Med. (Berl.) 1999, 77, 306–315. [Google Scholar] [CrossRef]
- He, L.; Lu, Y.; Wang, P.; Zhang, J.; Yin, C.; Qu, S. Up-regulated expression of type II very low density lipoprotein receptor correlates with cancer metastasis and has a potential link to beta-catenin in different cancers. BMC Cancer 2010, 10, 601. [Google Scholar] [CrossRef]
- Li, Y.; Lu, W.; Marzolo, M.P.; Bu, G. Differential functions of members of the low density lipoprotein receptor family suggested by their distinct endocytosis rates. J. Biol. Chem. 2001, 276, 18000–18006. [Google Scholar] [CrossRef] [PubMed]
- Wagner, T.; Pietrzik, C.U. The role of lipoprotein receptors on the physiological function of APP. Exp. Brain. Res. 2012, 217, 377–387. [Google Scholar] [CrossRef] [PubMed]
- Krieger, M.; Herz, J. Structures and functions of multiligand lipoprotein receptors: Macrophage scavenger receptors and LDL receptor-related protein (LRP). Annu. Rev. Biochem. 1994, 63, 601–637. [Google Scholar] [CrossRef] [PubMed]
- Strickland, D.K.; Kounnas, M.Z.; Argraves, W.S. LDL receptor-related protein: A multiligand receptor for lipoprotein and proteinase catabolism. FASEB J. 1995, 9, 890–898. [Google Scholar] [CrossRef]
- Hussain, M.M.; Strickland, D.K.; Bakillah, A. The mammalian low-density lipoprotein receptor family. Annu. Rev. Nutr. 1999, 19, 141–172. [Google Scholar] [CrossRef]
- Herz, J.; Gotthardt, M.; Willnow, T.E. Cellular signalling by lipoprotein receptors. Curr. Opin. Lipidol. 2000, 11, 161–166. [Google Scholar] [CrossRef] [PubMed]
- Herz, J.; Bock, H.H. Lipoprotein receptors in the nervous system. Annu. Rev. Biochem. 2002, 71, 405–434. [Google Scholar] [CrossRef] [PubMed]
- Herz, J.; Strickland, D.K. LRP: A multifunctional scavenger and signaling receptor. J. Clin. Invest. 2001, 108, 779–784. [Google Scholar] [CrossRef]
- May, P.; Herz, J. LDL receptor-related proteins in neurodevelopment. Traffic 2003, 4, 291–301. [Google Scholar] [CrossRef] [PubMed]
- Schneider, W.J.; Nimpf, J. LDL receptor relatives at the crossroad of endocytosis and signaling. Cell Mol. Life. Sci. 2003, 60, 892–903. [Google Scholar] [CrossRef] [PubMed]
- Strickland, D.K.; Ranganathan, S. Diverse role of LDL receptor-related protein in the clearance of proteases and in signaling. J. Thromb. Haemost. 2003, 1, 1663–1670. [Google Scholar] [CrossRef]
- Rodenburg, K.W.; Van der Horst, D.J. Lipoprotein-mediated lipid transport in insects: Analogy to the mammalian lipid carrier system and novel concepts for the functioning of LDL receptor family members. Biochim. Biophys. Acta 2005, 1736, 10–29. [Google Scholar] [CrossRef]
- Forster, E.; Bock, H.H.; Herz, J.; Chai, X.; Frotscher, M.; Zhao, S. Emerging topics in Reelin function. Eur. J. Neurosci. 2010, 31, 1511–1518. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, A.; Tao, H.; Metrione, M.; Hajri, T. Very low density lipoprotein receptor (VLDLR) expression is a determinant factor in adipose tissue inflammation and adipocyte-macrophage interaction. J. Biol. Chem. 2014, 289, 1688–1703. [Google Scholar] [CrossRef]
- Yu, J.K.; Holland, L.Z. Cephalochordates (amphioxus or lancelets): A model for understanding the evolution of chordate characters. Cold Spring Harb. Protoc. 2009, 2009, pdb emo130. [Google Scholar] [CrossRef]
- Huang, S.; Yuan, S.; Guo, L.; Yu, Y.; Li, J.; Wu, T.; Liu, T.; Yang, M.; Wu, K.; Liu, H.; et al. Genomic analysis of the immune gene repertoire of amphioxus reveals extraordinary innate complexity and diversity. Genome Res. 2008, 18, 1112–1126. [Google Scholar] [CrossRef]
- Zhang, Q.J.; Zhong, J.; Fang, S.H.; Wang, Y.Q. Branchiostoma japonicum and B. belcheri are distinct lancelets (Cephalochordata) in Xiamen waters in China. Zoolog. Sci. 2006, 23, 573–579. [Google Scholar] [CrossRef]
- Xu, Q.S.; Ma, F.; Wang, Y.Q. Morphological and 12S rRNA gene comparison of two Branchiostoma species in Xiamen waters. J. Exp. Zool. B. Mol. Dev. Evol. 2005, 304, 259–267. [Google Scholar] [CrossRef]
- Li, W.Y.; Zhong, J.; Wang, Y.Q. Analysis of amphioxus geographic populations in the West Pacific Ocean based on COX I gene. Dongwuxue Yanjiu 2010, 31, 375–380. [Google Scholar] [PubMed]
- Jin, P.; Ji, X.; Wang, H.; Li-Ling, J.; Ma, F. AmphiEST: Enabling comparative analysis of ESTs from five developmental stages of amphioxus. Mar. Genom. 2010, 3, 151–155. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Madden, T.L.; Schaffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Copley, R.R.; Pils, B.; Pinkert, S.; Schultz, J.; Bork, P. SMART 5: Domains in the context of genomes and networks. Nucleic Acids Res. 2006, 34, D257–D260. [Google Scholar] [CrossRef]
- Campanella, J.J.; Bitincka, L.; Smalley, J. MatGAT: An application that generates similarity/identity matrices using protein or DNA sequences. BMC Bioinform. 2003, 4, 29. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Guindon, S.; Lethiec, F.; Duroux, P.; Gascuel, O. PHYML Online--a web server for fast maximum likelihood-based phylogenetic inference. Nucleic Acids Res. 2005, 33, W557–W559. [Google Scholar] [CrossRef]
- Nielsen, H. Predicting Secretory Proteins with SignalP. Methods Mol. Biol. 2017, 1611, 59–73. [Google Scholar]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Zidek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Mirdita, M.; Schutze, K.; Moriwaki, Y.; Heo, L.; Ovchinnikov, S.; Steinegger, M. ColabFold: Making protein folding accessible to all. Nat. Methods 2022, 19, 679–682. [Google Scholar] [CrossRef]
- Hsin, J.; Arkhipov, A.; Yin, Y.; Stone, J.E.; Schulten, K. Using VMD: An introductory tutorial. Curr. Protoc. Bioinform. 2008, 24, 5–7. [Google Scholar] [CrossRef]
- Knapp, B.; Lederer, N.; Omasits, U.; Schreiner, W. vmdICE: A plug-in for rapid evaluation of molecular dynamics simulations using VMD. J. Comput. Chem. 2010, 31, 2868–2873. [Google Scholar] [CrossRef]
- Kapustin, Y.; Souvorov, A.; Tatusova, T.; Lipman, D. Splign: Algorithms for computing spliced alignments with identification of paralogs. Biol. Direct 2008, 3, 20. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z. PAML 4: Phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 2007, 24, 1586–1591. [Google Scholar] [CrossRef]
- Paysan-Lafosse, T.; Blum, M.; Chuguransky, S.; Grego, T.; Pinto, B.L.; Salazar, G.A.; Bileschi, M.L.; Bork, P.; Bridge, A.; Colwell, L.; et al. InterPro in 2022. Nucleic Acids Res. 2023, 51, D418–D427. [Google Scholar] [CrossRef]
- Cantalapiedra, C.P.; Hernandez-Plaza, A.; Letunic, I.; Bork, P.; Huerta-Cepas, J. eggNOG-mapper v2: Functional Annotation, Orthology Assignments, and Domain Prediction at the Metagenomic Scale. Mol. Biol. Evol. 2021, 38, 5825–5829. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Marletaz, F.; Firbas, P.N.; Maeso, I.; Tena, J.J.; Bogdanovic, O.; Perry, M.; Wyatt, C.D.R.; de la Calle-Mustienes, E.; Bertrand, S.; Burguera, D.; et al. Amphioxus functional genomics and the origins of vertebrate gene regulation. Nature 2018, 564, 64–70. [Google Scholar] [CrossRef] [PubMed]
- Seo, S.J.; Cheon, H.M.; Sun, J.; Sappington, T.W.; Raikhel, A.S. Tissue- and stage-specific expression of two lipophorin receptor variants with seven and eight ligand-binding repeats in the adult mosquito. J. Biol. Chem. 2003, 278, 41954–41962. [Google Scholar] [CrossRef]
- Gopalapillai, R.; Kadono-Okuda, K.; Tsuchida, K.; Yamamoto, K.; Nohata, J.; Ajimura, M.; Mita, K. Lipophorin receptor of Bombyx mori: cDNA cloning, genomic structure, alternative splicing, and isolation of a new isoform. J. Lipid Res. 2006, 47, 1005–1013. [Google Scholar] [CrossRef]
- Jackson, S.; Cannone, J.; Lee, J.; Gutell, R.; Woodson, S. Distribution of rRNA introns in the three-dimensional structure of the ribosome. J. Mol. Biol. 2002, 323, 35–52. [Google Scholar] [CrossRef] [PubMed]
- Baier, T.; Wichmann, J.; Kruse, O.; Lauersen, K.J. Intron-containing algal transgenes mediate efficient recombinant gene expression in the green microalga Chlamydomonas reinhardtii. Nucleic Acids Res. 2018, 46, 6909–6919. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.L.; Luo, M.T.; Song, J.H.; Pang, W.; Zheng, Y.T. An Alu Element Insertion in Intron 1 Results in Aberrant Alternative Splicing of APOBEC3G Pre-mRNA in Northern Pig-Tailed Macaques (Macaca leonina) That May Reduce APOBEC3G-Mediated Hypermutation Pressure on HIV-1. J. Virol. 2020, 94, e01722-19. [Google Scholar] [CrossRef] [PubMed]
- Kondrashov, F.A.; Koonin, E.V. Evolution of alternative splicing: Deletions, insertions and origin of functional parts of proteins from intron sequences. Trends Genet. 2003, 19, 115–119. [Google Scholar] [CrossRef]
- Dwyer, K.; Agarwal, N.; Gega, A.; Ansari, A. Proximity to the Promoter and Terminator Regions Regulates the Transcription Enhancement Potential of an Intron. Front. Mol. Biosci. 2021, 8, 712639. [Google Scholar] [CrossRef]
- Sun, Z.; Means, A.R. An intron facilitates activation of the calspermin gene by the testis-specific transcription factor CREM tau. J. Biol. Chem. 1995, 270, 20962–20967. [Google Scholar] [CrossRef]
- Oka, K.; Ishimura-Oka, K.; Chu, M.J.; Sullivan, M.; Krushkal, J.; Li, W.H.; Chan, L. Mouse very-low-density-lipoprotein receptor (VLDLR) cDNA cloning, tissue-specific expression and evolutionary relationship with the low-density-lipoprotein receptor. Eur. J. Biochem. 1994, 224, 975–982. [Google Scholar] [CrossRef]
- Pinto, P.I.; Matsumura, H.; Thorne, M.A.; Power, D.M.; Terauchi, R.; Reinhardt, R.; Canario, A.V. Gill transcriptome response to changes in environmental calcium in the green spotted puffer fish. BMC Genom. 2010, 11, 476. [Google Scholar] [CrossRef]


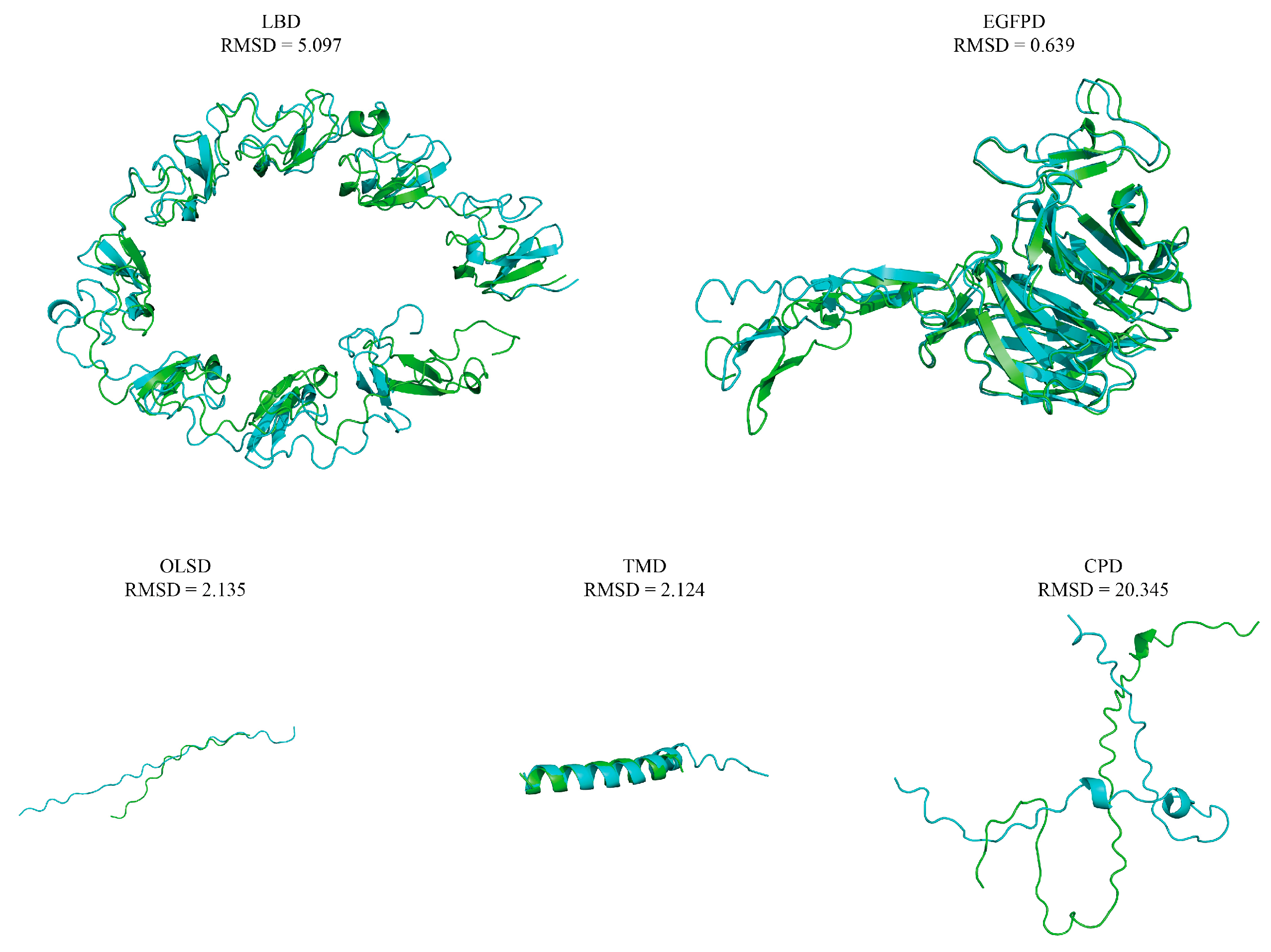

| Protein | Overall Protein | LBD | EGFPD | OLSD | TMD | CPD |
|---|---|---|---|---|---|---|
| Danio rerio VLDLR | 45.8 | 49.9 | 47.1 | 19.4 | 26.5 | 46.3 |
| Xenopus laevis VLDLR | 44.5 | 48.7 | 45.4 | 14.7 | 17.6 | 48.1 |
| Anas platyrhynchos VLDLR | 45.9 | 49.0 | 49.0 | 20.0 | 24.2 | 48.1 |
| Mus musculus VLDLR | 45.5 | 48.7 | 48.1 | 22.5 | 20.6 | 50.0 |
| Homo sapiens VLDLR | 45.3 | 48.7 | 47.8 | 20.5 | 20.6 | 50.0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cao, Y.; Wang, H.; Jin, P.; Ma, F.; Zhou, X. Identification and Characterization of the Very-Low-Density Lipoprotein Receptor Gene from Branchiostoma belcheri: Insights into the Origin and Evolution of the Low-Density Lipoprotein Receptor Gene Family. Animals 2023, 13, 2193. https://doi.org/10.3390/ani13132193
Cao Y, Wang H, Jin P, Ma F, Zhou X. Identification and Characterization of the Very-Low-Density Lipoprotein Receptor Gene from Branchiostoma belcheri: Insights into the Origin and Evolution of the Low-Density Lipoprotein Receptor Gene Family. Animals. 2023; 13(13):2193. https://doi.org/10.3390/ani13132193
Chicago/Turabian StyleCao, Yunpeng, Haili Wang, Ping Jin, Fei Ma, and Xue Zhou. 2023. "Identification and Characterization of the Very-Low-Density Lipoprotein Receptor Gene from Branchiostoma belcheri: Insights into the Origin and Evolution of the Low-Density Lipoprotein Receptor Gene Family" Animals 13, no. 13: 2193. https://doi.org/10.3390/ani13132193
APA StyleCao, Y., Wang, H., Jin, P., Ma, F., & Zhou, X. (2023). Identification and Characterization of the Very-Low-Density Lipoprotein Receptor Gene from Branchiostoma belcheri: Insights into the Origin and Evolution of the Low-Density Lipoprotein Receptor Gene Family. Animals, 13(13), 2193. https://doi.org/10.3390/ani13132193





