Conserved Evolution of MHC Supertypes among Japanese Frogs Suggests Selection for Bd Resistance
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals, Nucleic Acid Isolation, Next-Generation Sequencing, and De Novo Assembly
2.2. Isolation of MHC Sequences
2.3. MHC Phylogenetic Analyses
2.4. Supertyping Analyses
2.5. Preliminary MHC-II Binding Prediction
2.6. Mitochondrial DNA (mtDNA) Sequences and Species Tree
3. Results
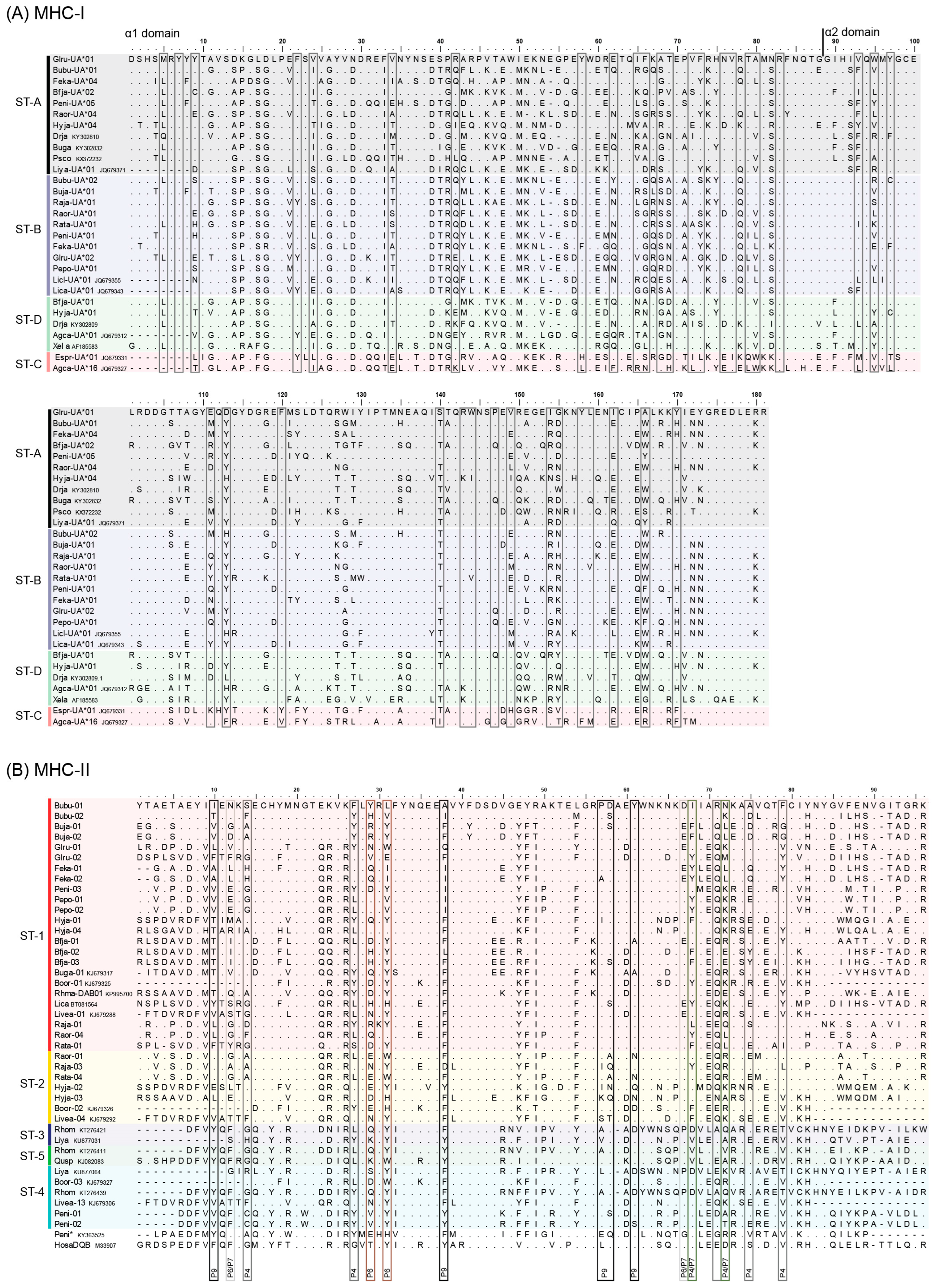
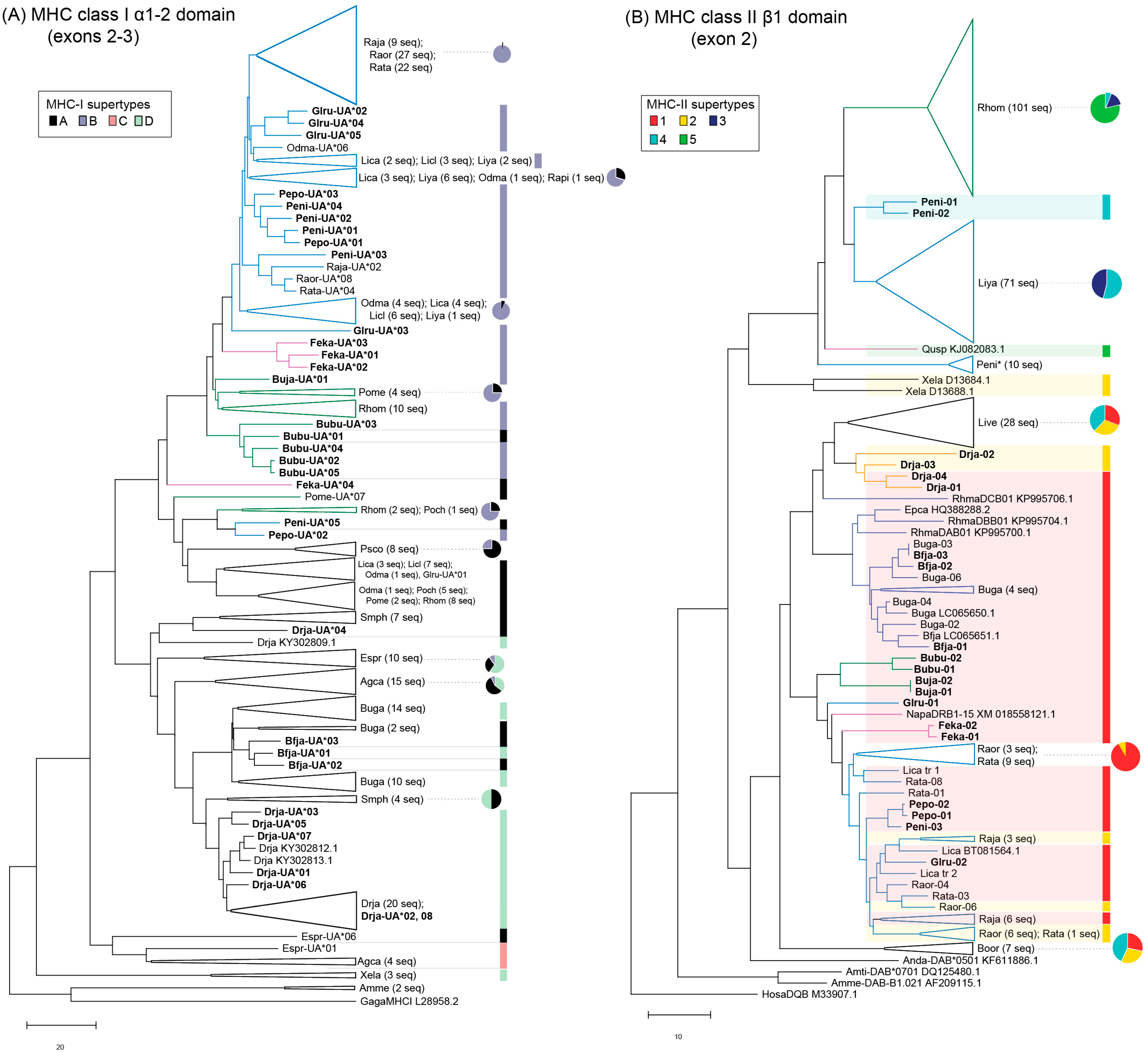
3.1. MHC Sequence and Phylogenetic Analyses
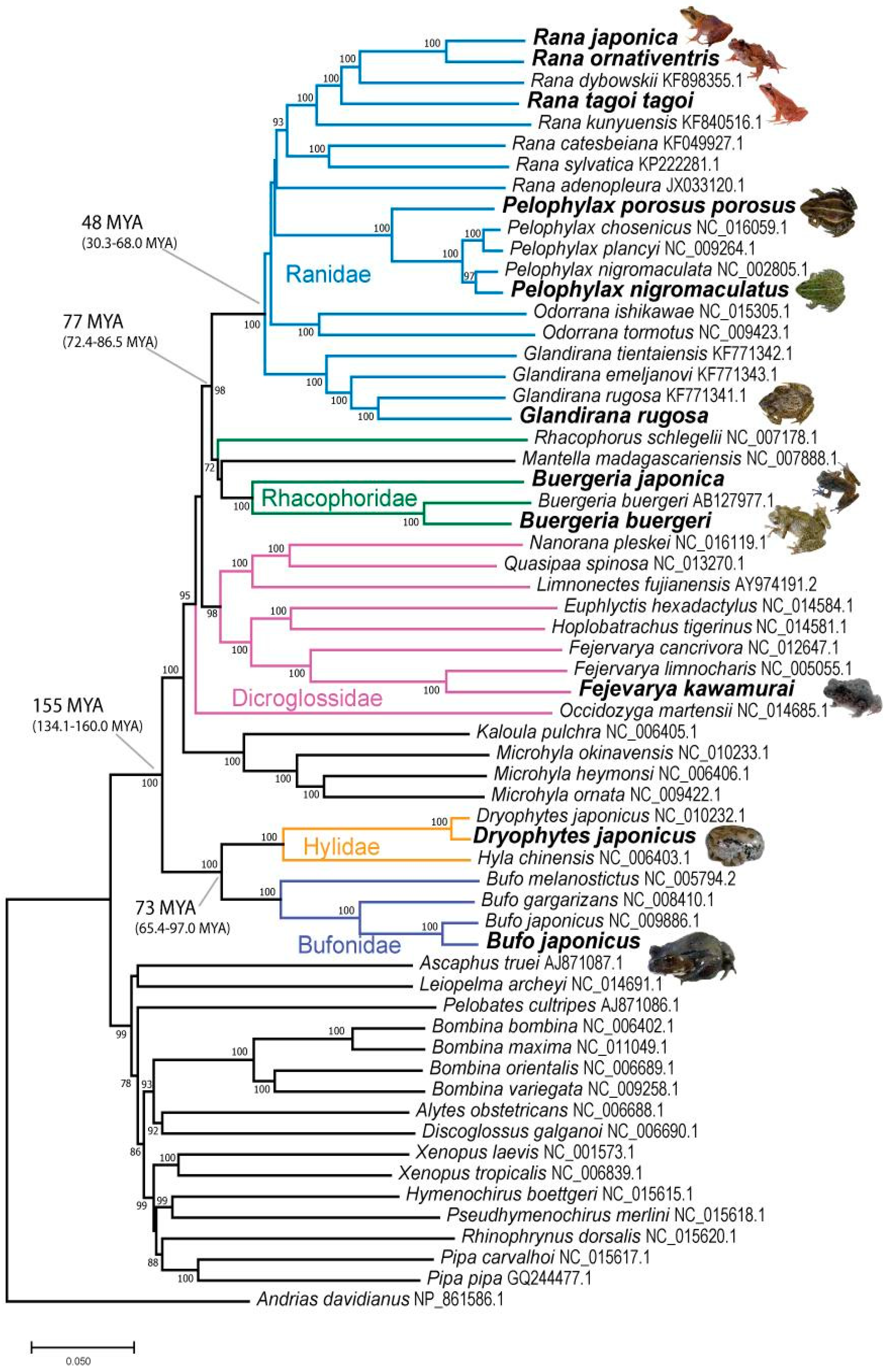
3.2. Species Tree
3.3. MHC Supertyping Analyses

3.4. MHC-II Binding Prediction
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Collins, E.J.; Garboczi, D.N.; Karpusas, M.N.; Wiley, D.C. The Three-Dimensional Structure of a Class I Major Histocompatibility Complex Molecule Missing the Alpha 3 Domain of the Heavy Chain. Proc. Natl. Acad. Sci. USA 1995, 92, 1218–1221. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cruz-Tapias, P.; Castiblanco, J.; Anaya, J.M. Major Histocompatibility Complex: Antigen Processing and Presentation. In Autoimmunity: From Bench to Bedside; El Rosario University Press: Bogota, Columbia, 2013. [Google Scholar]
- Barribeau, S.M.; Villinger, J.; Waldman, B. Major Histocompatibility Complex Based Resistance to a Common Bacterial Pathogen of Amphibians. PLoS ONE 2008, 3, e2692. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sommer, S. The Importance of Immune Gene Variability (MHC) in Evolutionary Ecology and Conservation. Front. Zool. 2005, 2, 16. [Google Scholar] [CrossRef] [Green Version]
- Kaufman, J. Generalists and Specialists: A New View of How MHC Class I Molecules Fight Infectious Pathogens. Trends Immunol. 2018, 39, 367–379. [Google Scholar] [CrossRef] [Green Version]
- Jones, E.Y.; Fugger, L.; Strominger, J.L.; Siebold, C. MHC Class II Proteins and Disease: A Structural Perspective. Nat. Rev. Immunol. 2006, 6, 271–282. [Google Scholar] [CrossRef] [PubMed]
- Wieczorek, M.; Abualrous, E.T.; Sticht, J.; Álvaro-Benito, M.; Stolzenberg, S.; Noé, F.; Freund, C. Major Histocompatibility Complex (MHC) Class I and MHC Class II Proteins: Conformational Plasticity in Antigen Presentation. Front. Immunol. 2017, 8, 292. [Google Scholar] [CrossRef] [Green Version]
- Bouvier, M.; Wiley, D.C. Importance of Peptide Amino and Carboxyl Termini to the Stability of MHC Class I Molecules. Science 1994, 265, 398–402. [Google Scholar] [CrossRef]
- Chicz, R.M.; Urban, R.G.; Lane, W.S.; Gorga, J.C.; Stern, L.J.; Vignali, D.A.A.; Strominger, J.L. Predominant Naturally Processed Peptides Bound to HLA-DR1 Are Derived from MHC-Related Molecules and Are Heterogeneous in Size. Nature 1992, 358, 764–768. [Google Scholar] [CrossRef]
- Densmore, C.L.; Green, D.E. Diseases of Amphibians. ILAR J. 2007, 48, 235–254. [Google Scholar] [CrossRef] [Green Version]
- Pennisi, E. Life and Death Play Out on the Skins of Frogs. Science 2009, 326, 507–508. [Google Scholar] [CrossRef]
- O’Hanlon, S.J.; Rieux, A.; Farrer, R.A.; Rosa, G.M.; Waldman, B.; Bataille, A.; Kosch, T.A.; Murray, K.A.; Brankovics, B.; Fumagalli, M.; et al. Recent Asian Origin of Chytrid Fungi Causing Global Amphibian Declines. Science 2018, 360, 621–627. [Google Scholar] [CrossRef] [Green Version]
- Byrne, A.Q.; Vredenburg, V.T.; Martel, A.; Pasmans, F.; Bell, R.C.; Blackburn, D.C.; Bletz, M.C.; Bosch, J.; Briggs, C.J.; Brown, R.M.; et al. Cryptic Diversity of a Widespread Global Pathogen Reveals Expanded Threats to Amphibian Conservation. Proc. Natl. Acad. Sci. USA 2019, 116, 20382–20387. [Google Scholar] [CrossRef]
- Fu, M.; Waldman, B. Ancestral Chytrid Pathogen Remains Hypervirulent Following Its Long Coevolution with Amphibian Hosts. Proc. R. Soc. B Biol. Sci. 2019, 286, 20190833. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goka, K.; Yokoyama, J.; Une, Y.; Kuroki, T.; Suzuki, K.; Nakahara, M.; Kobayashi, A.; Inaba, S.; Mizutani, T.; Hyatt, A.D. Amphibian Chytridiomycosis in Japan: Distribution, Haplotypes and Possible Route of Entry into Japan. Mol. Ecol. 2009, 18, 4757–4774. [Google Scholar] [CrossRef] [PubMed]
- Bataille, A.; Cashins, S.D.; Grogan, L.; Skerratt, L.F.; Hunter, D.; McFadden, M.; Scheele, B.; Brannelly, L.A.; Macris, A.; Harlow, P.S.; et al. Susceptibility of Amphibians to Chytridiomycosis Is Associated with MHC Class II Conformation. Proc. R. Soc. B Biol. Sci. 2015, 282, 20143127. [Google Scholar] [CrossRef] [Green Version]
- Kosch, T.A.; Silva, C.N.S.; Brannelly, L.A.; Roberts, A.A.; Lau, Q.; Marantelli, G.; Berger, L.; Skerratt, L.F. Genetic Potential for Disease Resistance in Critically Endangered Amphibians Decimated by Chytridiomycosis. Anim. Conserv. 2019, 22, 238–250. [Google Scholar] [CrossRef]
- Savage, A.E.; Zamudio, K.R. Adaptive Tolerance to a Pathogenic Fungus Drives Major Histocompatibility Complex Evolution in Natural Amphibian Populations. Proc. Biol. Sci. 2016, 283, 20153115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Savage, A.E.; Zamudio, K.R. MHC Genotypes Associate with Resistance to a Frog-Killing Fungus. Proc. Natl. Acad. Sci. USA 2011, 108, 16705–16710. [Google Scholar] [CrossRef] [Green Version]
- Trujillo, A.L.; Hoffman, E.A.; Becker, C.G.; Savage, A.E. Spatiotemporal Adaptive Evolution of an MHC Immune Gene in a Frog-Fungus Disease System. Heredity 2021, 126, 640–655. [Google Scholar] [CrossRef]
- Lau, Q.; Igawa, T.; Minei, R.; Kosch, T.A.; Satta, Y. Transcriptome Analyses of Immune Tissues from Three Japanese Frogs (Genus Rana) Reveals Their Utility in Characterizing Major Histocompatibility Complex Class II. BMC Genom. 2017, 18, 994. [Google Scholar] [CrossRef] [Green Version]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Trinity: Reconstructing a Full-Length Transcriptome without a Genome from RNA-Seq Data. Nat. Biotechnol. 2013, 29, 644–652. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lau, Q.; Igawa, T.; Komaki, S.; Satta, Y. Characterisation of Major Histocompatibility Complex Class I Genes in Japanese Ranidae Frogs. Immunogenetics 2016, 68, 797–806. [Google Scholar] [CrossRef] [Green Version]
- Hedrick, P.W. Evolutionary Genetics of the Major Histocompatibility Complex. Am. Nat. 1994, 143, 945–964. [Google Scholar] [CrossRef]
- Rozen, S.; Skaletsky, H.J. Primer3. In Bioinformatics Methods and Protocols Methods in Molecular Biology; Humana Press: Totowa, NJ, USA, 1998; Volume 3, pp. 1–41. [Google Scholar]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Sawai, H.; Go, Y.; Satta, Y. Biological Implication for Loss of Function at Major Histocompatibility Complex Loci. Immunogenetics 2008, 60, 295–302. [Google Scholar] [CrossRef] [PubMed]
- Yasukochi, Y.; Satta, Y. A Human-Specific Allelic Group of the MHC DRB1 Gene in Primates. J. Physiol. Anthropol. 2014, 33, 14. [Google Scholar] [CrossRef] [Green Version]
- Klein, J.; Satta, Y.; O’hUigin, C.; Takahata, N. The Molecular Descent of the Major Histocompatibility Complex. Annu. Rev. Immunol. 1993, 11, 269–295. [Google Scholar] [CrossRef]
- Bjorkman, P.J.; Saper, M.A.; Samraoui, B.; Bennett, W.S.; Strominger, J.L.; Wiley, D.C. The Foreign Antigen Binding Site and T Cell Recognition Regions of Class I Histocompatibility Antigens. Nature 1987, 329, 512–518. [Google Scholar] [CrossRef]
- Saper, M.A.; Bjorkman, P.J.; Wiley, D.C. Refined Structure of the Human Histocompatibility Antigen HLA-A2 at 2.6 Å Resolution. J. Mol. Biol. 1991, 219, 277–319. [Google Scholar] [CrossRef]
- Lillie, M.; Grueber, C.; Sutton, J.; Howitt, R.; Bishop, P.; Gleeson, D.; Belov, K. Selection on MHC Class II Supertypes in the New Zealand Endemic Hochstetter’s Frog. BMC Evol. Biol. 2015, 15, 63. [Google Scholar] [CrossRef] [Green Version]
- Didinger, C.; Eimes, J.A.; Lillie, M.; Waldman, B. Multiple Major Histocompatibility Complex Class I Genes in Asian Anurans: Ontogeny and Phylogeny of Expression. Dev. Comp. Immunol. 2016, 70, 69–79. [Google Scholar] [CrossRef] [PubMed]
- Matsumura, M.; Fremont, D.H.; Peterson, P.A.; Wilson, I.A. Emerging Principles for the Recognition of Peptide Antigens by MHC Class I Molecules. Science 1992, 257, 927–934. [Google Scholar] [CrossRef]
- Jombart, T.; Devillard, S.; Balloux, F. Discriminant Analysis of Principal Components: A New Method for the Analysis of Genetically Structured Populations. BMC Genet. 2010, 11, 94. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Becker, C.G.; Rodriguez, D.; Longo, A.V.; Talaba, A.L.; Zamudio, K.R. Disease Risk in Temperate Amphibian Populations Is Higher at Closed-Canopy Sites. PLoS ONE 2012, 7, e48205. [Google Scholar] [CrossRef] [PubMed]
- Crawford, A.J.; Lips, K.R.; Bermingham, E. Epidemic Disease Decimates Amphibian Abundance, Species Diversity, and Evolutionary History in the Highlands of Central Panama. Proc. Natl. Acad. Sci. USA 2010, 107, 13777–13782. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scheele, B.C.; Pasmans, F.; Skerratt, L.F.; Berger, L.; Martel, A.; Beukema, W.; Acevedo, A.A.; Burrowes, P.A.; Carvalho, T.; Catenazzi, A.; et al. Amphibian Fungal Panzootic Causes Catastrophic and Ongoing Loss of Biodiversity. Science 2019, 363, 1459–1463. [Google Scholar] [CrossRef]
- Brannelly, L.A.; Martin, G.; Llewelyn, J.; Skerratt, L.F.; Berger, L. Age- and Size-Dependent Resistance to Chytridiomycosis in the Invasive Cane Toad Rhinella Marina. Dis. Aquat. Organ. 2018, 131, 107–120. [Google Scholar] [CrossRef] [Green Version]
- Daszak, P.; Strieby, A.; Cunningham, A.A.; Longcore, J.E.; Brown, C.C.; Porter, D. Experimental Evidence That the Bullfrog (Rana Catesbeiana) Is a Potential Carrier of Chytridiomycosis, an Emerging Fungal Disease of Amphibians. Proc. Herpetol. J. 2004, 14, 201–207. [Google Scholar]
- Chen, H.; Tan, X.; Han, F.; Yao, Y.; Xu, H.; Zhang, M. Evolution by Gene Duplication, Recombination and Selection in MHC Class I Genes of Odorrana Margaretae. Sains Malays. 2018, 47, 1979–1989. [Google Scholar] [CrossRef]
- Zhao, M.; Wang, Y.; Shen, H.; Li, C.; Chen, C.; Luo, Z.; Wu, H. Evolution by Selection, Recombination, and Gene Duplication in MHC Class I Genes of Two Rhacophoridae Species. BMC Evol. Biol. 2013, 13, 113. [Google Scholar] [CrossRef] [Green Version]
- Chen, H.; Huang, S.; Jiang, Y.; Han, F.; Ni, Q.; Yao, Y.; Xu, H.; Sudhanshu, M.; Zhang, M. The MHC Class Ia Genes in Chenfu’s Treefrog (Zhangixalus Chenfui) Evolved via Gene Duplication, Recombination, and Selection. Animals 2020, 10, 34. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kiemnec-Tyburczy, K.M.; Richmond, J.Q.; Savage, E.; Lips, K.R.; Zamudio, K.R. Genetic Diversity of MHC Class I Loci in Six Non-Model Frogs Is Shaped by Positive Selection and Gene Duplication. Heredity 2012, 109, 146–155. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, L.; Zhao, M.; Luo, Z.; Wu, H. Allelic Polymorphism, Gene Duplication and Balancing Selection of MHC Class IIB Genes in the Omei Treefrog (Rhacophorus Omeimontis). Asian Herpetol. Res. 2016, 10, 53–64. [Google Scholar] [CrossRef]
- Liu, H.Y.; Xue, F.; Gong, J.; Wan, Q.H.; Fang, S.G. Limited Polymorphism of the Functional MHC Class II B Gene in the Black-Spotted Frog (Pelophylax Nigromaculatus) Identified by Locus-Specific Genotyping. Ecol. Evol. 2017, 7, 9860–9868. [Google Scholar] [CrossRef] [PubMed]
- Reynisson, B.; Alvarez, B.; Paul, S.; Peters, B.; Nielsen, M. NetMHCpan-4.1 and NetMHCIIpan-4.0: Improved Predictions of MHC Antigen Presentation by Concurrent Motif Deconvolution and Integration of MS MHC Eluted Ligand Data. Nucleic Acids Res. 2021, 48, W449–W454. [Google Scholar] [CrossRef] [PubMed]
- Hoof, I.; Peters, B.; Sidney, J.; Pedersen, L.E.; Sette, A.; Lund, O.; Buus, S.; Nielsen, M. NetMHCpan, a Method for MHC Class i Binding Prediction beyond Humans. Immunogenetics 2009, 61, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Lau, Q.; Yasukochi, Y.; Satta, Y. A Limit to the Divergent Allele Advantage Model Supported by Variable Pathogen Recognition across HLA-DRB1 Allele Lineages. Tissue Antigens 2015, 86, 343–352. [Google Scholar] [CrossRef] [Green Version]
- Impens, F.; Colaert, N.; Helsens, K.; Ghesquière, B.; Timmerman, E.; De Bock, P.J.; Chain, B.M.; Vandekerckhove, J.; Gevaert, K. A Quantitative Proteomics Design for Systematic Identification of Protease Cleavage Events. Mol. Cell. Proteom. 2010, 9, 2327–2333. [Google Scholar] [CrossRef] [Green Version]
- Bennett, K.; Levine, T.; Ellis, J.S.; Peanasky, R.J.; Michael Samloff, I.; Kay, J.; Chain, B.M. Antigen Processing for Presentation by Class II Major Histocompatibility Complex Requires Cleavage by Cathepsin E. Eur. J. Immunol. 1992, 22, 1519–1524. [Google Scholar] [CrossRef]
- Fisher, M.C.; Bosch, J.; Yin, Z.; Stead, D.A.; Walker, J.; Selway, L.; Brown, A.J.P.; Walker, L.A.; Gow, N.A.R.; Stajich, J.E.; et al. Proteomic and Phenotypic Profiling of the Amphibian Pathogen Batrachochytrium Dendrobatidis Shows That Genotype Is Linked to Virulence. Mol. Ecol. 2009, 18, 415–429. [Google Scholar] [CrossRef]
- Dillon, M.J.; Bowkett, A.E.; Bungard, M.J.; Beckman, K.M.; O’Brien, M.F.; Bates, K.; Fisher, M.C.; Stevens, J.R.; Thornton, C.R. Tracking the Amphibian Pathogens Batrachochytrium Dendrobatidis and Batrachochytrium Salamandrivorans Using a Highly Specific Monoclonal Antibody and Lateral-Flow Technology. Microb. Biotechnol. 2017, 10, 381–394. [Google Scholar] [CrossRef] [PubMed]
- Rosenblum, E.B.; Poorten, T.J.; Joneson, S.; Settles, M. Substrate-Specific Gene Expression in Batrachochytrium Dendrobatidis, the Chytrid Pathogen of Amphibians. PLoS ONE 2012, 8, e49924. [Google Scholar] [CrossRef] [Green Version]
- Bacher, P.; Kniemeyer, O.; Teutschbein, J.; Thön, M.; Vödisch, M.; Wartenberg, D.; Scharf, D.H.; Koester-Eiserfunke, N.; Schütte, M.; Dübel, S.; et al. Identification of Immunogenic Antigens from Aspergillus Fumigatus by Direct Multiparameter Characterization of Specific Conventional and Regulatory CD4 + T Cells. J. Immunol. 2014, 193, 3332–3343. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, S.; Stecher, G.; Suleski, M.; Hedges, S.B. TimeTree: A Resource for Timelines, Timetrees, and Divergence Times. Mol. Biol. Evol. 2017, 34, 1812–1819. [Google Scholar] [CrossRef] [PubMed]
- Byrne, A.Q.; Waddle, A.W.; Saenz, V.; Ohmer, M.; Jaeger, J.R.; Richards-Zawacki, C.L.; Voyles, J.; Rosenblum, E.B. Host Species Is Linked to Pathogen Genotype for the Amphibian Chytrid Fungus (Batrachochytrium Dendrobatidis). PLoS ONE 2022, 17, e0261047. [Google Scholar] [CrossRef] [PubMed]
- Greenspan, S.E.; Lambertini, C.; Carvalho, T.; James, T.Y.; Toledo, L.F.; Haddad, C.F.B.; Becker, C.G. Hybrids of Amphibian Chytrid Show High Virulence in Native Hosts. Sci. Rep. 2018, 8, 9600. [Google Scholar] [CrossRef] [Green Version]
- Fong, J.J.; Cheng, T.L.; Bataille, A.; Pessier, A.P.; Waldman, B.; Vredenburg, V.T. Early 1900s Detection of Batrachochytrium Dendrobatidis in Korean Amphibians. PLoS ONE 2015, 10, e0115656. [Google Scholar] [CrossRef]

| Target Gene | Target Species | Forward Primer (5′–3′) | Reverse Primer (5′–3′) | Amplicon Length (bp) |
|---|---|---|---|---|
| MHC class I | All study species | GAYRGHCACWBYYTSCGBTAYT | CTCYGGRCGYACTCTCYTYT | 552–555 |
| MHC class II | Buergeria spp., Glandirana rugosa, Pelophylax spp. | RVATTAYHWGCAGMAASATG a | CACWCCRGCAAYRATAARYA a | 752–761 |
| Fejervarya kawamurai | AGGAGAAKCCGCTGATTATG | CACAGSTGAAGDYRTCTCCTTTYK | 524 | |
| Pelophylax nigromaculatus | TACTATCCGCCTCCCATCC | GAGCCCAACAATGAAGAAA | 663 | |
| Dryophytes japonicus | GYTCSTCACCAGATGTCAGA | CCAGGATCTGGWAAGTCCAR | 486–489 | |
| Bufo japonicus b | (i) CTTTCTGAACGGGACTCAGC (ii) AAGCCGAGAACCAGAAGACA | (i) YTSRACTCTCCGGTCWGYWR (ii) MGAKGGGTAGAARCCAARCA | 244 466–469 |
| Species | No. of MHC-I Variants | No. of MHC-II Variants |
|---|---|---|
| Buergeria buergeri [Bubu] | 4 (2) | 2 (1) |
| Buergeria japonica [Buja] | 1 (1) | 2 (1) |
| Bufo japonicus [Bfja] | 3 (2) | 3 (2) |
| Dryophytes japonicus [Drja] | 8 (4) | 4 (2) |
| Fejervarya kawamurai [Feka] | 4 (2) | 2 (1) |
| Glandirana rugosa [Glru] | 5 (3) | 2 (1) |
| Pelophylax nigromaculatus [Peni] | 4 (2) | 3 (2) |
| Pelophylax porosus porosus [Pepo] | 3 (2) | 2 (1) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lau, Q.; Igawa, T.; Kosch, T.A.; Dharmayanthi, A.B.; Berger, L.; Skerratt, L.F.; Satta, Y. Conserved Evolution of MHC Supertypes among Japanese Frogs Suggests Selection for Bd Resistance. Animals 2023, 13, 2121. https://doi.org/10.3390/ani13132121
Lau Q, Igawa T, Kosch TA, Dharmayanthi AB, Berger L, Skerratt LF, Satta Y. Conserved Evolution of MHC Supertypes among Japanese Frogs Suggests Selection for Bd Resistance. Animals. 2023; 13(13):2121. https://doi.org/10.3390/ani13132121
Chicago/Turabian StyleLau, Quintin, Takeshi Igawa, Tiffany A. Kosch, Anik B. Dharmayanthi, Lee Berger, Lee F. Skerratt, and Yoko Satta. 2023. "Conserved Evolution of MHC Supertypes among Japanese Frogs Suggests Selection for Bd Resistance" Animals 13, no. 13: 2121. https://doi.org/10.3390/ani13132121
APA StyleLau, Q., Igawa, T., Kosch, T. A., Dharmayanthi, A. B., Berger, L., Skerratt, L. F., & Satta, Y. (2023). Conserved Evolution of MHC Supertypes among Japanese Frogs Suggests Selection for Bd Resistance. Animals, 13(13), 2121. https://doi.org/10.3390/ani13132121





