Identification of SNPs Associated with Goose Meat Quality Traits Using a Genome-Wide Association Study Approach
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Animals and Phenotypic Traits
2.2. Whole-Genome Resequencing and SNP Calling
2.3. GWAS
2.4. SNP Annotation
3. Results
3.1. Phenotypic Description of the Goose Population
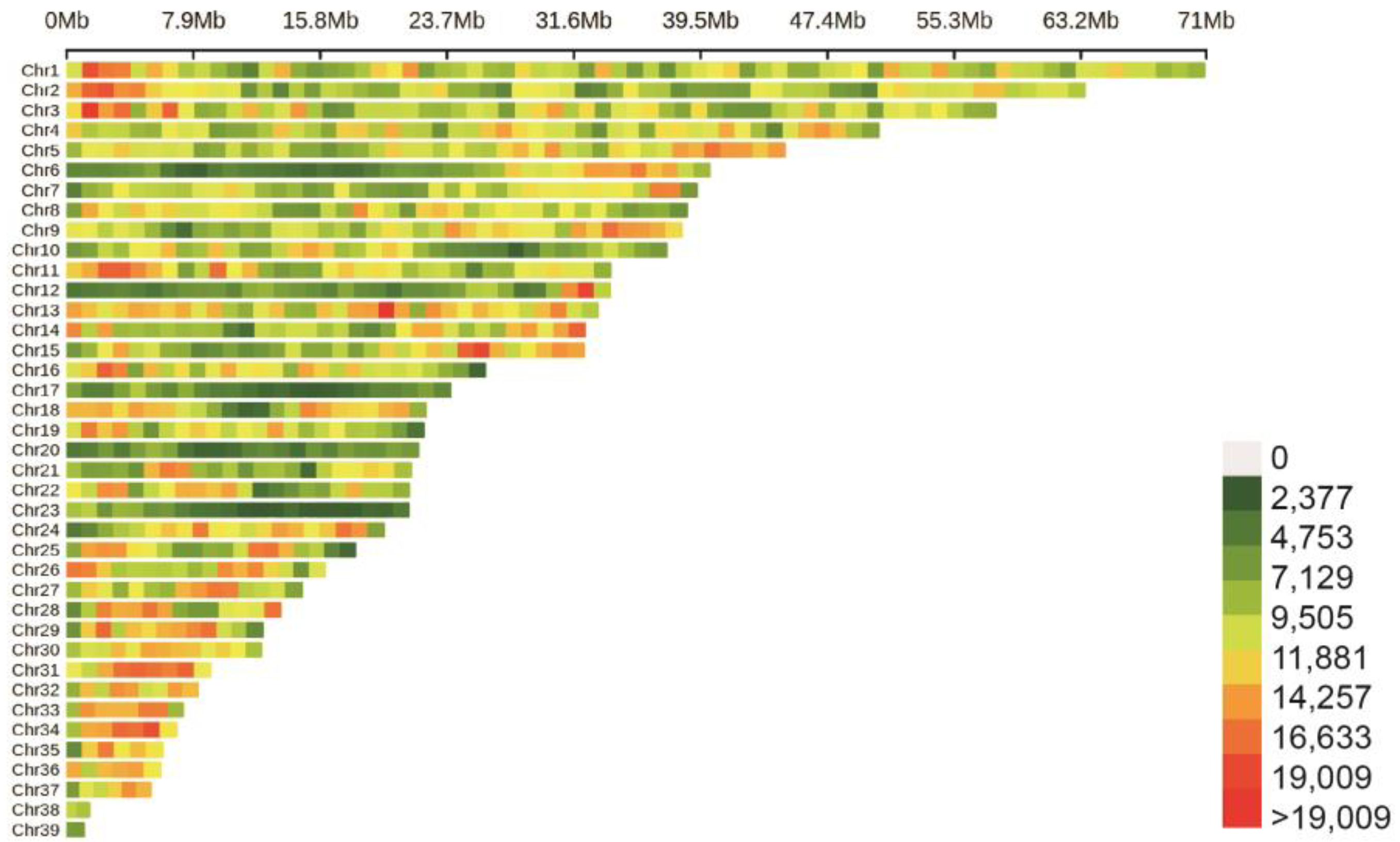
3.2. Whole-Genome Resequencing and SNP Calling
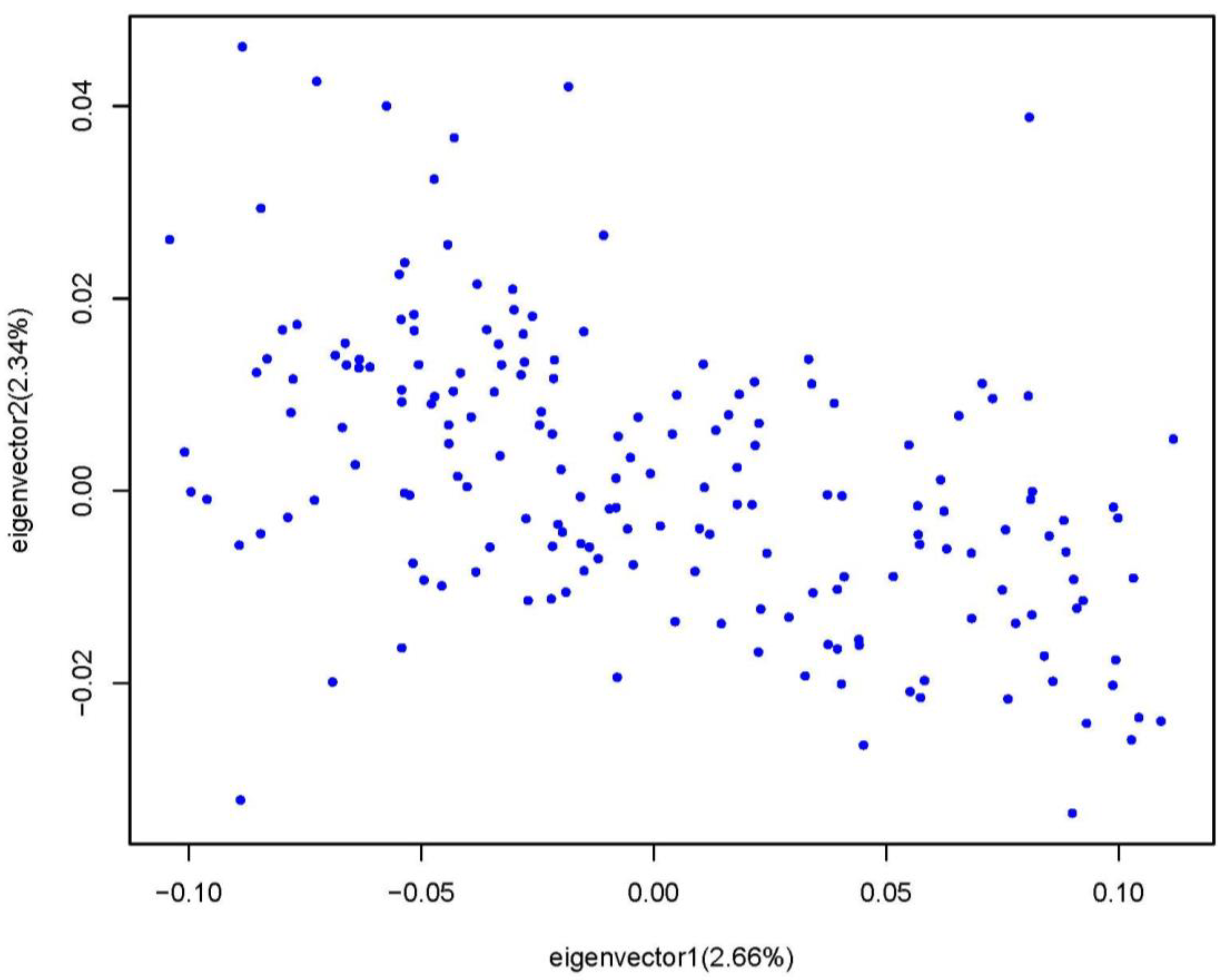
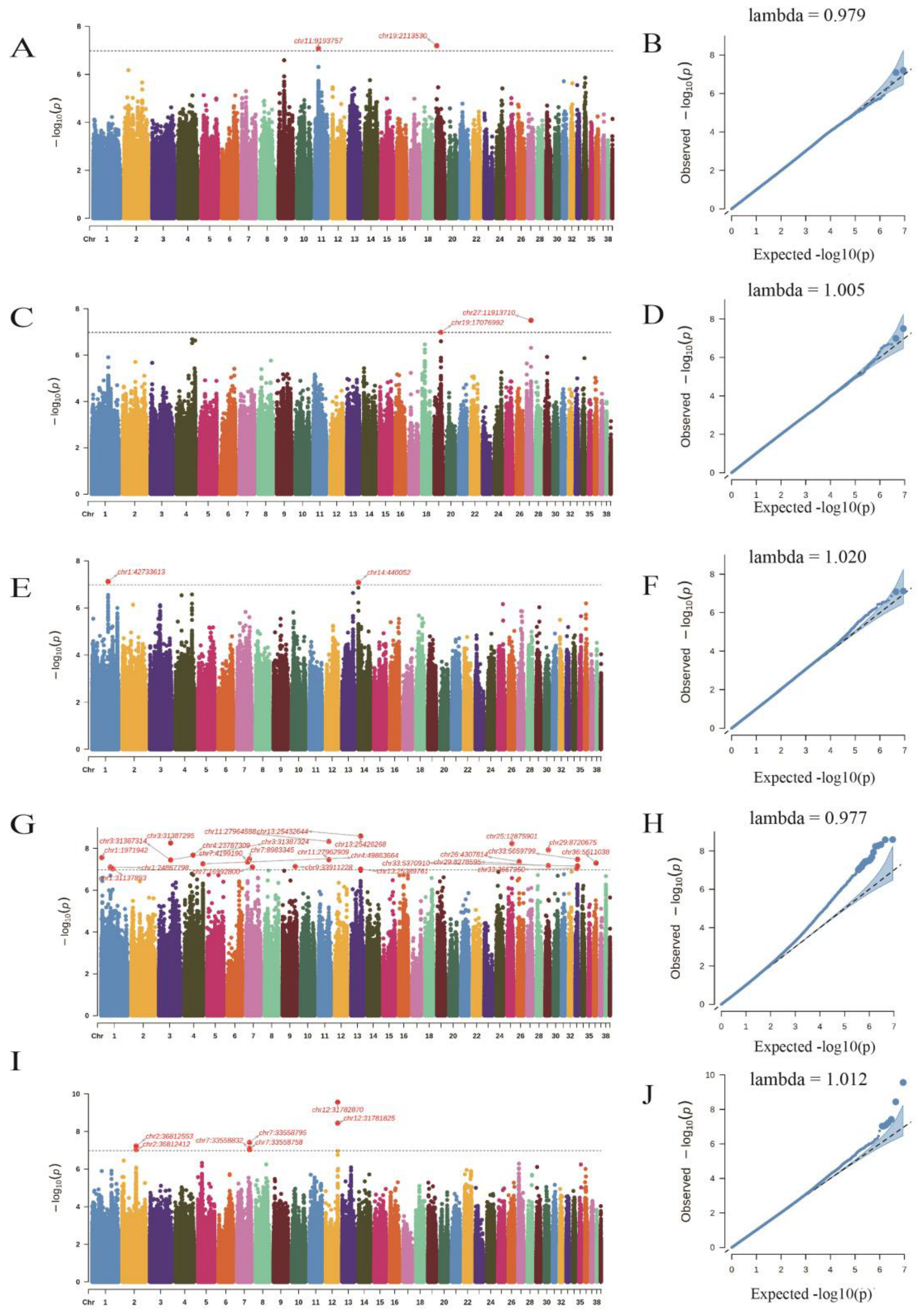
3.3. The Goose Population Structure and the GWAS
3.4. The Genotypes of the Selected SNPs
3.5. The Gene Annotation within 1 Mb of the Potentially Significant SNPs
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kozák, J. Goose production and goose products. World’s Poult. Sci. J. 2021, 77, 403–414. [Google Scholar] [CrossRef]
- Tang, J.; Ouyang, H.; Chen, X.; Jiang, D.; Tian, Y.; Huang, Y.; Shen, X. Comparative Transcriptome Analyses of Leg Muscle during Early Growth between Geese (Anser cygnoides) Breeds Differing in Body Size Characteristics. Genes 2023, 14, 1048. [Google Scholar] [CrossRef]
- Windhorst, H. Asia dominates global waterfowl production. World Poult. 2011, 27, 6–9. [Google Scholar]
- Wołoszyn, J.; Wereńska, M.; Goluch, Z.; Haraf, G.; Okruszek, A.; Teleszko, M.; Król, B. The selected goose meat quality traits in relation to various types of heat treatment. Poult. Sci. 2020, 99, 7214–7224. [Google Scholar] [CrossRef] [PubMed]
- Uhlířová, L.; Tůmová, E.; Chodová, D.; Vlčková, J.; Ketta, M.; Volek, Z.; Skřivanová, V. The effect of age, genotype and sex on carcass traits, meat quality and sensory attributes of geese. Asian-Australas. J. Anim. Sci. 2018, 31, 421. [Google Scholar] [CrossRef]
- Hoa, V.-B.; Seol, K.-H.; Seo, H.-W.; Seong, P.-N.; Kang, S.-M.; Kim, Y.-S.; Moon, S.-S.; Kim, J.-H.; Cho, S.-H. Meat quality characteristics of pork bellies in relation to fat level. Anim. Biosci. 2021, 34, 1663. [Google Scholar] [CrossRef]
- Wen, C.; Jiang, X.; Ding, L.; Wang, T.; Zhou, Y. Effects of dietary methionine on growth performance, meat quality and oxidative status of breast muscle in fast-and slow-growing broilers. Poult. Sci. 2017, 96, 1707–1714. [Google Scholar] [CrossRef]
- Wu, P.; Su, Y.; Feng, L.; Jiang, W.; Kuang, S.; Tang, L.; Jiang, J.; Liu, Y.; Zhou, X. Optimal DL-Methionyl-DL-Methionine Supplementation Improved Intestinal Physical Barrier Function by Changing Antioxidant Capacity, Apoptosis and Tight Junction Proteins in the Intestine of Juvenile Grass Carp (Ctenopharyngodon idella). Antioxidants 2022, 11, 1652. [Google Scholar] [CrossRef]
- Zeitz, J.O.; Fleischmann, A.; Ehbrecht, T.; Most, E.; Friedrichs, S.; Whelan, R.; Gessner, D.K.; Failing, K.; Lütjohann, D.; Eder, K. Effects of supplementation of DL-methionine on tissue and plasma antioxidant status during heat-induced oxidative stress in broilers. Poult. Sci. 2020, 99, 6837–6847. [Google Scholar] [CrossRef]
- Jeon, J.-J.; Kim, H.-J.; Kang, H.-K.; Kim, C.-H.; Kim, H.-S.; Hong, E.-C.; Jang, A.; Kim, S.-H. Effects of Dietary Thraustochytrid Schizochytrium sp. and Other Omega-3 Sources on Growth Performance, Carcass Characteristics, and Meat Quality of Broilers. Animals 2022, 12, 1166. [Google Scholar] [CrossRef]
- Long, S.; Liu, S.; Wu, D.; Mahfuz, S.; Piao, X. Effects of dietary fatty acids from different sources on growth performance, meat quality, muscle fatty acid deposition, and antioxidant capacity in broilers. Animals 2020, 10, 508. [Google Scholar] [CrossRef]
- Warner, R.D.; Wheeler, T.L.; Ha, M.; Li, X.; Bekhit, A.E.-D.; Morton, J.; Vaskoska, R.; Dunshea, F.R.; Liu, R.; Purslow, P. Meat tenderness: Advances in biology, biochemistry, molecular mechanisms and new technologies. Meat Sci. 2022, 185, 108657. [Google Scholar] [CrossRef]
- Purslow, P.; Archile-Contreras, A.; Cha, M. Meat Science and Muscle Biology Symposium: Manipulating meat tenderness by increasing the turnover of intramuscular connective tissue. J. Anim. Sci. 2012, 90, 950–959. [Google Scholar] [CrossRef]
- Sermyagin, A.; Bykova, O.; Loretts, O.; Kostyunina, O.; Zinovieva, N.J.S.s.B. Genomic variability assess for breeding traits in holsteinizated Russian Black-and-White cattle using GWAS analysis and ROH patterns. Agric. Biol. 2020, 55, 257–274. [Google Scholar] [CrossRef]
- Gebreselassie, G.; Berihulay, H.; Jiang, L.; Ma, Y.J.A. Review on genomic regions and candidate genes associated with economically important production and reproduction traits in sheep (Ovies aries). Animals 2020, 10, 33. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, J.; Gong, H.; Cui, L.; Zhang, W.; Ma, J.; Chen, C.; Ai, H.; Xiao, S.; Huang, L.J.M.s. Genetic correlation of fatty acid composition with growth, carcass, fat deposition and meat quality traits based on GWAS data in six pig populations. Meat Sci. 2019, 150, 47–55. [Google Scholar] [CrossRef]
- Zhang, H.; Shen, L.-Y.; Xu, Z.-C.; Kramer, L.M.; Yu, J.-Q.; Zhang, X.-Y.; Na, W.; Yang, L.-L.; Cao, Z.-P.; Luan, P. Haplotype-based genome-wide association studies for carcass and growth traits in chicken. Poult. Sci. 2020, 99, 2349–2361. [Google Scholar] [CrossRef]
- Gao, G.; Gao, D.; Zhao, X.; Xu, S.; Zhang, K.; Wu, R.; Yin, C.; Li, J.; Xie, Y.; Hu, S.; et al. Genome-Wide association study-based identification of SNPs and haplotypes associated with goose reproductive performance and egg quality. Front. Genet. 2021, 12, 360. [Google Scholar] [CrossRef]
- Guo, Q.; Huang, L.; Bai, H.; Wang, Z.; Bi, Y.; Chen, G.; Jiang, Y.; Chang, G. Genome-Wide Association Study of Potential Meat Quality Trait Loci in Ducks. Genes 2022, 13, 986. [Google Scholar] [CrossRef]
- Leal-Gutiérrez, J.D.; Rezende, F.M.; Reecy, J.M.; Kramer, L.M.; Peñagaricano, F.; Mateescu, R.G. Whole genome sequence data provides novel insights into the genetic architecture of meat quality traits in beef. Front. Genet. 2020, 11, 538640. [Google Scholar] [CrossRef]
- Liu, R.; Sun, Y.; Zhao, G.; Wang, F.; Wu, D.; Zheng, M.; Chen, J.; Zhang, L.; Hu, Y.; Wen, J. Genome-wide association study identifies loci and candidate genes for body composition and meat quality traits in Beijing-You chickens. PLoS ONE 2013, 8, e61172. [Google Scholar] [CrossRef] [PubMed]
- Zhu, F.; Yin, Z.-T.; Wang, Z.; Smith, J.; Zhang, F.; Martin, F.; Ogeh, D.; Hincke, M.; Lin, F.-B.; Burt, D.W. Three chromosome-level duck genome assemblies provide insights into genomic variation during domestication. Nat. Commun. 2021, 12, 5932. [Google Scholar] [CrossRef] [PubMed]
- Zhu, F.; Cui, Q.-Q.; Hou, Z.-C. SNP discovery and genotyping using Genotyping-by-Sequencing in Pekin ducks. Sci. Rep. 2016, 6, 36223. [Google Scholar] [CrossRef] [PubMed]
- Patel, R.K.; Jain, M. NGS QC Toolkit: A toolkit for quality control of next generation sequencing data. PLoS ONE 2012, 7, e30619. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Gao, G.; Lin, Y.; Hu, S.; Luo, Y.; Wang, G.; Jin, L.; Wang, Q.; Wang, J.; Tang, Q. Pacific Biosciences assembly with Hi-C mapping generates an improved, chromosome-level goose genome. GigaScience 2020, 9, giaa114. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.; Bender, D.; Maller, J.; Sklar, P.; De Bakker, P.I.; Daly, M.J. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef]
- Zhou, X.; Stephens, M. Genome-wide efficient mixed-model analysis for association studies. Nat. Genet. 2012, 44, 821–824. [Google Scholar] [CrossRef]
- Zhao, J.H. gap: Genetic analysis package. J. Stat. Softw. 2008, 23, 1–18. [Google Scholar] [CrossRef]
- Turner, S.D. qqman: An R package for visualizing GWAS results using QQ and manhattan plots. Biorxiv 2014, 005165. [Google Scholar]
- Karssen, L.C.; van Duijn, C.M.; Aulchenko, Y.S. The GenABEL Project for statistical genomics. FResearch 2016, 5, 914. [Google Scholar] [CrossRef]
- Quinlan, A.R. BEDTools: The Swiss-army tool for genome feature analysis. Curr. Protoc. Bioinform. 2014, 47, 11.12.11–11.12.34. [Google Scholar] [CrossRef]
- Wang, K.; Li, M.; Hakonarson, H. ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010, 38, e164. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhou, B.; Pache, L.; Chang, M.; Khodabakhshi, A.H.; Tanaseichuk, O.; Benner, C.; Chanda, S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019, 10, 1523. [Google Scholar] [CrossRef]
- Uffelmann, E.; Huang, Q.Q.; Munung, N.S.; De Vries, J.; Okada, Y.; Martin, A.R.; Martin, H.C.; Lappalainen, T.; Posthuma, D. Genome-wide association studies. Nat. Rev. Methods Prim. 2021, 1, 59. [Google Scholar] [CrossRef]
- Nishimura, N.; Gotoh, T.; Oike, Y.; Yano, M. TMEM65 is a mitochondrial inner-membrane protein. PeerJ 2014, 2, e349. [Google Scholar] [CrossRef]
- Nazli, A.; Safdar, A.; Saleem, A.; Akhtar, M.; Brady, L.I.; Schwartzentruber, J.; Tarnopolsky, M.A. A mutation in the TMEM65 gene results in mitochondrial myopathy with severe neurological manifestations. Eur. J. Hum. Genet. 2017, 25, 744–751. [Google Scholar] [CrossRef]
- Urushima, Y.; Haraguchi, M.; Yano, M. Depletion of TMEM65 leads to oxidative stress, apoptosis, induction of mitochondrial unfolded protein response, and upregulation of mitochondrial protein import receptor TOMM22. Biochem. Biophys. Rep. 2020, 24, 100870. [Google Scholar] [CrossRef]
- Eelen, G.; Dubois, C.; Cantelmo, A.R.; Goveia, J.; Brüning, U.; DeRan, M.; Jarugumilli, G.; van Rijssel, J.; Saladino, G.; Comitani, F. Role of glutamine synthetase in angiogenesis beyond glutamine synthesis. Nature 2018, 561, 63–69. [Google Scholar] [CrossRef]
- Sancak, Y.; Peterson, T.R.; Shaul, Y.D.; Lindquist, R.A.; Thoreen, C.C.; Bar-Peled, L.; Sabatini, D.M. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science 2008, 320, 1496–1501. [Google Scholar] [CrossRef] [PubMed]
- Lei, H.-T.; Mu, X.; Hattne, J.; Gonen, T. A conformational change in the N terminus of SLC38A9 signals mTORC1 activation. Structure 2021, 29, 426–432.e428. [Google Scholar] [CrossRef] [PubMed]
- Han, J.; Wang, Y. mTORC1 signaling in hepatic lipid metabolism. Protein Cell 2018, 9, 145–151. [Google Scholar] [CrossRef] [PubMed]
- Sethna, S.; Scott, P.A.; Giese, A.P.; Duncan, T.; Jian, X.; Riazuddin, S.; Randazzo, P.A.; Redmond, T.M.; Bernstein, S.L.; Riazuddin, S.J.N.c. CIB2 regulates mTORC1 signaling and is essential for autophagy and visual function. Nat. Commun. 2021, 12, 3906. [Google Scholar] [CrossRef]
- Schiaffino, S.; Reggiani, C.; Akimoto, T.; Blaauw, B. Molecular mechanisms of skeletal muscle hypertrophy. J. Neuromuscul. Dis. 2021, 8, 169–183. [Google Scholar] [CrossRef]
- Pang, B.; Yu, X.; Bowker, B.; Zhang, J.; Yang, Y.; Zhuang, H. Effect of meat temperature on moisture loss, water properties, and protein profiles of broiler pectoralis major with the woody breast condition. Poult. Sci. 2021, 100, 1283–1290. [Google Scholar] [CrossRef]
- Certo, M.; Moore, V.D.G.; Nishino, M.; Wei, G.; Korsmeyer, S.; Armstrong, S.A.; Letai, A. Mitochondria primed by death signals determine cellular addiction to antiapoptotic BCL-2 family members. Cancer Cell 2006, 9, 351–365. [Google Scholar] [CrossRef]
- Carrasco-Rozas, A.; Fernández-Simón, E.; Suárez-Calvet, X.; Piñol-Jurado, P.; Alonso-Pérez, J.; de Luna, N.; Schoser, B.; Meinke, P.; Domínguez-González, C.; Hernández-Laín, A. BNIP3 is involved in muscle fiber atrophy in late-onset pompe disease patients. Am. J. Pathol. 2022, 192, 1151–1166. [Google Scholar] [CrossRef]
- Antonicka, H.; Lin, Z.-Y.; Janer, A.; Aaltonen, M.J.; Weraarpachai, W.; Gingras, A.-C.; Shoubridge, E.A. A high-density human mitochondrial proximity interaction network. Cell Metab. 2020, 32, 479–497.e479. [Google Scholar] [CrossRef]
- Zhang, S.; Cheng, Z.M.; Yu, J.L.; Lu, K.; Xu, S.J.; Lu, Y.; Liu, T.; Xia, B.J.; Huang, Z.; Zhao, X.Y. Malic enzyme 2 promotes the progression of hepatocellular carcinoma via increasing triglyceride production. Cancer Med. 2021, 10, 6795–6806. [Google Scholar] [CrossRef]
- Kozák, J. Variations of geese under domestication. Poult. Sci. J. 2019, 75, 247–260. [Google Scholar] [CrossRef]
- Muroya, S.; Ueda, S.; Komatsu, T.; Miyakawa, T.; Ertbjerg, P. MEATabolomics: Muscle and meat metabolomics in domestic animals. Metabolites 2020, 10, 188. [Google Scholar] [CrossRef]
- Lonergan, E.H.; Zhang, W.; Lonergan, S.M. Biochemistry of postmortem muscle—Lessons on mechanisms of meat tenderization. Meat Sci. 2010, 86, 184–195. [Google Scholar] [CrossRef]
- Suman, S.P.; Wang, Y.; Gagaoua, M.; Kiyimba, F.; Ramanathan, R. Proteomic approaches to characterize biochemistry of fresh beef color. J. Proteom. 2023, 281, 104893. [Google Scholar] [CrossRef]
- He, J.; Zhang, P.; Shen, L.; Niu, L.; Tan, Y.; Chen, L.; Zhao, Y.; Bai, L.; Hao, X.; Li, X.; et al. Short-chain fatty acids and their association with signalling pathways in inflammation, glucose and lipid metabolism. Int. J. Mol. Sci. 2020, 21, 6356. [Google Scholar] [CrossRef]
- Patra, A.K. Agriculture. Meta-analyses of effects of phytochemicals on digestibility and rumen fermentation characteristics associated with methanogenesis. J. Sci. Food 2010, 90, 2700–2708. [Google Scholar] [CrossRef]

| SNPs | 2nd-PCRP | 2nd-PCRP | UEP_SEQ |
|---|---|---|---|
| chr13:25432644 | ACGTTGGATGGAAATACCCTGTTGTCTCCC | ACGTTGGATGACCTGTTTGACTCCTTTTGG | agaaCCTGTTGTCTCCCTCACTC |
| ctg930:11921 | ACGTTGGATGTGCCACCGCAGGGATCACG | ACGTTGGATGTGGCAGCAGGGTGGGGAAA | GCCCCCTCCTGCACCTT |
| chr36:5611038 | ACGTTGGATGTTCCCCCCTCGTTTGAATTG | ACGTTGGATGCCCAGTCTGAATTCCAACAC | tCGCTTTGATTTAGTTATTTTACTC |
| ctg2092:176333 | ACGTTGGATGGGAATATGTAGACTACGTCTG | ACGTTGGATGTTTTGGACAAACAGGAGACC | cccaTAGACTACGTCTGCCATCT |
| chr29:8278595 | ACGTTGGATGAAGGATTTGGGAAGCAGGAAC | ACGTTGGATGTGCACCGGGGAGGAGGAGA | CAGGAACCGAGGGAAATGC |
| chr9:33911228 | ACGTTGGATGCCTGGAGGCAATCAAAGATG | ACGTTGGATGCTTAAGTCGCCTTGGTACAC | CAGGAGTTAAGGGAGAAAAT |
| chr1:24867798 | ACGTTGGATGAGCCTGATGCAGTCACATCC | ACGTTGGATGGTGTGTGCCAGACAAAACTC | GCTGCCTGCCCAGAACT |
| chr7:16992800 | ACGTTGGATGTCACATTGGCAGGGTCCAAC | ACGTTGGATGTTATAGTCTGCTCTGGACTG | gtCAGGGTCCAACTCAGTCTCC |
| chr12:31782870 | ACGTTGGATGACAAAGAACACATCGCAAGG | ACGTTGGATGGGCAGCAGCTTTCAGCAAAC | CAATAATGTTTAACGTTAGACTC |
| chr12:31781825 | ACGTTGGATGTTCTGCAACGCTCGAAATCC | ACGTTGGATGTTTAACCTACGCATGCCTCC | CCAAAGACCTGTTGAGA |
| chr2:36812553 | ACGTTGGATGGTGGAAGAAGACATCACTGG | ACGTTGGATGGCAGGAGAAAAAAGCATAAG | tcGATCTTACTTTTTTATCTTCCATTA |
| Traits | Number | Mean | STDV | Minimum | Maximum | CV (%) |
|---|---|---|---|---|---|---|
| CFC | 199 | 9.92 | 1.89 | 4.69 | 15.52 | 0.19 |
| MCFD | 199 | 75.07 | 0.94 | 72.26 | 77.54 | 0.01 |
| CLR | 194 | 13.13 | 3.01 | 4.66 | 22.36 | 0.23 |
| L* (meat lightness) | 205 | 23.6 | 3.65 | 15.53 | 37.29 | 0.15 |
| a* (meat redness) | 205 | 48.77 | 3.02 | 39.76 | 56.06 | 0.06 |
| b* (meat yellowness) | 205 | 19.5 | 2.32 | 13.05 | 30.8 | 0.12 |
| SF (kgf) | 197 | 3.81 | 0.83 | 1.66 | 5.82 | 0.22 |
| SNP | Chr | Position (bp) | Allele1 | Traits | p Value | Gene |
|---|---|---|---|---|---|---|
| chr11:9193757 | chr11 | 9193757 | G/A | CFC | 8.12 × 10−8 | TMEM65 |
| chr19:2113530 | chr19 | 2113530 | C/T | CFC | 6.34 × 10−8 | SMAD6 |
| chr19:17076992 | chr19 | 17076992 | T/G | CLR | 1.03 × 10−7 | SYTC2 |
| chr27:11913710 | chr27 | 11913710 | G/A | CLR | 3.16 × 10−8 | MSI1H |
| chr1:42733613 | chr1 | 42733613 | T/C | L* (meat lightness) | 7.56 × 10−8 | LIN1 |
| chr14:440052 | chr14 | 440052 | T/C | L* (meat lightness) | 8.15 × 10−8 | GSS |
| chr1:1971942 | chr1 | 1971942 | C/T | b* (meat yellowness) | 2.74 × 10−8 | EXTL3 |
| chr1:24867798 | chr1 | 24867798 | C/G | b* (meat yellowness) | 7.82 × 10−8 | PRSS55 |
| chr1:31137893 | chr1 | 31137893 | C/T | b* (meat yellowness) | 9.31 × 10−8 | PTP4A1 |
| chr3:31387295 | chr3 | 31387295 | G/A | b* (meat yellowness) | 5.43 × 10−9 | TMEM19 |
| chr3:31387314 | chr3 | 31387314 | G/A | b* (meat yellowness) | 3.57 × 10−8 | TMEM19 |
| chr3:31387324 | chr3 | 31387324 | A/G | b* (meat yellowness) | 3.57 × 10−8 | TMEM19 |
| chr7:8983345 | chr7 | 8983345 | A/G | b* (meat yellowness) | 3.20 × 10−8 | ACKR3 |
| chr7:16992800 | chr7 | 16992800 | C/T | b* (meat yellowness) | 7.84 × 10−8 | CHIN |
| chr7:4199190 | chr7 | 4199190 | A/G | b* (meat yellowness) | 4.47 × 10−8 | GULP1 |
| chr9:33911228 | chr9 | 33911228 | A/G | b* (meat yellowness) | 7.35 × 10−8 | DOCK1 |
| chr11:27962909 | chr11 | 27962909 | A/G | b* (meat yellowness) | 3.43 × 10−8 | PP4R1 |
| chr11:27964588 | chr11 | 27964588 | G/T | b* (meat yellowness) | 4.66 × 10−9 | SLCO5A1 |
| chr13:25432644 | chr13 | 25432644 | C/T | b* (meat yellowness) | 2.55 × 10−9 | CATC |
| chr13:25426268 | chr13 | 25426268 | G/A | b* (meat yellowness) | 2.62 × 10−9 | CATC |
| chr13:25389761 | chr13 | 25389761 | T/G | b* (meat yellowness) | 9.87 × 10−8 | CATC |
| chr26:4307814 | chr26 | 4307814 | T/C | b* (meat yellowness) | 4.26 × 10−8 | PP4R1 |
| chr29:8720675 | chr29 | 8720675 | T/C | b* (meat yellowness) | 1.16 × 10−8 | MRPS2 |
| chr33:5370910 | chr33 | 5370910 | A/C | b* (meat yellowness) | 6.90 × 10−8 | NO40 |
| chr33:5659799 | chr33 | 5659799 | A/C | b* (meat yellowness) | 3.33 × 10−8 | PUM1 |
| chr33:3667950 | chr33 | 3667950 | C/T | b* (meat yellowness) | 8.99 × 10−8 | RRAGC |
| chr36:5611038 | chr36 | 5611038 | G/T | b* (meat yellowness) | 4.99 × 10−8 | GOGA7 |
| ctg2092:176333 | ctg2092 | 176333 | C/T | b* (meat yellowness) | 6.16 × 10−8 | RXRA |
| ctg745:127150 | ctg745 | 127150 | A/G | b* (meat yellowness) | 5.53 × 10−8 | NCOA2 |
| chr4:23787309 | chr4 | 23787309 | A/G | b* (meat yellowness) | 2.08 × 10−8 | None |
| chr4:49863664 | chr4 | 49863664 | T/A | b* (meat yellowness) | 5.49 × 10−8 | None |
| chr25:12875901 | chr25 | 12875901 | A/G | b* (meat yellowness) | 5.77 × 10−9 | None |
| chr29:8278595 | chr29 | 8278595 | C/T | b* (meat yellowness) | 6.64 × 10−8 | None |
| ctg834:24329 | ctg834 | 24329 | C/T | b* (meat yellowness) | 5.15 × 10−8 | None |
| ctg930:11921 | ctg930 | 11921 | G/A | b* (meat yellowness) | 5.30 × 10−9 | None |
| ctg956:92804 | ctg956 | 92804 | C/T | b* (meat yellowness) | 2.73 × 10−8 | None |
| chr2:36812553 | chr2 | 36812553 | C/T | SF (kgf) | 5.94 × 10−8 | BMF |
| chr2:36812412 | chr2 | 36812412 | G/C | SF (kgf) | 9.22 × 10−8 | BMF |
| chr7:33558795 | chr7 | 33558795 | A/T | SF (kgf) | 3.84 × 10−8 | NEMP2 |
| chr7:33558758 | chr7 | 33558758 | C/A | SF (kgf) | 8.04 × 10−8 | NEMP2 |
| chr7:33558832 | chr7 | 33558832 | T/C | SF (kgf) | 9.30 × 10−8 | NEMP2 |
| chr12:31782870 | chr12 | 31782870 | A/G | SF (kgf) | 2.78 × 10−10 | NAD-ME |
| chr12:31781825 | chr12 | 31781825 | G/A | SF (kgf) | 3.62 × 10−9 | NAD-ME |
| Traits | Number | SNPs | Genotypes (Frequency %) | ||
|---|---|---|---|---|---|
| b* (meat yellowness) | 203 | chr13:25432644 | CC | CT | TT |
| 17.61 ± 2.07 b (1.97) | 17.56 ± 2.92 b (0.66) | 19.31 ± 0.25 a (97.37) | |||
| b* (meat yellowness) | 203 | ctg930:11921 | AA | GA | GG |
| 19.18 ± 0.23 b (72.37) | 20.28 ± 0.41 a (23.03) | 19.03 ± 0.88 b (4.61) | |||
| b* (meat yellowness) | 203 | chr36:5611038 | GG | GT | TT |
| 19.83 ± 2.76 a (0.58) | 17.71 ± 1.04 b (4.62) | 19.44 ± 0.22 a (94.80) | |||
| b* (meat yellowness) | 203 | ctg2092:176333 | CC | TC | TT |
| 18.04 ± 1.15 b (3.49) | 19.24 ± 0.45 a (13.56) | 19.48 ± 0.26 a (70.93) | |||
| b* (meat yellowness) | 203 | chr29:8278595 | CC | CT | TT |
| 17.99 ± 2.80 b (0.56) | 19.21 ± 0.60 a (13.56) | 19.39 ± 0.23 a (85.88) | |||
| b* (meat yellowness) | 203 | chr9:33911228 | AA | GA | GG |
| 20.32 ± 1.62 a (2.27) | 19.52 ± 0.75 b (8.52) | 19.31 ± 0.23 b (89.20) | |||
| b* (meat yellowness) | 203 | chr1:24867798 | CC | GC | GG |
| 16.33 ± 2.79 b (0.57) | 19.08 ± 0.72 a (9.09) | 19.41 ± 0.23 a (90.34) | |||
| b* (meat yellowness) | 203 | chr7:16992800 | CC | CT | TT |
| 15.43 ± 2.77 b (1.12) | 19.30 ± 0.57 a (13.48) | 19.40 ± 0.23 a (85.39) | |||
| SF | 203 | chr12:31782870 | AA | GA | GG |
| 3.12 ± 0.97 b (1.12) | 3.82 ± 0.25 a (8.43) | 3.74 ± 0.08 a (90.45) | |||
| SF | 203 | chr12:31781825 | AA | GA | GG |
| 3.72 ± 0.08 b (87.01) | 3.89 ± 0.22 a (12.99) | \ | |||
| SF | 203 | chr2:36812553 | CC | CT | TT |
| \ | 3.60 ± 0.22 b (15.49) | 3.73 ± 0.09 a (84.51) | |||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gao, G.; Zhang, K.; Huang, P.; Zhao, X.; Li, Q.; Xie, Y.; Yin, C.; Li, J.; Wang, Z.; Zhong, H.; et al. Identification of SNPs Associated with Goose Meat Quality Traits Using a Genome-Wide Association Study Approach. Animals 2023, 13, 2089. https://doi.org/10.3390/ani13132089
Gao G, Zhang K, Huang P, Zhao X, Li Q, Xie Y, Yin C, Li J, Wang Z, Zhong H, et al. Identification of SNPs Associated with Goose Meat Quality Traits Using a Genome-Wide Association Study Approach. Animals. 2023; 13(13):2089. https://doi.org/10.3390/ani13132089
Chicago/Turabian StyleGao, Guangliang, Keshan Zhang, Ping Huang, Xianzhi Zhao, Qin Li, Youhui Xie, Chunhui Yin, Jing Li, Zhen Wang, Hang Zhong, and et al. 2023. "Identification of SNPs Associated with Goose Meat Quality Traits Using a Genome-Wide Association Study Approach" Animals 13, no. 13: 2089. https://doi.org/10.3390/ani13132089
APA StyleGao, G., Zhang, K., Huang, P., Zhao, X., Li, Q., Xie, Y., Yin, C., Li, J., Wang, Z., Zhong, H., Xue, J., Chen, Z., Wu, X., & Wang, Q. (2023). Identification of SNPs Associated with Goose Meat Quality Traits Using a Genome-Wide Association Study Approach. Animals, 13(13), 2089. https://doi.org/10.3390/ani13132089








