Goat Pleomorphic Adenoma Gene 1 (PLAG1): mRNA Expression, CNV Detection and Associations with Growth Traits
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animal Welfare Explanation
2.2. Identification and Bioinformatics Analysis of the PLAG1 Gene
2.3. Extraction of Genomic DNA and RNA
2.4. Designing of Primers
2.5. Detection of the PLAG1 Gene mRNA Expression Levels
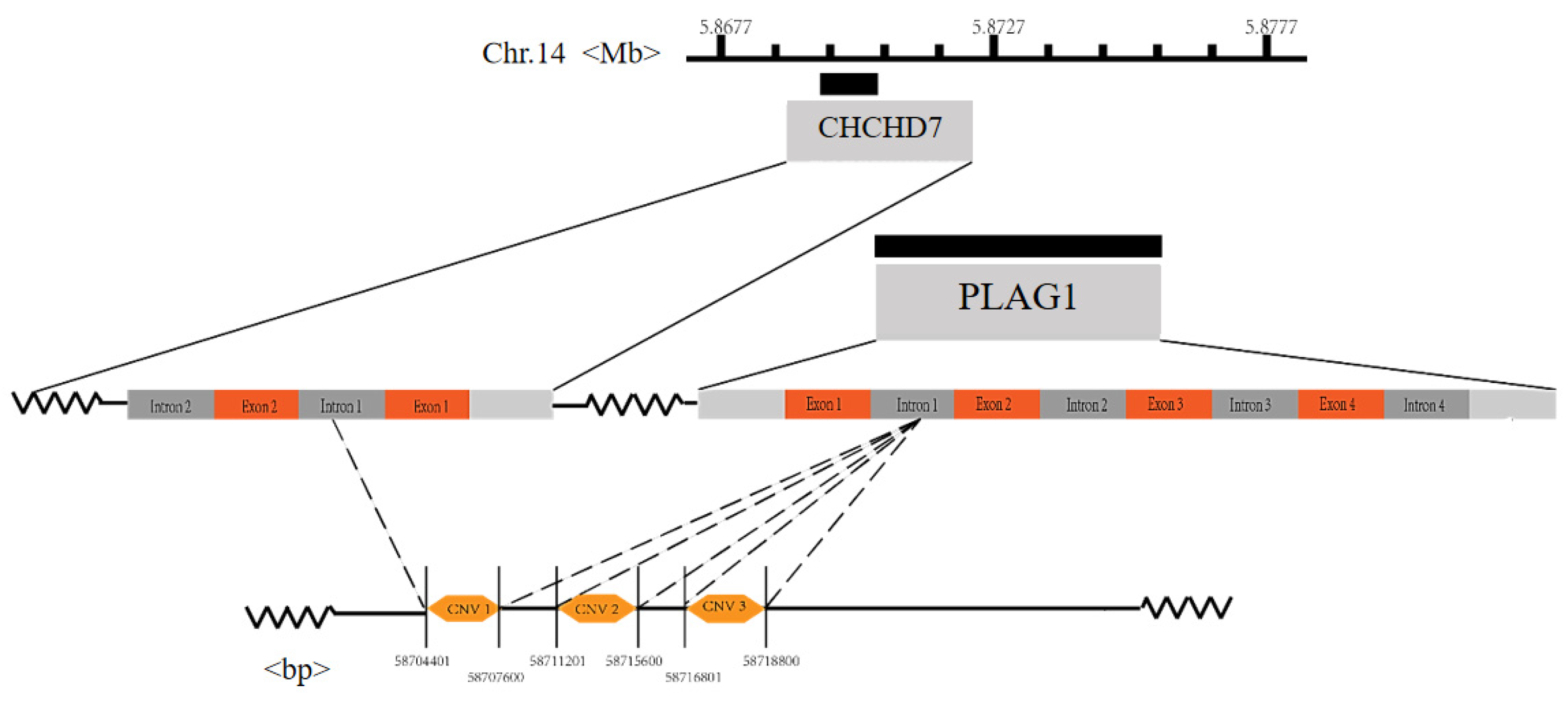
2.6. CNV Genotyping of the PLAG1 Gene
2.7. Statistical Analyses
3. Results
3.1. Bioinformatics Analysis of PLAG1 Gene
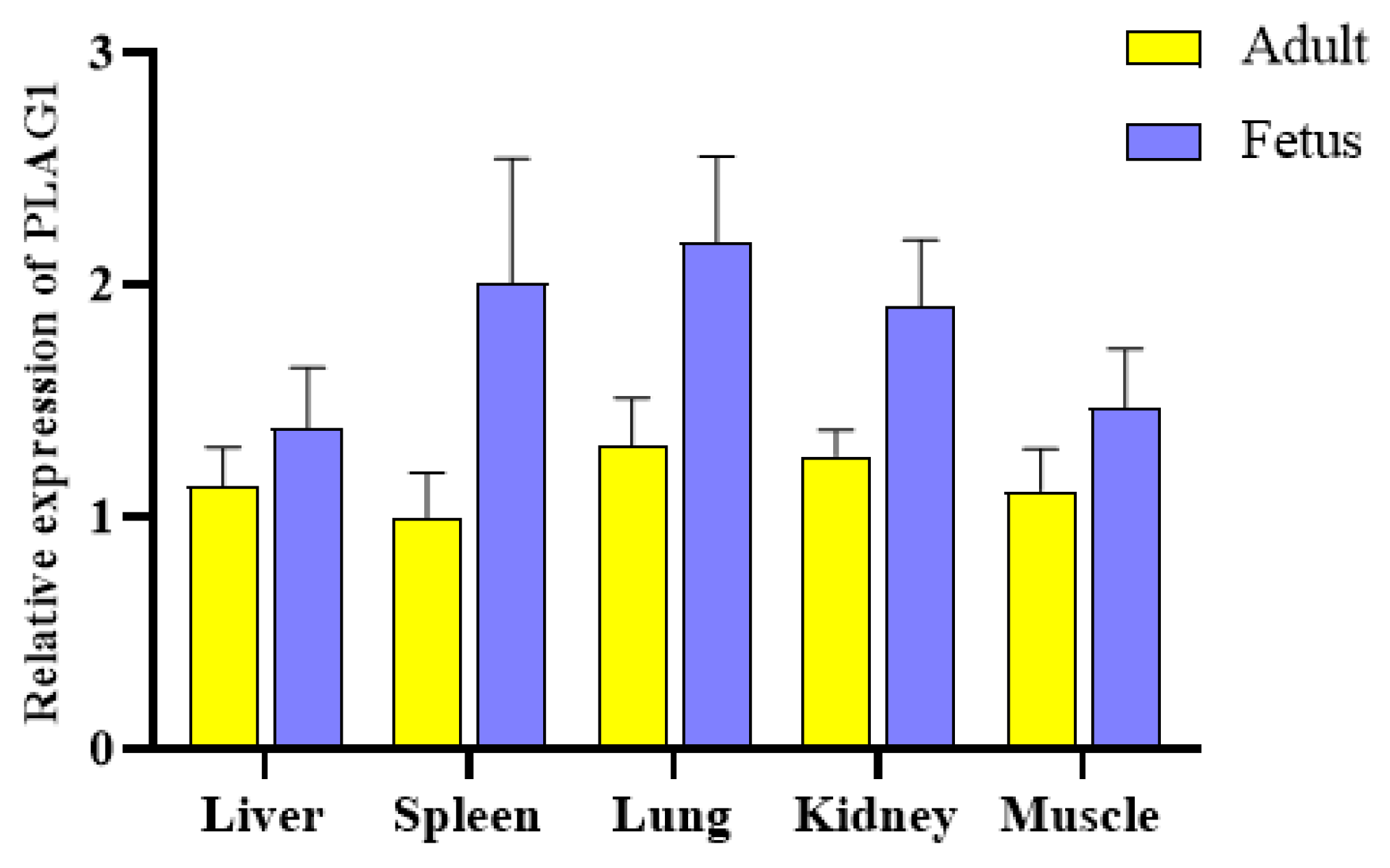
3.2. mRNA Expression Levels of Fetal and Adult Goats
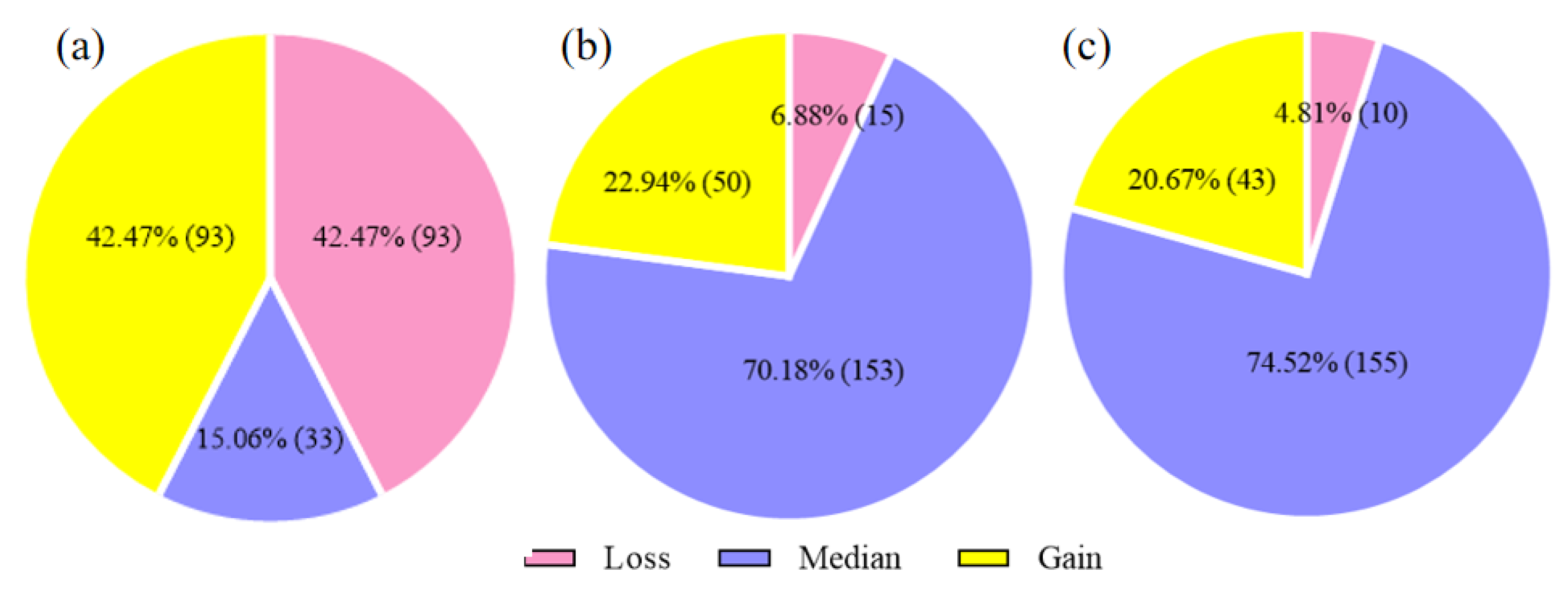
3.3. Frequency of CNV Genotypes
3.4. Association Analyses between CNV and the Goat PLAG1 Gene
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Corva, P.M.; Medrano, J.F. Quantitative trait loci (QTLs) mapping for growth traits in the mouse: A review. Genet. Sel. Evol. 2001, 33, 105–132. [Google Scholar] [CrossRef] [PubMed]
- Pös, O.; Radvanszky, J.; Buglyó, G.; Pös, Z.; Rusnakova, D.; Nagy, B.; Szemes, T. DNA copy number variation: Main characteristics, evolutionary significance, and pathological aspects. Biomed. J. 2021, 44, 548–559. [Google Scholar] [CrossRef] [PubMed]
- Takasuga, A. PLAG1 and NCAPG-LCORL in livestock. Anim. Sci. J. 2016, 87, 159–167. [Google Scholar] [CrossRef] [PubMed]
- Juma, A.R.; Damdimopoulou, P.E.; Grommen, S.V.; Van de Ven, W.J.; De Groef, B. Emerging role of PLAG1 as a regulator of growth and reproduction. J. Endocrinol. 2016, 228, R45–R56. [Google Scholar] [CrossRef]
- Wang, J.; Huang, Y.; Xu, J.; Yue, B.; Wen, Y.; Wang, X.; Lei, C.; Chen, H. Pleomorphic adenoma gene 1 (PLAG1) promotes proliferation and inhibits apoptosis of bovine primary myoblasts through the PI3K-Akt signaling pathway. J. Anim. Sci. 2022, 100, skac098. [Google Scholar] [CrossRef]
- Dadousis, C.; Somavilla, A.; Ilska, J.J.; Johnsson, M.; Batista, L.; Mellanby, R.J.; Headon, D.; Gottardo, P.; Whalen, A.; Wilson, D.; et al. A genome-wide association analysis for body weight at 35 days measured on 137,343 broiler chickens. Genet. Sel. Evol. 2021, 53, 70. [Google Scholar] [CrossRef]
- Song, Y.; Xu, L.; Chen, Y.; Zhang, L.; Gao, H.; Zhu, B.; Niu, H.; Zhang, W.; Xia, J.; Gao, X.; et al. Genome-wide association study reveals the PLAG1 gene for knuckle, biceps and shank weight in Simmental beef cattle. PLoS ONE 2016, 11, e0168316. [Google Scholar] [CrossRef]
- Hoshiba, H.; Setoguchi, K.; Watanabe, T.; Kinoshita, A.; Mizoshita, K.; Sugimoto, Y.; Takasuga, A. Comparison of the effects explained by variations in the bovine PLAG1 and NCAPG genes on daily body weight gain, linear skeletal measurements and carcass traits in Japanese Black steers from a progeny testing program. Anim. Sci. J. 2013, 84, 529–534. [Google Scholar] [CrossRef]
- Pan, Y.; Wang, M.; Wu, H.; Akhatayeva, Z.; Lan, X.; Fei, P.; Mao, C.; Jiang, F. Indel mutations of sheep PLAG1 gene and their associations with growth traits. Anim. Biotechnol. 2022, 33, 1459–1465. [Google Scholar] [CrossRef]
- Zhou, Z.; Huang, B.; Lai, Z.; Li, S.; Wu, F.; Qu, K.; Jia, Y.; Hou, J.; Liu, J.; Lei, C.; et al. The distribution characteristics of a 19-bp Indel of the PLAG1 gene in Chinese cattle. Animals 2019, 9, 1082. [Google Scholar] [CrossRef]
- Xu, W.; He, H.; Zheng, L.; Xu, J.W.; Lei, C.Z.; Zhang, G.M.; Dang, R.H.; Niu, H.; Qi, X.L.; Chen, H.; et al. Detection of 19-bp deletion within PLAG1 gene and its effect on growth traits in cattle. Gene 2018, 675, 144–149. [Google Scholar] [CrossRef]
- Zhong, J.L.; Xu, J.W.; Wang, J.; Wen, Y.F.; Niu, H.; Zheng, L.; He, H.; Peng, K.; He, P.; Shi, S.Y.; et al. A novel SNP of PLAG1 gene and its association with growth traits in Chinese cattle. Gene 2019, 689, 166–171. [Google Scholar] [CrossRef]
- Zhang, R.; Miao, J.; Song, Y.; Zhang, W.; Xu, L.; Chen, Y.; Zhang, L.; Gao, H.; Zhu, B.; Li, J.; et al. Genome-wide association study identifies the PLAG1-OXR1 region on BTA14 for carcass meat yield in cattle. Physiol. Genom. 2019, 51, 137–144. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, M.; Yuan, J.; Zhou, X.; Xu, S.; Liu, B. Association of polymorphisms in NR6A1, PLAG1 and VRTN with the number of vertebrae in Chinese Tongcheng × Large White crossbred pigs. Anim. Genet. 2018, 49, 353–354. [Google Scholar] [CrossRef]
- Jin, L.; Chun, J.; Pan, C.; Kumar, A.; Zhang, G.; Ha, Y.; Li, D.; Alesi, G.N.; Kang, Y.; Zhou, L.; et al. The PLAG1-GDH1 axis promotes anoikis resistance and tumor metastasis through CamKK2-AMPK signaling in LKB1-Deficient lung cancer. Mol. Cell 2018, 69, 87–99. [Google Scholar] [CrossRef]
- Chiang, S. Recent advances in smooth muscle tumors with PGR and PLAG1 gene fusions and myofibroblastic uterine neoplasms. Genes Chromosom. Cancer 2021, 60, 138–146. [Google Scholar] [CrossRef]
- Thiryayi, S.A.; Turashvili, G.; Latta, E.K.; Swanson, D.; Zhang, L.; Antonescu, C.R.; Dickson, B.C. PLAG1-rearrangment in a uterine leiomyosarcoma with myxoid stroma and heterologous differentiation. Genes Chromosom. Cancer 2021, 60, 713–717. [Google Scholar] [CrossRef]
- Huang, W.; Li, B.R.; Feng, H. PLAG1 silencing promotes cell chemosensitivity in ovarian cancer via the IGF2 signaling pathway. Int. J. Mol. Med. 2020, 45, 703–714. [Google Scholar] [CrossRef]
- Zheng, Y.; Xu, L.; Hassan, M.; Zhou, X.; Zhou, Q.; Rakheja, D.; Skapek, S.X. Bayesian modeling identifies PLAG1 as a key regulator of proliferation and survival in rhabdomyosarcoma cells. Mol. Cancer Res. 2020, 18, 364–374. [Google Scholar] [CrossRef]
- Rajender, S.; Avery, K.; Agarwal, A. Epigenetics, spermatogenesis and male infertility. Mutat. Res. 2011, 727, 62–71. [Google Scholar] [CrossRef]
- Wong, J.; Juma, A.R.; Tran, S.C.; Gasperoni, J.G.; Grommen, S.V.H.; De Groef, B. Deficiency of the transcription factor PLAG1 results in aberrant coiling and morphology of the epididymis. Asian J. Androl. 2020, 22, 342–347. [Google Scholar] [PubMed]
- Juma, A.R.; Grommen, S.V.H.; O’Bryan, M.K.; O’Connor, A.E.; Merriner, D.J.; Hall, N.E.; Doyle, S.R.; Damdimopoulou, P.E.; Barriga, D.; Hart, A.H.; et al. PLAG1 deficiency impairs spermatogenesis and sperm motility in mice. Sci. Rep. 2017, 7, 5317. [Google Scholar] [CrossRef] [PubMed]
- Juma, A.R.; Hall, N.E.; Wong, J.; Gasperoni, J.G.; Watanabe, Y.; Sahota, A.; Damdimopoulou, P.E.; Grommen, S.V.H.; De Groef, B. PLAG1 expression and target genes in the hypothalamo-pituitary system in male mice. Mol. Cell. Endocrinol. 2018, 478, 77–83. [Google Scholar] [CrossRef] [PubMed]
- Sakai, H.; Fujii, Y.; Kuwayama, N.; Kawaji, K.; Gotoh, Y.; Kishi, Y. Plag1 regulates neuronal gene expression and neuronal differentiation of neocortical neural progenitor cells. Genes Cells 2019, 24, 650–666. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 2019, 20, 1160–1166. [Google Scholar] [CrossRef]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; von Haeseler, A.; Lanfear, R. IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef]
- Kang, Z.; Jiang, E.; Wang, K.; Pan, C.; Chen, H.; Yan, H.; Zhu, H.; Liu, J.; Qu, L.; Lan, X. Goat membrane associated ring-CH-type finger 1 (MARCH1) mRNA expression and association with litter size. Theriogenology 2019, 128, 8–16. [Google Scholar] [CrossRef]
- Aljanabi, S.M.; Martinez, I. Universal and rapid salt-extraction of high quality genomic DNA for PCR-based techniques. Nucleic Acids Res. 1997, 25, 4692–4693. [Google Scholar] [CrossRef]
- Fu, W.; Wang, R.; Yu, J.; Hu, D.; Cai, Y.; Shao, J.; Jiang, Y. GGVD: A goat genome variation database for tracking the dynamic evolutionary process of selective signatures and ancient introgressions. J. Genet. Genom. 2021, 48, 248–256. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, S.; Tang, Q.; Jiang, E.; Wang, K.; Lan, X.; Pan, C. Goat sperm associated antigen 17 protein gene (SPAG17): Small and large fragment genetic variation detection, association analysis, and mRNA expression in gonads. Genomics 2020, 112, 5115–5121. [Google Scholar] [CrossRef]
- Hui, Y.; Zhang, Y.; Wang, K.; Pan, C.; Chen, H.; Qu, L.; Song, X.; Lan, X. Goat DNMT3B: An indel mutation detection, association analysis with litter size and mRNA expression in gonads. Theriogenology 2020, 147, 108–115. [Google Scholar] [CrossRef]
- Pei, S.W.; Qin, F.; Li, W.H.; Li, F.D.; Yue, X.P. Copy number variation of ZNF280AY across 21 cattle breeds and its association with the reproductive traits of Holstein and Simmental bulls. J. Dairy Sci. 2019, 102, 7226–7236. [Google Scholar] [CrossRef]
- Yang, Q.; Zhang, S.; Li, J.; Wang, X.; Peng, K.; Lan, X.; Pan, C. Development of a touch-down multiplex PCR method for simultaneously rapidly detecting three novel insertion/deletions (indels) within one gene: An example for goat GHR gene. Anim. Biotechnol. 2019, 30, 366–371. [Google Scholar] [CrossRef]
- Declercq, J.; Van Dyck, F.; Braem, C.V.; Van Valckenborgh, I.C.; Voz, M.; Wassef, M.; Schoonjans, L.; Van Damme, B.; Fiette, L.; Van de Ven, W.J. Salivary gland tumors in transgenic mice with targeted PLAG1 proto-oncogene overexpression. Cancer Res. 2005, 65, 4544–4553. [Google Scholar] [CrossRef]
- Li, Z.; Wu, M.; Zhao, H.; Fan, L.; Zhang, Y.; Yuan, T.; He, S.; Wang, P.; Zhang, Y.; Sun, X.; et al. The PLAG1 mRNA expression analysis among genetic variants and relevance to growth traits in Chinese cattle. Anim. Biotechnol. 2020, 31, 504–511. [Google Scholar] [CrossRef]
- Hensen, K.; Braem, C.; Declercq, J.; Van Dyck, F.; Dewerchin, M.; Fiette, L.; Denef, C.; Van de Ven, W.J. Targeted disruption of the murine Plag1 proto-oncogene causes growth retardation and reduced fertility. Dev. Growth Differ. 2004, 46, 459–470. [Google Scholar] [CrossRef]
- Madissoon, E.; Damdimopoulos, A.; Katayama, S.; Krjutškov, K.; Einarsdottir, E.; Mamia, K.; De Groef, B.; Hovatta, O.; Kere, J.; Damdimopoulou, P. Pleomorphic adenoma gene 1 is needed for timely zygotic genome activation and early embryo development. Sci. Rep. 2019, 9, 8411. [Google Scholar] [CrossRef]
- Fink, T.; Tiplady, K.; Lopdell, T.; Johnson, T.; Snell, R.G.; Spelman, R.J.; Davis, S.R.; Littlejohn, M.D. Functional confirmation of PLAG1 as the candidate causative gene underlying major pleiotropic effects on body weight and milk characteristics. Sci. Rep. 2017, 7, 44793. [Google Scholar] [CrossRef]
- Nishimura, S.; Watanabe, T.; Mizoshita, K.; Tatsuda, K.; Fujita, T.; Watanabe, N.; Sugimoto, Y.; Takasuga, A. Genome-wide association study identified three major QTL for carcass weight including the PLAG1-CHCHD7 QTN for stature in Japanese Black cattle. BMC Genet. 2012, 13, 40. [Google Scholar] [CrossRef]
- Michael, A.K.; Thomä, N.H. Reading the chromatinized genome. Cell 2021, 184, 3599–3611. [Google Scholar] [CrossRef]
- Li, M.; Yin, C.; Zhao, F.; Liu, Y. Copy number variation association studies for sheep tail-relevant traits in Hulunbuir sheep. Anim. Genet. 2022, 53, 897–900. [Google Scholar] [CrossRef] [PubMed]
- Jiang, R.; Cheng, J.; Cao, X.K.; Ma, Y.L.; Chaogetu, B.; Huang, Y.Z.; Lan, X.Y.; Lei, C.Z.; Hu, L.Y.; Chen, H. Copy number variation of the SHE gene in sheep and its association with economic traits. Animals 2019, 9, 531. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Cao, X.; Ma, Y.; Cheng, J.; Song, C.; Jiang, R.; Wang, X.; Huang, Y.; Buren, C.; Lan, X.; et al. Novel copy number variation of the BAG4 gene is associated with growth traits in three Chinese sheep populations. Anim. Biotechnol. 2021, 32, 461–469. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Zhou, Y.; Rosen, B.D.; Van Tassell, C.P.; Stella, A.; Tosser-Klopp, G.; Rupp, R.; Palhière, I.; Colli, L.; Sayre, B.; et al. Diversity of copy number variation in the worldwide goat population. Heredity 2019, 122, 636–646. [Google Scholar] [CrossRef]
- Shi, S.Y.; Li, L.J.; Zhang, Z.J.; Wang, E.Y.; Wang, J.; Xu, J.W.; Liu, H.B.; Wen, Y.F.; He, H.; Lei, C.Z.; et al. Copy number variation of MYLK4 gene and its growth traits of capra hircus (goat). Anim. Biotechnol. 2020, 31, 532–537. [Google Scholar] [CrossRef]
- Zhang, Z.; Peng, M.; Wen, Y.; Chai, Y.; Liang, J.; Yang, P.; Liu, X.; Li, J.; Huang, Y.; Li, L.; et al. Copy number variation of EIF4A2 loci related to phenotypic traits in Chinese cattle. Vet. Med. Sci. 2022, 8, 2147–2156. [Google Scholar] [CrossRef]
- Liang, J.; Liu, X.; Yang, P.; Yao, Z.; Qu, K.; Wang, H.; Zhang, Z.; Liang, H.; Cheng, B.; Li, Z.; et al. Copy number variation of GAL3ST1 gene is associated with growth traits of Chinese cattle. Anim. Biotechnol. 2022, 8, 672–678. [Google Scholar] [CrossRef]
- Li, X.; Ding, X.; Liu, L.; Yang, P.; Yao, Z.; Lei, C.; Chen, H.; Huang, Y.; Liu, W. Copy number variation of bovine DYNC1I2 gene is associated with body conformation traits in chinese beef cattle. Gene 2022, 810, 146060. [Google Scholar] [CrossRef]
- Skinner, B.M.; Robertson, L.B.; Tempest, H.G.; Langley, E.J.; Ioannou, D.; Fowler, K.E.; Crooijmans, R.P.; Hall, A.D.; Griffin, D.K.; Völker, M. Comparative genomics in chicken and Pekin duck using FISH mapping and microarray analysis. BMC Genom. 2009, 10, 357. [Google Scholar] [CrossRef]
- Lauer, S.; Gresham, D. An evolving view of copy number variants. Curr. Genet. 2019, 65, 1287–1295. [Google Scholar] [CrossRef]

| Primer Names | Primer Pairs (5′-3′) | Tm (°C) | Length (bp) |
|---|---|---|---|
| PLAG1-mRNA | F: ATTCCGCGGGCGGTGTAAA | 62.61 | 298 |
| R: TCACCAGGAATGACAGTGGC | 59.96 | ||
| GAPDH | F: AAAGTGGACATCGTTGCCAT | 60.04 | 116 |
| R: CCGTTCTCTGCCTTGACTGT | 59.97 | ||
| CNV1 | F: CTTTCGACGGCACCCAAGAA | 60.88 | 199 |
| R: TTGCCGTTGCGCTTCCT | 60.26 | ||
| CNV2 | F: CTCAAACATGAATTTGCTCCCC | 58.15 | 250 |
| R: AGACTGCTTCCACCACTCATA | 58.46 | ||
| CNV3 | F: CGTATAGGGGAAGCTCAG | 50.20 | 192 |
| R: GTTTGGATGTCTTCTATTTCTT | 50.10 | ||
| MC1R | F: GGCCTGAGAGGGGAATCACA | 61.27 | 126 |
| R: AGTGGGTCTCTGGATGGAGG | 60.33 |
| Trait-Types | Typic Frequencies (AVG ± SE) | p-Values | ||
|---|---|---|---|---|
| Loss | Median | Gain | ||
| body weight (kg) | 52.42 ± 1.58 A (n = 70) | 43.35 ± 3.38 AB (n = 17) | 40.68 ± 1.05 B (n = 65) | 2.701 × 10−8 |
| body height (cm) | 57.51 ± 0.50 A (n = 93) | 56.89 ± 0.81 AB (n = 33) | 55.15 ± 0.42 B (n = 92) | 0.002 |
| height at hip cross (cm) | 61.17 ± 0.54 A (n = 92) | 59.83 ± 0.95 AB (n = 33) | 57.95 ± 0.38 B (n = 91) | 8.000 × 10−6 |
| body length (cm) | 67.75 ± 0.51 A (n = 93) | 64.77 ± 0.99 B (n = 33) | 67.73 ± 0.51 A (n = 91) | 0.008 |
| hip width (cm) | 17.37 ± 0.33 A (n = 87) | 16.98 ± 0.61 A (n = 26) | 15.11 ± 0.19 B (n = 77) | 8.252 × 10−8 |
| heart girth (cm) | 90.00 ± 1.09 A (n= 93) | 86.10 ± 1.60 B (n = 31) | 84.70 ± 0.88 B (n = 92) | 0.001 |
| cannon circumference (cm) | 8.45 ± 0.08 A (n = 93) | 8.34 ± 0.11 A (n = 32) | 8.03 ± 0.07 B (n = 93) | 4.660 × 10−4 |
| chest width (cm) | 20.66 ± 0.34 (n = 93) | 20.00 ± 0.50 (n = 33) | 19.71 ± 0.41 (n = 92) | 0.183 |
| chest depth (cm) | 28.78 ± 0.28 (n = 93) | 27.98 ± 0.53 (n = 32) | 28.66 ± 0.30 (n = 92) | 0.381 |
| Trait-Types | Typic Frequencies (AVG ± SE) | p-Values | ||
|---|---|---|---|---|
| Loss | Median | Gain | ||
| body weight (kg) | 35.91 ± 3.68 b (n = 8) | 48.50 ± 1.29 A (n = 105) | 41.61 ± 1.62 B (n = 38) | 4.023 × 10−3 |
| body height (cm) | 56.07 ± 1.08 (n = 15) | 56.86 ± 0.39 (n = 153) | 55.65 ± 0.54 (n = 50) | 0.254 |
| height at hip cross (cm) | 56.93 ± 0.99 B (n = 15) | 60.382 ± 0.42 A (n = 152) | 58.12 ± 0.49 B (n = 49) | 0.002 |
| body length (cm) | 64.04 ± 1.71 (n = 14) | 67.21 ± 0.38 (n = 153) | 67.68 ± 0.75 (n = 50) | 0.510 |
| hip width (cm) | 13.94 ± 0.61 b (n = 9) | 16.97 ± 2.98 A (n = 136) | 15.16 ± 0.23 B (n = 44) | 2.000 × 10−6 |
| heart girth (cm) | 82.40 ± 2.51 B (n = 15) | 88.72 ± 0.79 A (n = 151) | 83.18 ± 1.24 B (n = 50) | 3.270 × 10−4 |
| cannon circumference (cm) | 8.13 ± 0.14 (n = 15) | 8.34 ± 0.06 (n = 153) | 8.06 ± 0.10 (n = 50) | 0.059 |
| chest width (cm) | 19.18 ± 0.90 (n = 15) | 20.20 ± 0.26 (n = 153) | 19.83 ± 0.53 (n = 50) | 0.481 |
| chest depth (cm) | 26.63 ± 0.57 c (n = 15) | 28.61 ± 0.20 b (n = 152) | 29.05 ± 0.51 a (n = 50) | 0.013 |
| Trait-Types | Typic Frequencies (AVG ± SE) | p-Values | ||
|---|---|---|---|---|
| Loss | Median | Gain | ||
| body weight (kg) | 35.37 ± 4.22 b (n = 6) | 48.09 ± 1.31 a (n =104) | 42.34 ± 1.60 b (n = 38) | 0.020 |
| body height (cm) | 55.60 ± 1.41 (n = 10) | 56.67 ± 0.40 (n = 154) | 55.99 ± 0.56 (n = 43) | 0.685 |
| height at hip cross (cm) | 57.55 ± 0.65 B (n = 10) | 60.23 ± 0.43 A (n = 152) | 58.04 ± 0.47 B (n = 43) | 0.003 |
| body length (cm) | 63.65 ± 1.45 B (n = 10) | 67.26 ± 0.39 ab (n = 153) | 68.45 ± 0.81 a (n = 43) | 0.022 |
| hip width (cm) | 14.50 ± 0.73 B (n = 6) | 16.82 ± 0.26 A (n = 138) | 15.25 ± 0.27 B (n = 42) | 1.540 × 10−4 |
| heart girth (cm) | 82.70 ± 2.70 AB (n = 10) | 88.36 ± 0.74 A (n = 154) | 82.26 ± 1.47 B (n = 43) | 3.480 × 10−4 |
| cannon circumference (cm) | 7.75 ± 0.25 b (n = 10) | 8.35 ± 0.06 A (n = 155) | 8.13 ± 0.10 Ab (n = 43) | 0.020 |
| chest width (cm) | 17.67 ± 1.03 (n = 10) | 20.24 ± 0.24 (n = 154) | 20.29 ± 0.69 (n = 43) | 0.098 |
| chest depth (cm) | 27.25 ± 0.67 (n = 10) | 28.44 ± 0.19 (n = 153) | 29.48 ± 0.61 (n = 43) | 0.062 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Q.; Wei, Z.; Zhu, H.; Pan, C.; Akhatayeva, Z.; Song, X.; Lan, X. Goat Pleomorphic Adenoma Gene 1 (PLAG1): mRNA Expression, CNV Detection and Associations with Growth Traits. Animals 2023, 13, 2023. https://doi.org/10.3390/ani13122023
Wang Q, Wei Z, Zhu H, Pan C, Akhatayeva Z, Song X, Lan X. Goat Pleomorphic Adenoma Gene 1 (PLAG1): mRNA Expression, CNV Detection and Associations with Growth Traits. Animals. 2023; 13(12):2023. https://doi.org/10.3390/ani13122023
Chicago/Turabian StyleWang, Qian, Zhenyu Wei, Haijing Zhu, Chuanying Pan, Zhanerke Akhatayeva, Xiaoyue Song, and Xianyong Lan. 2023. "Goat Pleomorphic Adenoma Gene 1 (PLAG1): mRNA Expression, CNV Detection and Associations with Growth Traits" Animals 13, no. 12: 2023. https://doi.org/10.3390/ani13122023
APA StyleWang, Q., Wei, Z., Zhu, H., Pan, C., Akhatayeva, Z., Song, X., & Lan, X. (2023). Goat Pleomorphic Adenoma Gene 1 (PLAG1): mRNA Expression, CNV Detection and Associations with Growth Traits. Animals, 13(12), 2023. https://doi.org/10.3390/ani13122023




