Novel Gene Rearrangement Pattern in Pachycrepoideus vindemmiae Mitochondrial Genome: New Gene Order in Pteromalidae (Hymenoptera: Chalcidoidea)
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Preparation and DNA Extraction
2.2. Sequencing and Assembly
2.3. Mitochondrial Genome Annotation
2.4. Comparative Analysis
2.5. Phylogenetic Analysis
2.6. Ancestral Character Reconstruction and Gene Rearrangement in Pteromalidae
3. Results
3.1. Genome Structure
3.2. Characteristics of Base Composition
3.3. Overlap and Gap
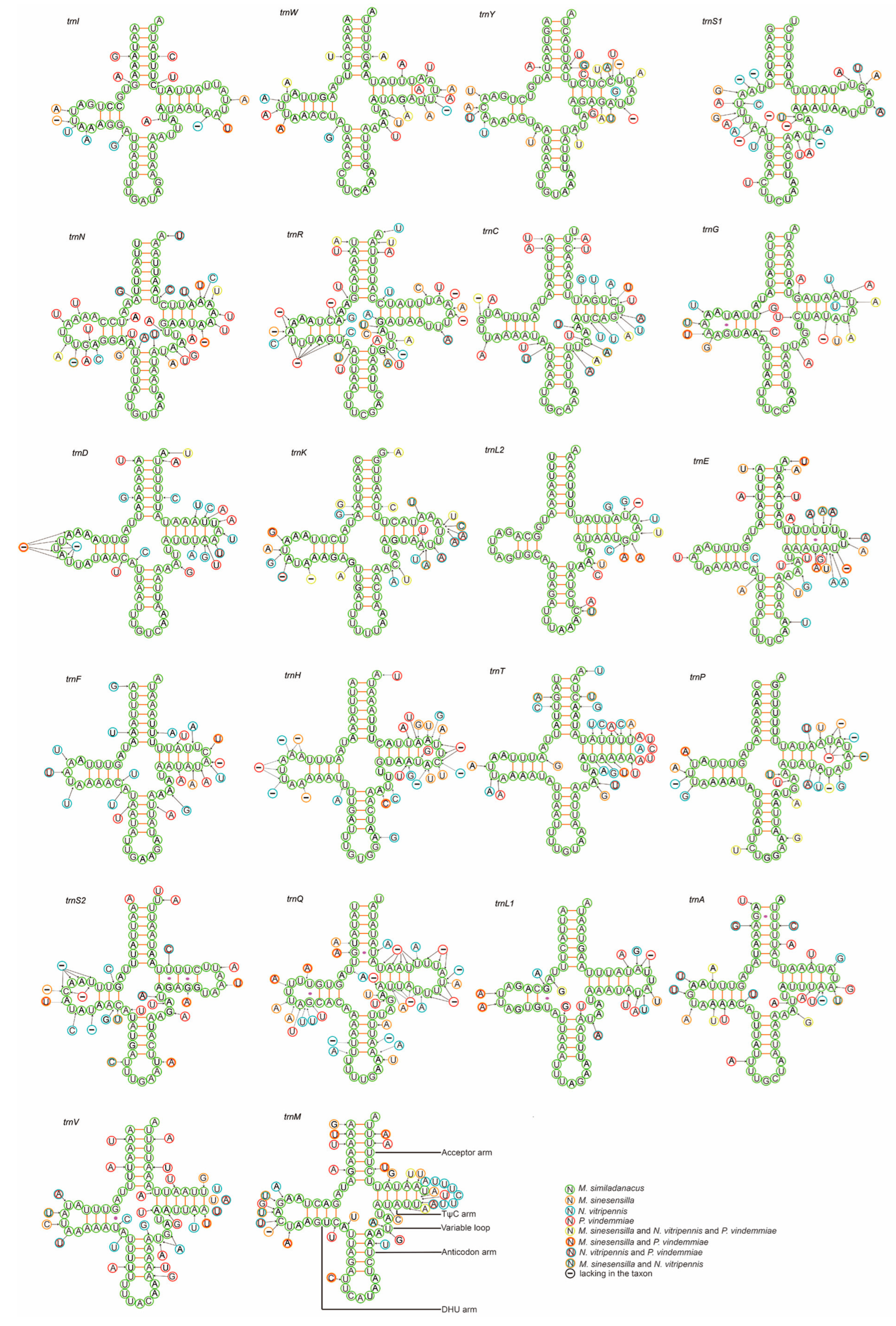
3.4. Transfer RNA and Ribosomal RNA Genes
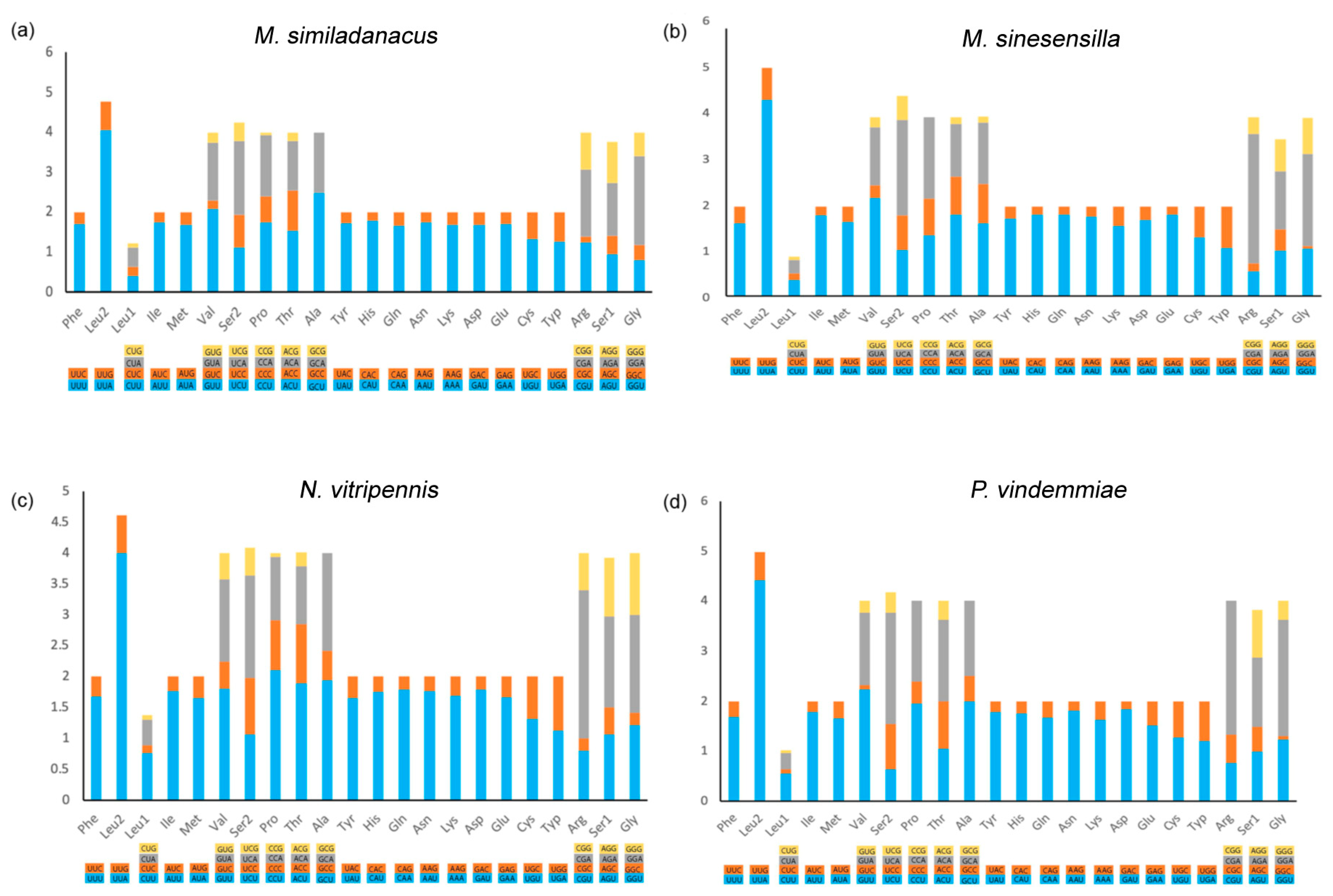
3.5. Protein-Coding Genes
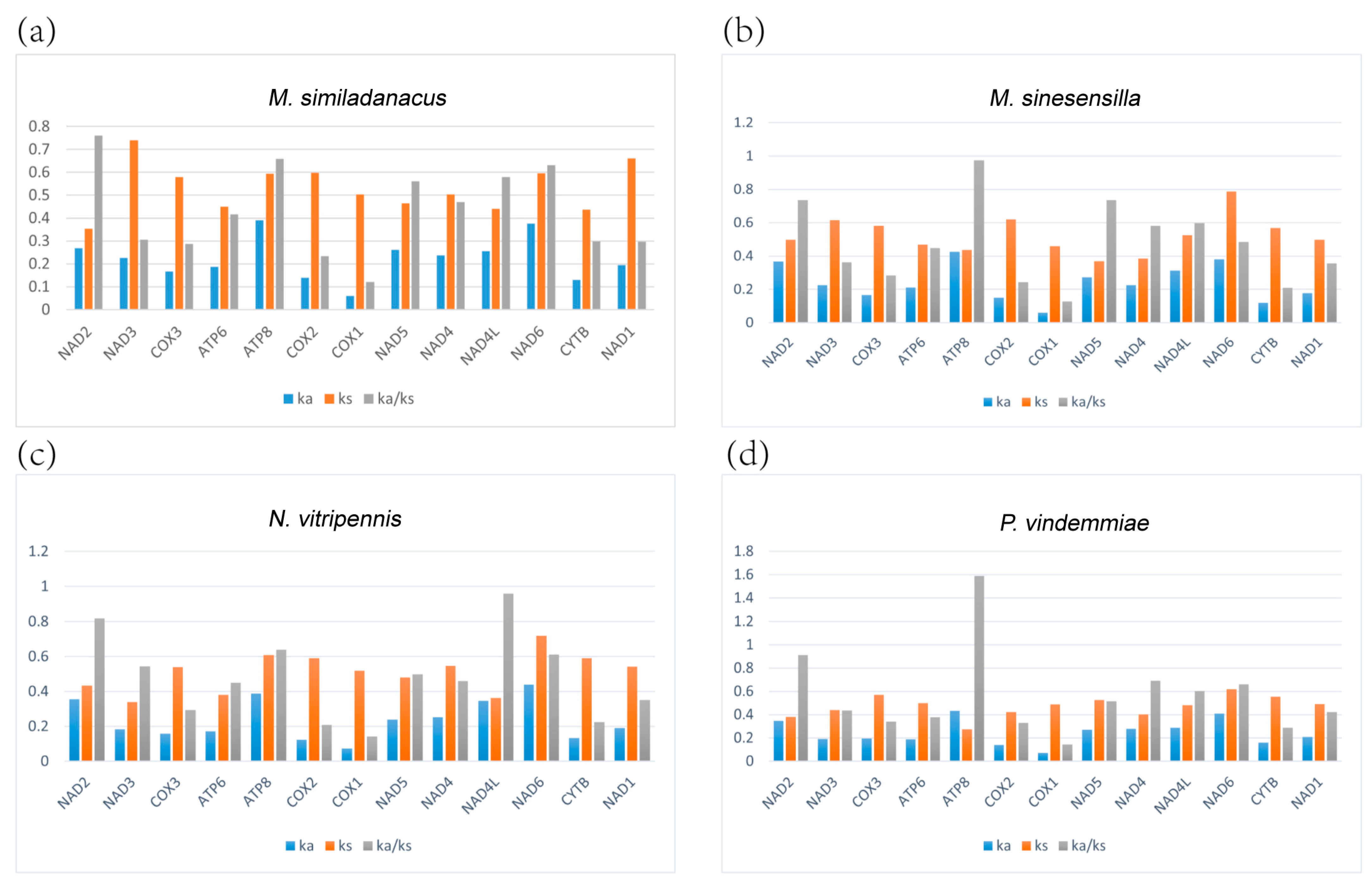
3.6. Evolutionary Rate Analysis
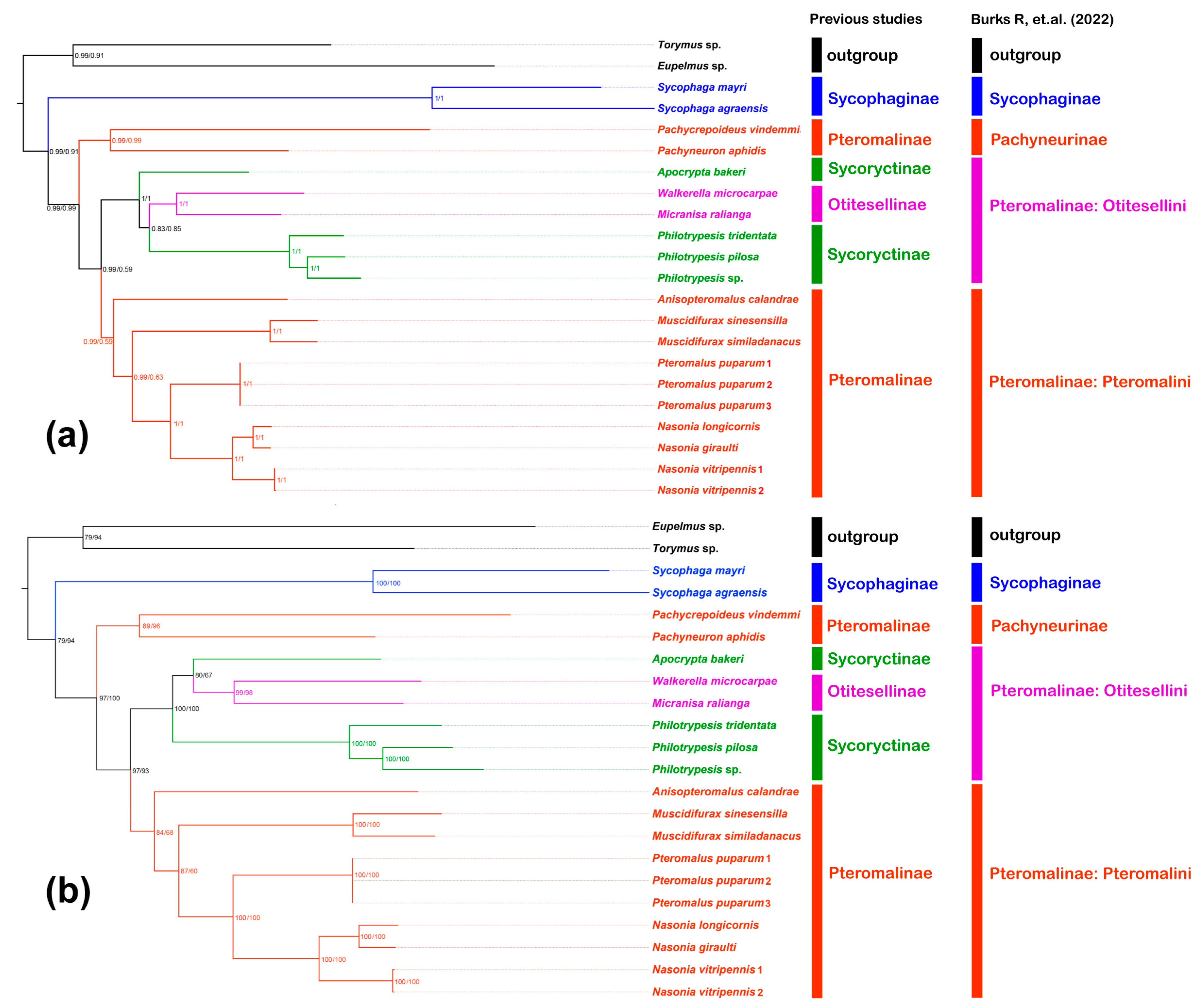
3.7. Phylogenetic Analysis
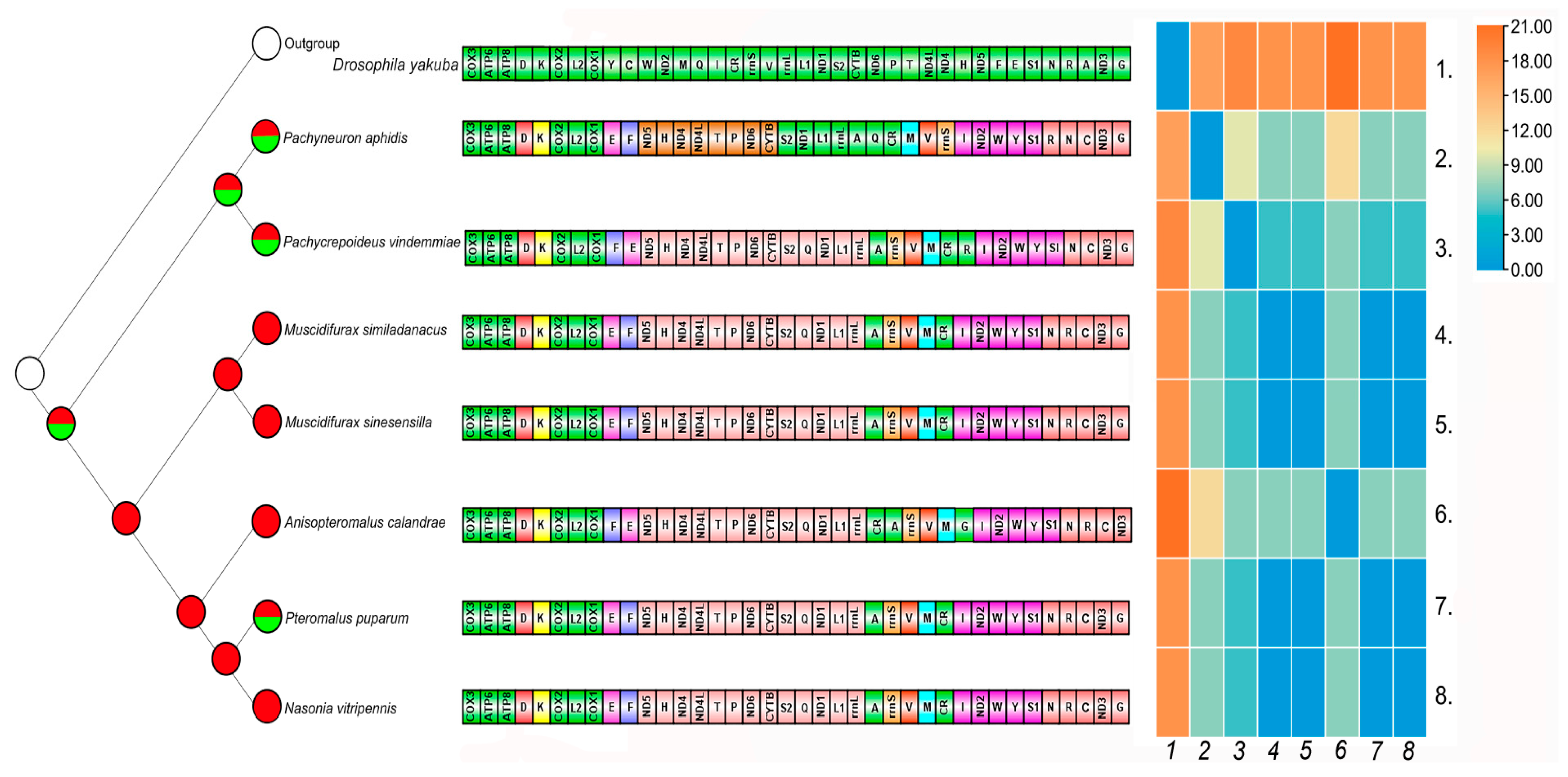
3.8. Ancestral Character Reconstruction and Gene Rearrangement
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wolstenholme, D. Animal Mitochondrial DNA: Structure and Evolution. In International Review of Cytology; Academic Press: Cambridge, MA, USA, 1992; Volume 141, pp. 173–216. [Google Scholar]
- Boore, J.L. Animal mitochondrial genomes. Nucleic Acids Res. 1999, 27, 1767–1780. [Google Scholar] [CrossRef] [PubMed]
- Cameron, S.L. Insect mitochondrial genomics: Implications for evolution and phylogeny. Annu. Rev. Entomol. 2014, 59, 95–117. [Google Scholar] [CrossRef] [PubMed]
- Wei, S.J.; Shi, M.; Sharkey, M.J.; Cornelisvan, C.A.; Chen, X.X. Comparative mitogenomics of Braconidae (Insecta: Hymenoptera) and the phylogenetic utility of mitochondrial genomes with special reference to Holometabolous insects. BMC Genom. 2010, 11, 371. [Google Scholar] [CrossRef]
- Hua, J.; Li, M.; Dong, P.; Cui, Y.; Xie, Q.; Bu, W. Phylogenetic analysis of the true water bugs (Insecta: Hemiptera: Heteroptera: Nepomorpha): Evidence from mitochondrial genomes. BMC Evol. Biol. 2009, 9, 134. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.X.; Feng, J.R.; Bartlett, C.R.; Wei, Y.S.; Qin, D.Z. Contribution to the mitogenome diversity in Delphacinae: Phylogenetic and ecological implications. Genomics 2020, 112, 1363–1370. [Google Scholar] [CrossRef] [PubMed]
- Ma, C.; Yang, P.; Jiang, F.; Chapuis, M.P.; Shali, Y.; Sword, G.A.; Kang, L. Mitochondrial genomes reveal the global phylogeography and dispersal routes of the migratory locust. Mol. Ecol. 2012, 21, 4344–4358. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Chen, J.; Jiang, L.Y.; Qiao, G.X. Hemipteran mitochondrial genomes: Features, structures and implications for phylogeny. Int. J. Mol. Sci. 2015, 16, 12382–12404. [Google Scholar] [CrossRef]
- Nelson, L.A.; Lambkin, C.L.; Batterham, P.; Wallman, J.F.; Dowton, M.; Whiting, M.F.; Yeates, D.K.; Cameron, S.L. Beyond barcoding: A mitochondrial genomics approach to molecular phylogenetics and diagnostics of blowflies (Diptera: Calliphoridae). Gene 2012, 511, 131–142. [Google Scholar] [CrossRef]
- Cameron, S.L.; Lambkin, C.L.; Barker, S.C.; Whiting, M.F. A mitochondrial genome phylogeny of Diptera: Whole genome sequence data accurately resolve relationships over broad timescales with high precision. Syst. Entomol. 2007, 32, 40–59. [Google Scholar] [CrossRef]
- Cameron, S.L.; Whiting, M.F. Mitochondrial genomic comparisons of the subterranean termites from the Genus Reticulitermes (Insecta: Isoptera: Rhinotermitidae). Genome 2007, 50, 188–202. [Google Scholar] [CrossRef]
- Ingman, M.; Kaessmann, H.; Pääbo, S.; Gyllensten, U. Mitochondrial genome variation and the origin of modern humans. Nature 2000, 408, 708–713. [Google Scholar] [CrossRef] [PubMed]
- Reyes, A.; Gissi, C.; Pesole, G.; Saccone, C. Asymmetrical directional mutation pressure in the mitochondrial genome of mammals. Mol. Biol. Evol. 1998, 15, 957–966. [Google Scholar] [CrossRef] [PubMed]
- Xiao, J.H.; Jia, J.G.; Murphy, R.W.; Huang, D.W. Rapid evolution of the mitochondrial genome in Chalcidoid wasps (Hymenoptera: Chalcidoidea) driven by parasitic lifestyles. PLoS ONE 2011, 6, e26645. [Google Scholar] [CrossRef]
- Chen, L.; Chen, P.Y.; Xue, X.F.; Hua, H.Q.; Li, Y.X.; Zhang, F.; Wei, S.J. Extensive gene rearrangements in the mitochondrial genomes of two egg parasitoids, Trichogramma japonicum and Trichogramma ostriniae (Hymenoptera: Chalcidoidea: Trichogrammatidae). Sci. Rep. 2018, 8, 7034. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.F.; Yang, H.L.; Feng, Z.B.; Li, B.Y.; Zhou, W.B.; Song, F.; Li, H.; Zhang, L.; Cai, W.Z. Novel gene rearrangement in the mitochondrial genome of Pachyneuron aphidis (Hymenoptera: Pteromalidae). Int. J. Biol. Macromol. 2020, 149, 1207–1212. [Google Scholar] [CrossRef]
- Gibson, G.J.Z.S. Sister-group relationships of the Platygastroidea and Chalcidoidea (Hymenoptera)—An alternate hypothesis to Rasnitsyn (1998). Zool. Scr. 1999, 28, 125–138. [Google Scholar] [CrossRef]
- Munro, J.B.; Heraty, J.M.; Burks, R.A.; Hawks, D.; Mottern, J.; Cruaud, A.; Rasplus, J.Y.; Jansta, P. A molecular phylogeny of the Chalcidoidea (Hymenoptera). PLoS ONE 2011, 6, e27023. [Google Scholar] [CrossRef]
- Heraty, J.M.; Burks, R.A.; Cruaud, A.; Gibson, G.A.P.; Liljeblad, J.; Munro, J.; Rasplus, J.Y.; Delvare, G.; Janšta, P.; Gumovsky, A.; et al. A phylogenetic analysis of the megadiverse Chalcidoidea (Hymenoptera). Cladistics Int. J. Willi Hennig Soc. 2013, 29, 466–542. [Google Scholar] [CrossRef]
- Peters, R.S.; Niehuis, O.; Gunkel, S.; Bläser, M.; Mayer, C.; Podsiadlowski, L.; Kozlov, A.; Donath, A.; Noort, S.V.; Liu, S.; et al. Transcriptome sequence-based phylogeny of chalcidoid wasps (Hymenoptera: Chalcidoidea) reveals a history of rapid radiations, convergence, and evolutionary success. Mol. Phylogenetics Evol. 2018, 120, 286–296. [Google Scholar] [CrossRef]
- Zhang, Y.Z.; Xiong, M.; Zhou, Q.S.; Jiang, G.C.; Zhu, C.D. The mitochondrial gensome of Platencyrtus parkeri Feriere (Hymenoptera: Encyrtidae). Mitochondrial DNA Part B Resour. 2019, 4, 3479–3481. [Google Scholar] [CrossRef]
- Zhang, D.; Gao, F.; Jakovlić, I.; Zou, H.; Zhang, J.; Li, W.X.; Wang, G.T. PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 2020, 20, 348–355. [Google Scholar] [CrossRef] [PubMed]
- Rasplus, J.Y.; Kerdelhué, C.; Le Clainche, I.; Mondor, G. Molecular phylogeny of fig wasps Agaonidae are not monophyletic. In Comptes Rendus de l’Académie des Sciences-Series III-Sciences de la Vie; Elsevier: Amsterdam, The Netherlands, 1998; Volume 321, pp. 517–526. [Google Scholar] [CrossRef]
- Dzhanokmen, K.A. Phylogenetic relations between Palaearctic Pteromalidae (Hymenoptera, Chalcidoidea) subfamilies. Zool. Zhurnal 2000, 79, 564–571. [Google Scholar] [CrossRef]
- Desjardins, C.A.; Regier, J.C.; Mitter, C. Phylogeny of pteromalid parasitic wasps (Hymenoptera: Pteromalidae): Initial evidence from four protein-coding nuclear genes. Mol. Phylogenetics Evol. 2007, 45, 454–469. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, D.C.S.G.; Raychoudhury, R.; Lavrov, D.V.; Werren, J.H. Rapidly evolving mitochondrial genome and directional selection in mitochondrial genes in the parasitic wasp nasonia (hymenoptera: Pteromalidae). Mol. Biol. Evol. 2008, 25, 2167–2180. [Google Scholar] [CrossRef] [PubMed]
- Tang, P.; Zhu, J.C.; Zheng, B.Y.; Wei, S.J.; Sharkey, M.; Chen, X.X.; Vogler, A.P. Mitochondrial phylogenomics of the Hymenoptera. Mol. Phylogenetics Evol. 2019, 131, 8–18. [Google Scholar] [CrossRef] [PubMed]
- Bernt, M.; Merkle, D.; Ramsch, K.; Fritzsch, G.; Perseke, M.; Bernhard, D.; Schlegel, M.; Stadler, P.F.; Middendorf, M. CREx: Inferring genomic rearrangements based on common intervals. Bioinformatics 2007, 23, 2957–2958. [Google Scholar] [CrossRef]
- Xiao, H.; Zhou, S.Y.; Tong, Y.F. A taxonomic study of Muscidifurax Girault & Sanders from China (Hymenoptera, Chalcidoidea, Pteromalidae). ZooKeys 2018, 776, 91–103. [Google Scholar] [CrossRef]
- Chen, W.; He, Z.; Ji, X.L.; Tang, S.T.; Hu, H.Y. Hyperparasitism in a Generalist Ectoparasitic Pupal Parasitoid, Pachycrepoideus vindemmiae (Hymenoptera: Pteromalidae), on Its Own Conspecifics: When the Lack of Resource Lead to Cannibalism. PLoS ONE 2015, 10, e0124305. [Google Scholar] [CrossRef]
- Doğanlar, M. A new species of Muscidifurax Girault & Sanders, 1910 (Hymenoptera: Pteromalidae) from Adana province, Turkey. Turk. J. Entomol. 2007, 31, 243–252. [Google Scholar]
- Gao, M.Q.; Cheng, J.Q.; Hu, W.; Pan, D.; Xiao, H.; Hu, H.Y. Life tables of two newly discovered parasitoid wasps, Muscidifura similadanacus and M. sinesensilla (Hymenoptera: Pteromalidae), reared on Musca domestica (Diptera: Muscidae). Biocontrol Sci. Technol. 2020, 30, 779–794. [Google Scholar] [CrossRef]
- Girault, A.A.; Sanders, G.E. The chalcidoid parasites of the common house or typhoid fly (|Musca domestica| Linn.) and its allies. ii. Reconstruction of the genus |Pachycrepoideus| Ashmead of the family Pteromalidae. Psyche 1910, 17, 108–117. [Google Scholar] [CrossRef]
- Legner, E.F. Some Effects of the Ambient Arthropod Complex on the Density and Potential Parasitization of Muscoid Diptera in Poultry Wastes. J. Econ. Entomol. 1971, 64, 1406–1417. [Google Scholar] [CrossRef]
- Noyes, J.S. Universal Chalcidoidea Database. World Wide Web Electronic Publication. Available online: http://www.nhm.ac.uk/our-science/data/chalcidoids/familyindex.html (accessed on 9 December 2021).
- Zhou, C.Q.; Zhan, H.X.; Xiao, C.; Zhang, J.P. The development, fecundity, and functional response of Pachycrepoideus vindemmiae on the pupae of Drosophila melanogaster. J. Environ. Entomol. 2019, 41, 599–604. [Google Scholar]
- Museum, N.H. Available online: https://www.nhm.ac.uk/our-science/data/chalcidoids/pteromalidae1.html (accessed on 24 January 2022).
- Shahjahan, R.M.; Hughes, K.J.; Leopold, R.A.; DeVault, J.D. Lower incubation temperature increases yield of insect genomic DNA isolated by the CTAB method. BioTechniques 1995, 19, 332–334. [Google Scholar] [PubMed]
- Wang, J.X.; Liu, J.; Miao, Y.H.; Huang, D.W.; Xiao, J.H. Tracking the Distribution and Burst of Nuclear Mitochondrial DNA Sequences (NUMTs) in Fig Wasp Genomes. Insects 2020, 11, 680. [Google Scholar] [CrossRef]
- Zhao, D.; Xin, Z.Z.; Hou, H.X.; Zhou, Y.; Wang, J.X.; Xiao, J.H.; Huang, D.W. Inferring the Phylogenetic Positions of Two Fig Wasp Subfamilies of Epichrysomallinae and Sycophaginae Using Transcriptomes and Mitochondrial Data. Life 2021, 11, 40. [Google Scholar] [CrossRef]
- Babraham Institute. Available online: http://www.bioinformatics.babraham.ac.uk/projects/ (accessed on 10 February 2020).
- Schubert, M.; Lindgreen, S.; Orlando, L. AdapterRemoval v2: Rapid adapter trimming, identification, and read merging. BMC Res. Notes 2016, 9, 88. [Google Scholar] [CrossRef]
- Kearse, M.; Moir, R.; Wilson, A.; Stones, H.S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef]
- Lin, Z.J.; Wang, X.; Wang, J.; Tan, Y.; Tang, X.; Werren, J.H.; Zhang, D.; Wang, X. Comparative analysis reveals the expansion of mitochondrial DNA control region containing unusually high G-C tandem repeat arrays in Nasonia vitripennis. Int. J. Biol. Macromol. 2021, 166, 1246–1257. [Google Scholar] [CrossRef]
- Bernt, M.; Donath, A.; Jühling, F.; Externbrink, F.; Florentz, C.; Fritzsch, G.; Pütz, J.; Middendorf, M.; Stadler, P.F. MITOS: Improved de novo metazoan mitochondrial genome annotation. Mol. Phylogenetics Evol. 2013, 69, 313–319. [Google Scholar] [CrossRef]
- Lohse, M.; Drechsel, O.; Kahlau, S.; Bock, R. OrganellarGenomeDRAW—A suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res. 2013, 41, W575–W581. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Librado, P.; Rozas, J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 2009, 25, 1451–1452. [Google Scholar] [CrossRef]
- Ma, Y.; Zheng, B.Y.; Zhu, J.C.; Tang, P.; Chen, X.X. The mitochondrial genome of Aenasius arizonensis (Hymenoptera: Encyrtidae) with novel gene order. Mitochondrial DNA Part B 2019, 4, 2023–2024. [Google Scholar] [CrossRef]
- Perna, N.T.; Kocher, T.D. Patterns of nucleotide composition at fourfold degenerate sites of animal mitochondrial genomes. J. Mol. Evol. 1995, 41, 353–358. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. A simple method to control over-alignment in the MAFFT multiple sequence alignment program. Bioinformatics 2016, 32, 1933–1942. [Google Scholar] [CrossRef]
- Trifinopoulos, J.; Nguyen, L.T.; von Haeseler, A.; Minh, B.Q. W-IQ-TREE: A fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 2016, 44, W232–W235. [Google Scholar] [CrossRef]
- Minh, B.Q.; Nguyen, M.A.T.; Haeseler, A.V. Ultrafast approximation for phylogenetic bootstrap. Mol. Biol. Evol. 2013, 30, 1188–1195. [Google Scholar] [CrossRef]
- Guindon, S.; Dufayard, J.F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef] [PubMed]
- Lartillot, N.; Rodrigue, N.; Stubbs, D.; Richer, J. PhyloBayes MPI: Phylogenetic reconstruction with infinite mixtures of profiles in a parallel environment. Syst. Biol. 2013, 62, 611–615. [Google Scholar] [CrossRef] [PubMed]
- Rambaut, A. FigTree v1.3.1. Available online: http://tree.bio.ed.ac.uk/software/figtree (accessed on 10 February 2018).
- Polme, S.; Bahram, M.; Yamanaka, T.; Nara, K.; Dai, Y.C.; Grebenc, T.; Kraigher, H.; Toivonen, M.; Wang, P.H.; Matsuda, Y.; et al. Biogeography of ectomycorrhizal fungi associated with alders (Alnus spp.) in relation to biotic and abiotic variables at the global scale. New Phytol. 2013, 198, 1239–1249. [Google Scholar] [CrossRef]
- Liu, W.; Xie, Y.; Ma, J.; Luo, X.; Nie, P.; Zuo, Z.; Lahrmann, U.; Zhao, Q.; Zheng, Y.; Zhao, Y.; et al. IBS: An illustrator for the presentation and visualization of biological sequences. Bioinformatics 2015, 31, 3359–3361. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Crozier, R.H.; Crozier, Y.C. The mitochondrial genome of the honeybee Apis mellifera: Complete sequence and genome organization. Genetics 1993, 133, 97–117. [Google Scholar] [CrossRef] [PubMed]
- Cameron, S.L.; Dowton, M.; Castro, L.R.; Ruberu, K.; Whiting, M.F.; Austin, A.D.; Diement, K.; Stevens, J. Mitochondrial genome organization and phylogeny of two vespid wasps. Genome 2008, 51, 800–808. [Google Scholar] [CrossRef] [PubMed]
- Mao, M.; Dowto, M. Complete mitochondrial genomes of Ceratobaeus sp. and Idris sp. (Hymenoptera: Scelionidae): Shared gene rearrangements as potential phylogenetic markers at the tribal level. Mol. Biol. Rep. 2014, 41, 6419–6427. [Google Scholar] [CrossRef] [PubMed]
- Doublet, V.; Ubrig, E.; Alioua, A.; Bouchon, D.; Marcadé, I.; Maréchal-Drouard, L. Large gene overlaps and tRNA processing in the compact mitochondrial genome of the crustacean Armadillidium vulgare. RNA Biol. 2015, 12, 1159–1168. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.F.; Li, Y.P.; Jakovlic, I.; Yuan, X.Q. Tandem duplication of two tRNA genes in the mitochondrial genome of Tagiades vajuna (Lepidoptera: Hesperiidae). Eur. J. Entomol. 2017, 114, 407–415. [Google Scholar] [CrossRef]
- Ojala, D.; Montoya, J.; Attardi, G. tRNA punctuation model of RNA processing in human mitochondria. Nature 1981, 290, 470–474. [Google Scholar] [CrossRef]
- Beckenbach, A.T.; Joy, J.B. Evolution of the mitochondrial genomes of gall midges (Diptera: Cecidomyiidae): Rearrangement and severe truncation of tRNA genes. Genome Biol. Evol. 2009, 1, 278–287. [Google Scholar] [CrossRef]
- Foster, P.G.; Jermiin, L.S.; Hickey, D.A. Nucleotide Composition Bias Affects Amino Acid Content in Proteins Coded by Animal Mitochondria. J. Mol. Evol. 1997, 44, 282–288. [Google Scholar] [CrossRef]
- Burks, R.; Mitroiu, M.D.; Fusu, L.; Heraty, J.M. From hell’s heart I stab at thee! A determined approach towards a monophyletic Pteromalidae and reclassification of Chalcidoidea (Hymenoptera). J. Hymenoptrea Res. 2022, 94, 13–88. [Google Scholar] [CrossRef]
- Clary, D.O.; Wolstenholme, D.R. The mitochondrial DNA molecular of Drosophila yakuba: Nucleotide sequence, gene organization, and genetic code. J. Mol. Evol. 1985, 22, 252–271. [Google Scholar] [CrossRef] [PubMed]
- Simon, S.; Hadrys, H. A comparative analysis of complete mitochondrial genomes among Hexapoda. Mol. Phylogenetics Evol. 2013, 69, 393–403. [Google Scholar] [CrossRef] [PubMed]
- Shen, Z.C.; Chen, L.; Chen, L.; Li, Y.X. Information from the mitochondrial genomes of two egg parasitoids, Gonatocerus sp. and Telenomus sp., reveals a controversial phylogenetic relationship between Mymaridae and Scelionidae. Genomics 2019, 111, 1059–1065. [Google Scholar] [CrossRef] [PubMed]
- Zheng, B.Y.; Cao, L.J.; Tang, P.; Achterberg, K.V.; Hoffmann, A.A.; Chen, H.Y.; Chen, X.X.; Wei, S.J. Gene arrangement and sequence of mitochondrial genomes yield insights into the phylogeny and evolution of bees and sphecid wasps (Hymenoptera: Apoidea). Mol. Phylogenetics Evol. 2018, 124, 1–9. [Google Scholar] [CrossRef]
- Covacin, C.; Shao, R.; Cameron, S.; Barker, S.C. Extraordinary number of gene rearrangements in the mitochondrial genomes of lice (Phthiraptera: Insecta). Insect Mol. Biol. 2006, 15, 63–68. [Google Scholar] [CrossRef]
- Li, Q.; Wei, S.J.; Shi, M.; Chen, X.X. Complete mitochondrial genome of Neochauliodes bowringi (MacLachlan) (Megaloptera: Corydalidae). Mitochondrial DNA 2015, 26, 112–113. [Google Scholar] [CrossRef]
- Chen, W.J.; Bu, Y.; Carapelli, A.; Dallai, R.; Li, S.; Yin, W.Y.; Luan, Y.X. The mitochondrial genome of Sinentomon erythranum (Arthropoda: Hexapoda: Protura): An example of highly divergent evolution. BMC Evol. Biol. 2011, 11, 246. [Google Scholar] [CrossRef]
- Feng, Z.B.; Wu, Y.F.; Yang, C.; Gu, X.H.; Wilson, J.J.; Li, H.; Cai, W.Z.; Yang, H.L.; Song, F. Evolution of tRNA gene rearrangement in the mitochondrial genome of ichneumonoid wasps (Hymenoptera: Ichneumonoidea). Int. J. Biol. Macromol. 2020, 164, 540–547. [Google Scholar] [CrossRef]
- Song, F.; Li, H.; Liu, G.H.; Wang, W.; James, P.; Colwell, D.D.; Tran, A.; Gong, S.; Cai, W.; Shao, R. Mitochondrial Genome Fragmentation Unites the Parasitic Lice of Eutherian Mammals. Syst. Biol. 2019, 68, 430–440. [Google Scholar] [CrossRef] [PubMed]
- Cruaud, A.; Rasplus, J.Y.; Zhang, J.; Burks, R.; Delvare, G.; Fusu, L.; Gumovsky, A.; Huber, J.T.; Janšta, P.; Mitroiu, M.D.; et al. The Chalcidoidea bush of life—A massive radiation blurred by mutational saturation. bioRxiv 2022. [Google Scholar] [CrossRef]

| No. | Family | Subfamily | Tribe | Taxa | GenBank Accession No. | Location/Reference |
|---|---|---|---|---|---|---|
| 1 | Eupelmidae | Eupelmus sp. | MG923493 | [27] | ||
| 2 | Torymidae | Torymus sp. | MG923516 | [27] | ||
| 3 | Pteromalidae | Pachyneurinae | Pachycrepoideus vindemmiae | MT712142 | This study, Anhui | |
| 4 | Pachyneuron aphidis | MK577639 | [16] | |||
| 5 | Pteromalinae | Pteromalini | Anisopteromalus calandrae | MW817149 | Unpublished | |
| 6 | Muscidifurax sinesensilla | MT712140 | This study, Xinjiang | |||
| 7 | Muscidifurax similadanacus | MT712139 | This study, Xinjiang | |||
| 8 | Nasonia giraulti | EU746611, EU746614 | [26] | |||
| 9 | Nasonia longicornis | EU746612, EU746616 | [26] | |||
| 10 | Nasonia vitripennis | EU746609, EU746613 | [26] | |||
| 11 | Nasonia vitripennis | MT712141 | This study, Shandong | |||
| 12 | Pteromalus puparum | MH051556 | Unpublished | |||
| 13 | Pteromalus puparum | NC039656 | Unpublished | |||
| 14 | Pteromalus puparum | MG923513 | [27] | |||
| 15 | Otitesellini | Apocrypta bakeri | MT906648 | [39] | ||
| 16 | Micranisa ralianga | MW167115 | [40] | |||
| 17 | Philotrypesis tridentata | MT947602 | [39] | |||
| 18 | Philotrypesis pilosa | JF808723 | [14] | |||
| 19 | Philotrypesis sp. | JF808722 | [14] | |||
| 20 | Walkerella microcarpae | MW167116 | [40] | |||
| 21 | Sycophaginae | Sycophaga agraensis | MT947599 | [39] | ||
| 22 | Sycophaga mayri | MW167114 | [40] |
| Position | Size (bp) | Intergenic Nucleotides | Codon | Strand | |||
|---|---|---|---|---|---|---|---|
| From | To | Start | Stop | ||||
| M. similadanacus/M. sinesensilla/N. vitripennis | |||||||
| trnI | 1/1/1 | 69/68/70 | 69/68/70 | +/+/+ | |||
| nad2 | 89/87/92 | 1096/1094/1102 | 1008/1008/1011 | 19/18/21 | ATT/ATT/ATA | TAA/TAA/TAA | −/−/− |
| trnW | 1095/1101/1105 | 1161/1167/1171 | 67/67/67 | −2/6/2 | −/−/− | ||
| trnY | 1160/1166/1170 | 1226/1230/1237 | 67/65/68 | −2/−2/−2 | +/+/+ | ||
| trnS1 | 1233/1240/1240 | 1295/1302/1299 | 63/63/60 | 6/9/2 | +/+/+ | ||
| trnN | 1299/1324/1302 | 1366/1391/1368 | 68/68/67 | 3/21/2 | −/−/− | ||
| trnR | 1362/1393/1375 | 1430/1463/1445 | 69/71/71 | −5/1/6 | +/+/+ | ||
| trnC | 1433/1475/1448 | 1496/1537/1514 | 64/63/67 | 2/11/2 | +/+/+ | ||
| nad3 | 1500/1537/1514 | 1850/1887/1864 | 351/351/351 | 3/−1/−1 | ATT/ATT/ATA | TAA/TAA/TAA | +/+/+ |
| trnG | 1851/1888/1865 | 1916/1954/1932 | 66/67/68 | +/+/+ | |||
| cox3 | 1917/1960/1935 | 2702/2745/2720 | 786/786/786 | −/5/2 | ATG/ATG/ATG | TAA/TAA/TAA | +/+/+ |
| atp6 | 2702/2745/2723 | 3376/3419/3397 | 675/675/675 | −1/−1/2 | ATG/ATG/ATG | TAA/TAA/TAA | +/+/+ |
| atp8 | 3367/3410/3391 | 3528/3571/3549 | 162/162/159 | −10/−10/−7 | ATT/ATT/ATA | TAA/TAA/TAA | +/+/+ |
| trnD | 3529/3572/3551 | 3597/3636/3615 | 69/65/65 | −/−/1 | +/+/+ | ||
| trnK | 3601/3652/3619 | 3669/3722/3692 | 69/71/74 | 3/15/3 | −/−/− | ||
| cox2 | 3670/3726/3696 | 4344/4400/4364 | 675/675/669 | −/3/3 | ATT/ATT/ATT | TAA/TAA/TAA | +/+/+ |
| trnL2 | 4345/4401/4365 | 4410/4468/4432 | 66/68/68 | +/+/+ | |||
| cox1 | 4413/4471/4433 | 5942/6000/5963 | 1530/1530/1531 | 2/2/− | ATG/ATG/ATG | TAA/TAA/T | +/+/+ |
| trnE | 5945/6003/5964 | 6010/6067/6033 | 66/65/70 | 2/2/− | −/−/− | ||
| trnF | 6009/6072/6045 | 6072/6136/6109 | 64/65/65 | −2/4/11 | +/+/+ | ||
| nad5 | 6073/6137/6109 | 7741/7808/7794 | 1669/1672/1686 | −/−/−1 | ATT/ATT/ATT | T/T/TAG | +/+/+ |
| trnH | 7750/7812/7795 | 7818/7877/7859 | 69/66/65 | 8/3/− | +/+/+ | ||
| nad4 | 7819/7879/7860 | 9159/9219/9198 | 1341/1341/1339 | −/1/− | ATG/ATG/ATG | TAA/TAA/T | +/+/+ |
| nad4l | 9153/9213/9192 | 9440/9500/9479 | 288/288/288 | −7/−7/−7 | ATT/ATT/ATT | TAA/TAA/TAA | +/+/+ |
| trnT | 9441/9501/9479 | 9504/9564/9543 | 64/64/65 | −/−/−1 | −/−/− | ||
| trnP | 9507/9565/9544 | 9575/9629/9608 | 69/65/65 | 2/−/− | +/+/+ | ||
| nad6 | 9581/9633/9617 | 10126/10184/10165 | 546/552/549 | 5/3/8 | ATG/ATG/ATG | TAA/TAA/TAA | −/−/− |
| cytb | 10126/10185/10167 | 11271/11330/11306 | 1146/1146/1140 | −1/−/1 | ATG/ATG/ATG | TAA/TAA/TAA | −/−/− |
| trnS2 | 11271/11329/11305 | 11339/11396/11371 | 69/68/67 | −1/−2/−2 | −/−/− | ||
| trnQ | 11341/11409/11398 | 11408/11480/11468 | 68/72/71 | 1/12/26 | +/+/+ | ||
| nad1 | 11409/11484/11469 | 12338/12405/12401 | 930/922/933 | −/3/− | ATT/ATT/TTG | TAA/T/TAA | +/+/+ |
| trnL1 | 12339/12406/12402 | 12404/12472/12468 | 66/67/67 | +/+/+ | |||
| rrnL | 12405/12473/12469 | 13717/13780/13793 | 1313/1308/1325 | +/+/+ | |||
| trnA | 13718/13781/13794 | 13782/13845/13857 | 65/65/64 | +/+/+ | |||
| rrnS | 13783/13846/13858 | 14558/14625/14644 | 776/780/787 | +/+/+ | |||
| trnV | 14559/14626/14645 | 14623/14691/14712 | 65/66/68 | +/+/+ | |||
| trnM | 14626/14694/14716 | 14690/14758/14787 | 65/65/72 | 2/2/3 | +/+/+ | ||
| AT-rich | 14691/14759/14788 | 15080/15020/14791 | 390/262/4 | -/-/- | |||
| Position | Size (bp) | Intergenic Nucleotides | Codon | Strand | |||
|---|---|---|---|---|---|---|---|
| From | To | Start | Stop | ||||
| P. vindemmiae | |||||||
| trnR | 1 | 59 | 59 | − | |||
| trnI | 63 | 131 | 69 | 3 | + | ||
| nad2 | 167 | 1168 | 1002 | 35 | ATT | TAA | − |
| trnW | 1172 | 1241 | 70 | 3 | − | ||
| trnY | 1240 | 1305 | 66 | −2 | + | ||
| trnS1 | 1307 | 1366 | 60 | 1 | + | ||
| trnN | 1370 | 1438 | 69 | 3 | + | ||
| trnC | 1440 | 1504 | 65 | 1 | + | ||
| nad3 | 1505 | 1853 | 349 | ATT | T | + | |
| trnG | 1854 | 1918 | 65 | + | |||
| cox3 | 1919 | 2704 | 786 | ATG | TAA | + | |
| atp6 | 2704 | 3378 | 675 | −1 | ATG | TAA | + |
| atp8 | 3372 | 3530 | 159 | −7 | ATT | TAA | + |
| trnD | 3544 | 3608 | 65 | 13 | + | ||
| trnK | 3618 | 3689 | 72 | 9 | − | ||
| cox2 | 3691 | 4359 | 669 | 1 | ATT | TAA | + |
| trnL2 | 4360 | 4425 | 66 | + | |||
| cox1 | 4426 | 5959 | 1534 | ATG | T | + | |
| trnF | 5958 | 6020 | 63 | −2 | − | ||
| trnE | 6021 | 6085 | 65 | + | |||
| nad5 | 6086 | 7757 | 1672 | ATT | T | + | |
| trnH | 7760 | 7823 | 64 | 2 | + | ||
| nad4 | 7824 | 9156 | 1333 | ATG | T | + | |
| nad4l | 9150 | 9437 | 288 | −7 | ATT | TAA | + |
| trnT | 9437 | 9506 | 70 | −1 | − | ||
| trnP | 9514 | 9577 | 64 | 7 | + | ||
| nad6 | 9581 | 10,147 | 567 | 3 | ATG | TAA | − |
| cytb | 10,147 | 11,283 | 1137 | −1 | ATG | TAA | − |
| trnS2 | 11,282 | 11,348 | 67 | −2 | − | ||
| trnQ | 11,364 | 11,418 | 55 | 15 | + | ||
| nad1 | 11,420 | 12,340 | 921 | 1 | ATT | TAA | + |
| trnL1 | 12,341 | 12,405 | 65 | + | |||
| rrnL | 12,406 | 13,705 | 1300 | + | |||
| trnA | 13,706 | 13,770 | 65 | + | |||
| rrnS | 13,771 | 14,533 | 763 | + | |||
| trnV | 14,534 | 14,600 | 67 | + | |||
| trnM | 14,601 | 14,667 | 67 | + | |||
| AT-rich | 14,668 | 14,850 | 183 | - | |||
| Regions | Species | Size (bp) | T% | C% | A% | G% | AT (%) | GC (%) | AT Skew | GC Skew |
|---|---|---|---|---|---|---|---|---|---|---|
| Full genome | M. similadanacus | 15080 | 41.0 | 9.0 | 42.5 | 7.5 | 83.5 | 16.5 | 0.018 | −0.091 |
| M. sinesensilla | 15020 | 40.7 | 8.8 | 43.4 | 7.1 | 84.1 | 15.9 | 0.032 | −0.107 | |
| N. vitripennis | 14791 | 39.6 | 9.4 | 43.5 | 7.5 | 83.0 | 17.0 | 0.047 | −0.112 | |
| P. vindemmiae | 14850 | 41.1 | 8.2 | 44.2 | 6.6 | 85.3 | 14.7 | 0.036 | −0.108 | |
| PCGs | M. similadanacus | 11107 | 39.7 | 9.3 | 42.5 | 8.4 | 82.3 | 17.7 | 0.034 | −0.051 |
| M. sinesensilla | 11108 | 39.7 | 9.1 | 43.2 | 8.1 | 82.8 | 17.2 | 0.042 | −0.058 | |
| N. vitripennis | 11117 | 38.6 | 9.9 | 43.1 | 8.4 | 81.7 | 18.3 | 0.055 | −0.082 | |
| P. vindemmiae | 11092 | 40.0 | 8.6 | 44.2 | 7.3 | 84.2 | 15.8 | 0.050 | −0.082 | |
| tRNAs | M. similadanacus | 1467 | 44.0 | 6.5 | 44.1 | 5.3 | 88.1 | 11.9 | 0.001 | −0.102 |
| M. sinesensilla | 1464 | 43.7 | 6.6 | 44.9 | 4.8 | 88.6 | 11.4 | 0.014 | −0.158 | |
| N. vitripennis | 1484 | 42.2 | 7.7 | 44.9 | 5.2 | 87.1 | 12.9 | 0.031 | −0.194 | |
| P. vindemmiae | 1438 | 44.7 | 6.7 | 44.2 | 4.4 | 88.9 | 11.1 | −0.006 | −0.207 | |
| rRNAs | M. similadanacus | 2089 | 45.4 | 7.6 | 42.2 | 4.8 | 87.6 | 12.4 | −0.037 | −0.226 |
| M. sinesensilla | 2088 | 44.3 | 8.1 | 43.4 | 4.2 | 87.7 | 12.3 | −0.010 | −0.317 | |
| N. vitripennis | 2112 | 43.1 | 8.0 | 43.8 | 5.0 | 86.9 | 13.1 | 0.008 | −0.231 | |
| P. vindemmiae | 2063 | 44.8 | 7.5 | 43.2 | 4.5 | 88.0 | 12.0 | 0.018 | −0.250 | |
| Control region | M. similadanacus | 390 | 42.1 | 17.7 | 36.2 | 4.1 | 78.2 | 21.8 | −0.075 | −0.624 |
| M. sinesensilla | 262 | 36.3 | 16.0 | 42.4 | 5.3 | 78.6 | 21.4 | 0.076 | −0.502 | |
| N. vitripennis | 4 | 50.0 | 0.0 | 50.0 | 0.0 | 100.0 | 0.0 | 0.000 | 0.000 | |
| P. vindemmiae | 183 | 34.4 | 8.7 | 51.9 | 4.9 | 86.3 | 13.7 | 0.203 | −0.279 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, Y.; Yang, Y.; Qi, L.; Hu, H.; Rasplus, J.-Y.; Wang, X. Novel Gene Rearrangement Pattern in Pachycrepoideus vindemmiae Mitochondrial Genome: New Gene Order in Pteromalidae (Hymenoptera: Chalcidoidea). Animals 2023, 13, 1985. https://doi.org/10.3390/ani13121985
Huang Y, Yang Y, Qi L, Hu H, Rasplus J-Y, Wang X. Novel Gene Rearrangement Pattern in Pachycrepoideus vindemmiae Mitochondrial Genome: New Gene Order in Pteromalidae (Hymenoptera: Chalcidoidea). Animals. 2023; 13(12):1985. https://doi.org/10.3390/ani13121985
Chicago/Turabian StyleHuang, Yixin, Yuanhan Yang, Liqing Qi, Haoyuan Hu, Jean-Yves Rasplus, and Xu Wang. 2023. "Novel Gene Rearrangement Pattern in Pachycrepoideus vindemmiae Mitochondrial Genome: New Gene Order in Pteromalidae (Hymenoptera: Chalcidoidea)" Animals 13, no. 12: 1985. https://doi.org/10.3390/ani13121985
APA StyleHuang, Y., Yang, Y., Qi, L., Hu, H., Rasplus, J.-Y., & Wang, X. (2023). Novel Gene Rearrangement Pattern in Pachycrepoideus vindemmiae Mitochondrial Genome: New Gene Order in Pteromalidae (Hymenoptera: Chalcidoidea). Animals, 13(12), 1985. https://doi.org/10.3390/ani13121985






