An Intracellular Epitope of ASFV CD2v Protein Elicits Humoral and Cellular Immune Responses
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statements
2.2. Plasmids, Cells, Serum, and Experimental Animals
2.3. Prokaryotic Expression, Purification, and Identification of CD2v-IR Domain
2.4. Eukaryotic Expression of Full Length CD2v and CD2v-IR Domain
2.5. Preparation of Monoclonal Antibody
2.6. Production of the CD2v-IR Fragments and Oligopeptides
2.7. Western Blot
2.8. Indirect Immunofluorescence Assay (IFA)
2.9. Indirect ELISA and Dot Blot Assay
2.10. Conservation Analysis of ASFV Strains
2.11. Preparation of Peptide Vaccines
2.12. Animal Immunization
2.13. Detection of Anti-CD2v IgG, IgG1, and IgG2a Antibodies
2.14. Detection of T Cell Subtypes
2.15. Detection of Interleukin-4 (IL-4) and Interferon-γ (IFN-γ)
2.16. Statistical Analysis
3. Results
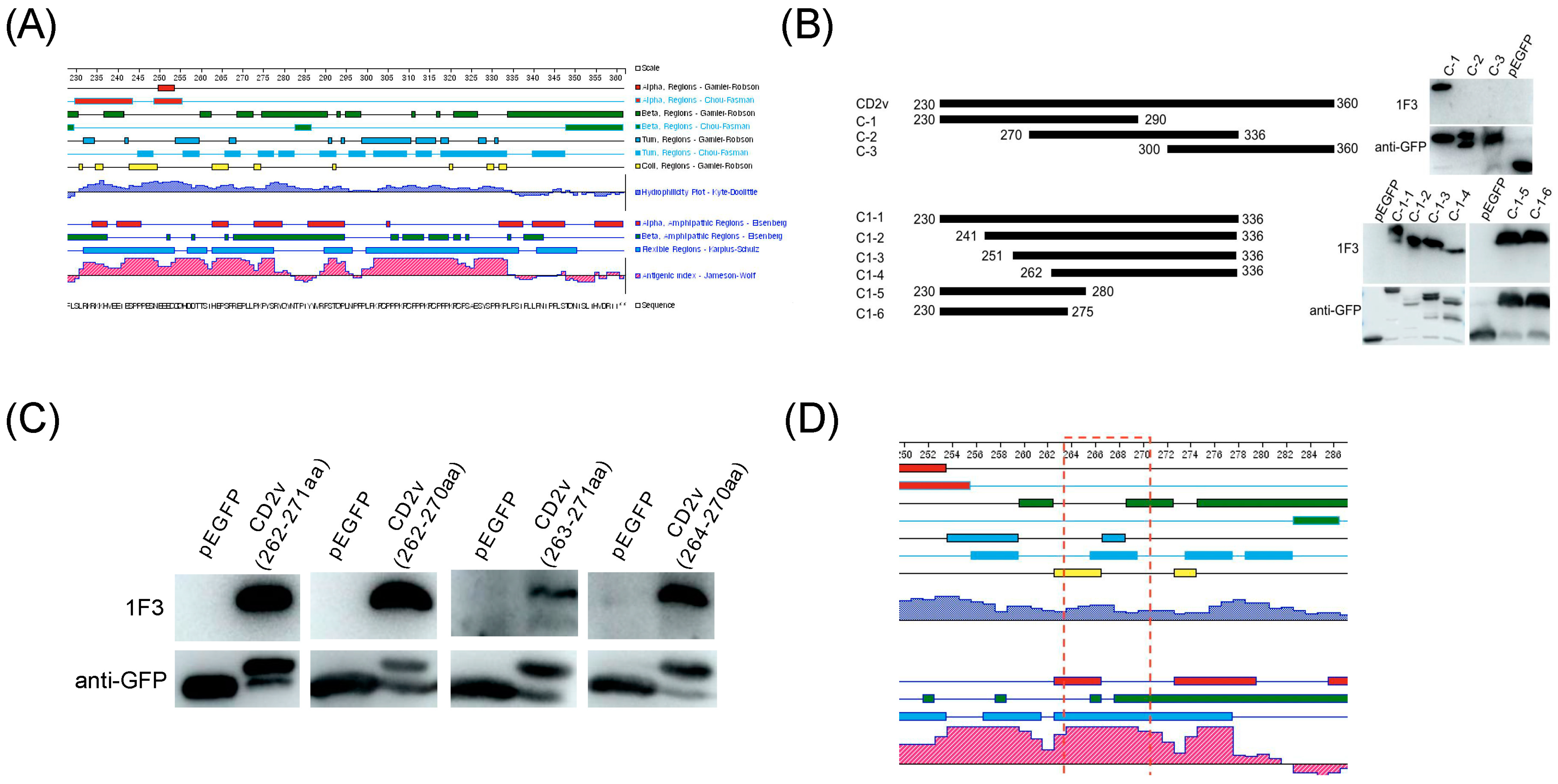
3.1. Expression and Purification of CD2v-IR Domain
3.2. Production and Characterization of mAb against the CD2v-IR Domain
3.3. Identification of a Highly Conserved Linear B Cell Epitope, 264EPSPREP270
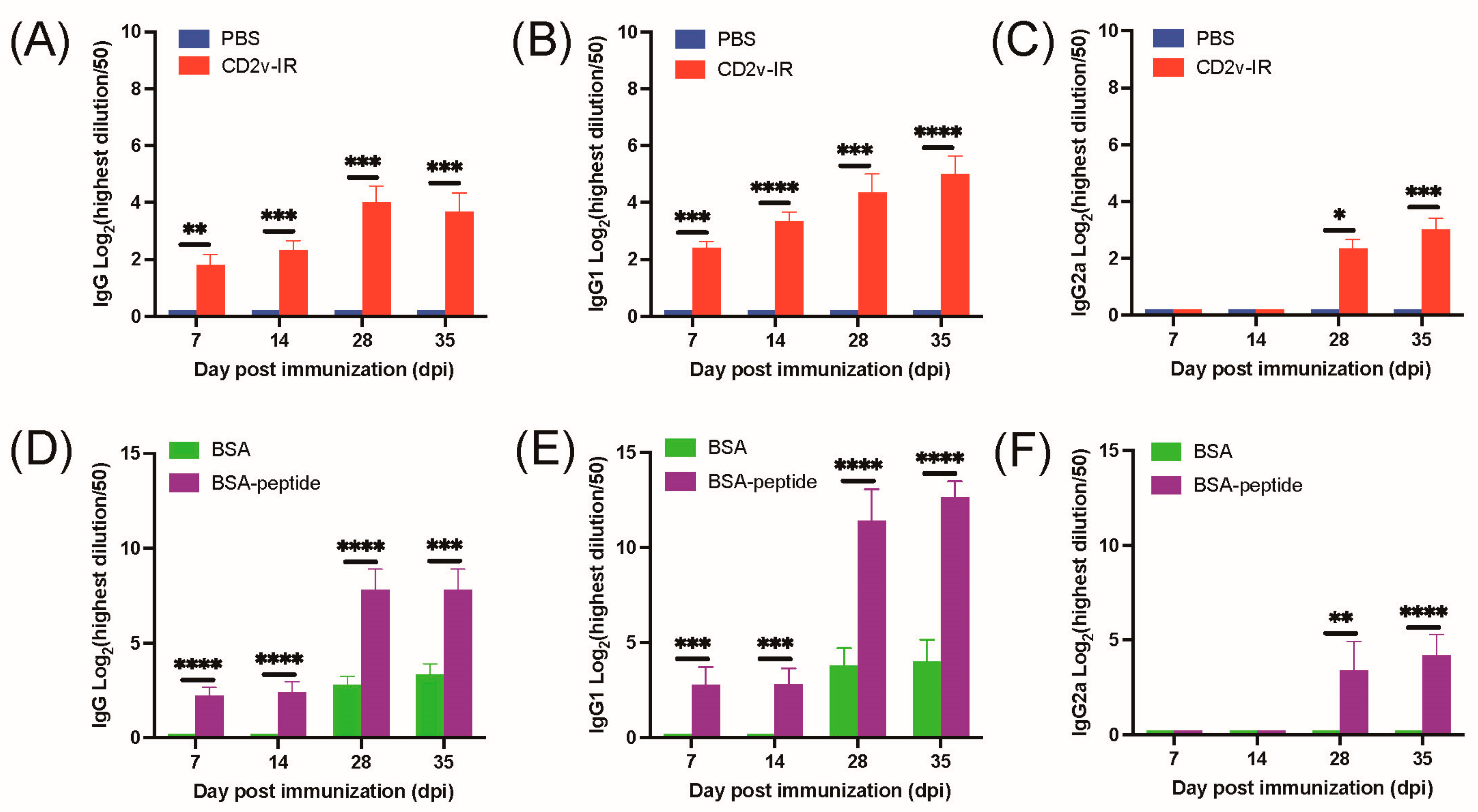
3.4. The Epitope 264EPSPREP270 Induced High Antibody Titer in Humoral Immune Response
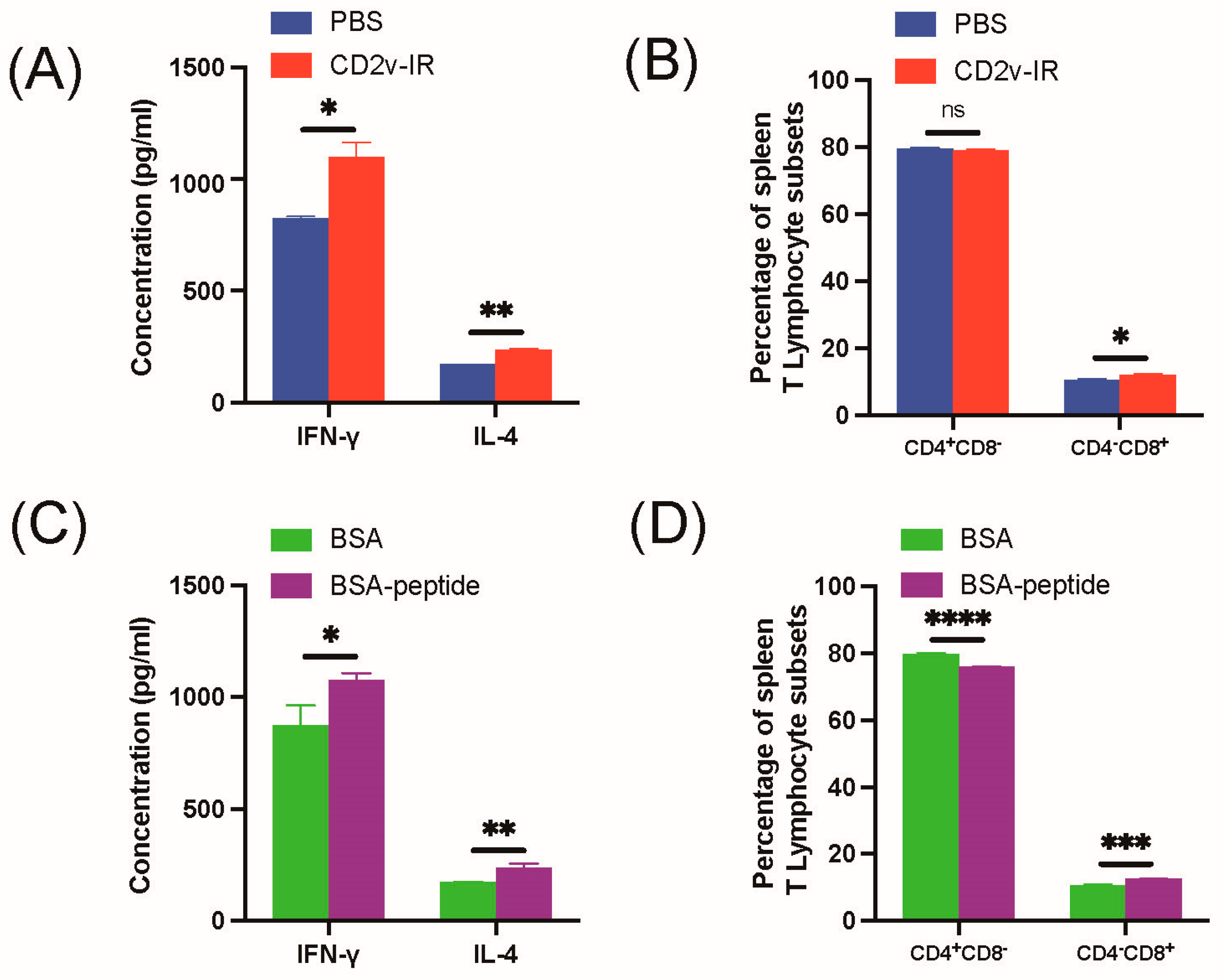
3.5. Effect of Epitope 264EPSPREP270 on Cellular Immune Response
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Dixon, L.K.; Sun, H.; Roberts, H. African swine fever. Antivir. Res. 2019, 165, 34–41. [Google Scholar] [CrossRef]
- Dixon, L.K.; Stahl, K.; Jori, F.; Vial, L.; Pfeiffer, D.U. African Swine Fever Epidemiology and Control. Annu. Rev. Anim. Biosci. 2020, 8, 221–246. [Google Scholar] [CrossRef]
- Blome, S.; Franzke, K.; Beer, M. African swine fever—A review of current knowledge. Virus Res. 2020, 287, 198099. [Google Scholar] [CrossRef]
- Malogolovkin, A.; Kolbasov, D. Genetic and antigenic diversity of African swine fever virus. Virus Res. 2019, 271, 197673. [Google Scholar] [CrossRef]
- Rowlands, R.J.; Michaud, V.; Heath, L.; Hutchings, G.; Oura, C.; Vosloo, W.; Dwarka, R.; Onashvili, T.; Albina, E.; Dixon, L.K. African swine fever virus isolate, Georgia, 2007. Emerg. Infect. Dis. 2008, 14, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Goller, K.V.; Malogolovkin, A.S.; Katorkin, S.; Kolbasov, D.; Titov, I.; Hoper, D.; Beer, M.; Keil, G.M.; Portugal, R.; Blome, S. Tandem repeat insertion in African swine fever virus, Russia, 2012. Emerg. Infect. Dis. 2015, 21, 731–732. [Google Scholar] [CrossRef]
- Smietanka, K.; Wozniakowski, G.; Kozak, E.; Niemczuk, K.; Fraczyk, M.; Bocian, L.; Kowalczyk, A.; Pejsak, Z. African Swine Fever Epidemic, Poland, 2014–2015. Emerg. Infect. Dis. 2016, 22, 1201–1207. [Google Scholar] [CrossRef]
- Zhou, X.; Li, N.; Luo, Y.; Liu, Y.; Miao, F.; Chen, T.; Zhang, S.; Cao, P.; Li, X.; Tian, K.; et al. Emergence of African Swine Fever in China, 2018. Transbound. Emerg. Dis. 2018, 65, 1482–1484. [Google Scholar] [CrossRef] [PubMed]
- Mighell, E.; Ward, M.P. African Swine Fever spread across Asia, 2018–2019. Transbound. Emerg. Dis. 2021, 68, 2722–2732. [Google Scholar] [CrossRef] [PubMed]
- Urbano, A.C.; Ferreira, F. African swine fever control and prevention: An update on vaccine development. Emerg. Microbes Infect. 2022, 11, 2021–2033. [Google Scholar] [CrossRef]
- Gladue, D.P.; Borca, M.V. Recombinant ASF Live Attenuated Virus Strains as Experimental Vaccine Candidates. Viruses 2022, 14, 878. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, X.; Qi, W.; Yang, Y.; Liu, Z.; An, T.; Wu, X.; Chen, J. Prevention and Control Strategies of African Swine Fever and Progress on Pig Farm Repopulation in China. Viruses 2021, 13, 2552. [Google Scholar] [CrossRef]
- Alonso, C.; Borca, M.; Dixon, L.; Revilla, Y.; Rodriguez, F.; Escribano, J.M.; Ictv Report, C. ICTV Virus Taxonomy Profile: Asfarviridae. J. Gen. Virol. 2018, 99, 613–614. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Xie, M.; Wu, W.; Chen, Z. Structures and Functional Diversities of ASFV Proteins. Viruses 2021, 13, 2124. [Google Scholar] [CrossRef]
- Duan, X.; Ru, Y.; Yang, W.; Ren, J.; Hao, R.; Qin, X.; Li, D.; Zheng, H. Research progress on the proteins involved in African swine fever virus infection and replication. Front. Immunol. 2022, 13, 947180. [Google Scholar] [CrossRef]
- Wang, N.; Zhao, D.; Wang, J.; Zhang, Y.; Wang, M.; Gao, Y.; Li, F.; Wang, J.; Bu, Z.; Rao, Z.; et al. Architecture of African swine fever virus and implications for viral assembly. Science 2019, 366, 640–644. [Google Scholar] [CrossRef] [PubMed]
- Alejo, A.; Matamoros, T.; Guerra, M.; Andres, G. A Proteomic Atlas of the African Swine Fever Virus Particle. J. Virol. 2018, 92, e01293-18. [Google Scholar] [CrossRef] [PubMed]
- Goatley, L.C.; Dixon, L.K. Processing and localization of the african swine fever virus CD2v transmembrane protein. J. Virol. 2011, 85, 3294–3305. [Google Scholar] [CrossRef] [PubMed]
- Kay-Jackson, P.C.; Goatley, L.C.; Cox, L.; Miskin, J.E.; Parkhouse, R.M.E.; Wienands, J.; Dixon, L.K. The CD2v protein of African swine fever virus interacts with the actin-binding adaptor protein SH3P7. J. Gen. Virol. 2004, 85, 119–130. [Google Scholar] [CrossRef]
- Perez-Nunez, D.; Garcia-Urdiales, E.; Martinez-Bonet, M.; Nogal, M.L.; Barroso, S.; Revilla, Y.; Madrid, R. CD2v Interacts with Adaptor Protein AP-1 during African Swine Fever Infection. PLoS ONE 2015, 10, e0123714. [Google Scholar] [CrossRef]
- Ruiz-Gonzalvo, F.; Rodriguez, F.; Escribano, J.M. Functional and immunological properties of the baculovirus-expressed hemagglutinin of African swine fever virus. Virology 1996, 218, 285–289. [Google Scholar] [CrossRef]
- Burmakina, G.; Malogolovkin, A.; Tulman, E.R.; Xu, W.; Delhon, G.; Kolbasov, D.; Rock, D.L. Identification of T-cell epitopes in African swine fever virus CD2v and C-type lectin proteins. J. Gen. Virol. 2019, 100, 259–265. [Google Scholar] [CrossRef]
- Jia, R.; Zhang, G.; Bai, Y.; Liu, H.; Chen, Y.; Ding, P.; Zhou, J.; Feng, H.; Li, M.; Tian, Y.; et al. Identification of Linear B Cell Epitopes on CD2V Protein of African Swine Fever Virus by Monoclonal Antibodies. Microbiol. Spectr. 2022, 10, e0105221. [Google Scholar] [CrossRef]
- Liu, H.; Wang, A.; Yang, W.; Liang, C.; Zhou, J.; Chen, Y.; Liu, Y.; Zhou, Y.; Zhang, G. Expression of extracellular domain of ASFV CD2v protein in mammalian cells and identification of B cell epitopes. Virus Res. 2022, 323, 199000. [Google Scholar] [CrossRef]
- Ren, D.; Ding, P.; Liu, S.; Zhang, N.; Chen, Y.; Li, Q.; Fan, L.; Chang, Z.; Zhang, G. Development and characterization of recombinant ASFV CD2v protein nanoparticle-induced monoclonal antibody. Int. J. Biol. Macromol. 2022, 209, 533–541. [Google Scholar] [CrossRef]
- Song, J.; Wang, M.; Du, Y.; Wan, B.; Zhang, A.; Zhang, Y.; Zhuang, G.; Ji, P.; Wu, Y.; Zhang, G. Identification of a linear B-cell epitope on the African swine fever virus CD2v protein. Int. J. Biol. Macromol. 2023, 232, 123264. [Google Scholar] [CrossRef] [PubMed]
- Herrera, L.R.M.; Bisa, E.P. In silico analysis of highly conserved cytotoxic T-cell epitopes in the structural proteins of African swine fever virus. Vet. World 2021, 14, 2625–2633. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Wang, X.; Zhou, J.; Shi, S.; Shen, T.; Chen, L.; Zhang, M.; Liao, C.; Wang, C. Development of PDA Nanoparticles for H9N2 Avian Influenza BPP-V/BP-IV Epitope Peptide Vaccines: Immunogenicity and Delivery Efficiency Improvement. Front. Immunol. 2021, 12, 693972. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Shen, T.; Zhou, J.; Chen, L.; Shi, S.; Wang, X.; Zhang, M.; Wang, C.; Liao, C. Bursal peptide BP-IV as a novel immunoadjuvant enhances the protective efficacy of an epitope peptide vaccine containing T and B cell epitopes of the H9N2 avian influenza virus. Microb. Pathog. 2021, 158, 105095. [Google Scholar] [CrossRef]
- Monteagudo, P.L.; Lacasta, A.; Lopez, E.; Bosch, L.; Collado, J.; Pina-Pedrero, S.; Correa-Fiz, F.; Accensi, F.; Navas, M.J.; Vidal, E.; et al. BA71DeltaCD2: A New Recombinant Live Attenuated African Swine Fever Virus with Cross-Protective Capabilities. J. Virol. 2017, 91, e01058-17. [Google Scholar] [CrossRef] [PubMed]
- Bosch-Camos, L.; Alonso, U.; Esteve-Codina, A.; Chang, C.Y.; Martin-Mur, B.; Accensi, F.; Munoz, M.; Navas, M.J.; Dabad, M.; Vidal, E.; et al. Cross-protection against African swine fever virus upon intranasal vaccination is associated with an adaptive-innate immune crosstalk. PLoS Pathog. 2022, 18, e1010931. [Google Scholar] [CrossRef] [PubMed]
- Borca, M.V.; O'Donnell, V.; Holinka, L.G.; Risatti, G.R.; Ramirez-Medina, E.; Vuono, E.A.; Shi, J.; Pruitt, S.; Rai, A.; Silva, E.; et al. Deletion of CD2-like gene from the genome of African swine fever virus strain Georgia does not attenuate virulence in swine. Sci. Rep. 2020, 10, 494. [Google Scholar] [CrossRef] [PubMed]
- Teklue, T.; Wang, T.; Luo, Y.; Hu, R.; Sun, Y.; Qiu, H.J. Generation and Evaluation of an African Swine Fever Virus Mutant with Deletion of the CD2v and UK Genes. Vaccines 2020, 8, 763. [Google Scholar] [CrossRef] [PubMed]
- Petrovan, V.; Rathakrishnan, A.; Islam, M.; Goatley, L.C.; Moffat, K.; Sanchez-Cordon, P.J.; Reis, A.L.; Dixon, L.K. Role of African Swine Fever Virus Proteins EP153R and EP402R in Reducing Viral Persistence in Blood and Virulence in Pigs Infected with BeninDeltaDP148R. J. Virol. 2022, 96, e0134021. [Google Scholar] [CrossRef]
- Malogolovkin, A.; Burmakina, G.; Tulman, E.R.; Delhon, G.; Diel, D.G.; Salnikov, N.; Kutish, G.F.; Kolbasov, D.; Rock, D.L. African swine fever virus CD2v and C-type lectin gene loci mediate serological specificity. J. Gen. Virol. 2015, 96, 866–873. [Google Scholar] [CrossRef] [PubMed]
- Luong, H.Q.; Lai, H.T.; Do, L.D.; Ha, B.X.; Nguyen, G.V.; Vu, H.L. Differential antibody responses in sows and finishing pigs naturally infected with African swine fever virus under field conditions. Virus Res. 2022, 307, 198621. [Google Scholar] [CrossRef]
- Lopera-Madrid, J.; Osorio, J.E.; He, Y.; Xiang, Z.; Adams, L.G.; Laughlin, R.C.; Mwangi, W.; Subramanya, S.; Neilan, J.; Brake, D.; et al. Safety and immunogenicity of mammalian cell derived and Modified Vaccinia Ankara vectored African swine fever subunit antigens in swine. Vet. Immunol. Immunopathol. 2017, 185, 20–33. [Google Scholar] [CrossRef]
- Chen, L.; Zhang, X.; Shao, G.; Shao, Y.; Hu, Z.; Feng, K.; Xie, Z.; Li, H.; Chen, W.; Lin, W.; et al. Construction and Evaluation of Recombinant Pseudorabies Virus Expressing African Swine Fever Virus Antigen Genes. Front. Vet. Sci. 2022, 9, 832255. [Google Scholar] [CrossRef] [PubMed]
- Argilaguet, J.M.; Perez-Martin, E.; Lopez, S.; Goethe, M.; Escribano, J.M.; Giesow, K.; Keil, G.M.; Rodriguez, F. BacMam immunization partially protects pigs against sublethal challenge with African swine fever virus. Antivir. Res. 2013, 98, 61–65. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Kang, W.; Yang, W.; Zhang, J.; Li, D.; Zheng, H. Structure of African Swine Fever Virus and Associated Molecular Mechanisms Underlying Infection and Immunosuppression: A Review. Front. Immunol. 2021, 12, 715582. [Google Scholar] [CrossRef]
- Heitmann, J.S.; Bilich, T.; Tandler, C.; Nelde, A.; Maringer, Y.; Marconato, M.; Reusch, J.; Jager, S.; Denk, M.; Richter, M.; et al. A COVID-19 peptide vaccine for the induction of SARS-CoV-2 T cell immunity. Nature 2022, 601, 617–622. [Google Scholar] [CrossRef] [PubMed]
- Qin, Y.; Tu, K.; Teng, Q.; Feng, D.; Zhao, Y.; Zhang, G. Identification of Novel T-Cell Epitopes on Infectious Bronchitis Virus N Protein and Development of a Multi-epitope Vaccine. J. Virol. 2021, 95, e0066721. [Google Scholar] [CrossRef] [PubMed]
- Han, K.; Zhao, D.; Liu, Y.; Huang, X.; Yang, J.; Liu, Q.; An, F.; Li, Y. Design and evaluation of a polytope construct with multiple B and T epitopes against Tembusu virus infection in ducks. Res. Vet. Sci. 2016, 104, 174–180. [Google Scholar] [CrossRef] [PubMed]
- Netherton, C.L.; Goatley, L.C.; Reis, A.L.; Portugal, R.; Nash, R.H.; Morgan, S.B.; Gault, L.; Nieto, R.; Norlin, V.; Gallardo, C.; et al. Identification and Immunogenicity of African Swine Fever Virus Antigens. Front. Immunol. 2019, 10, 1318. [Google Scholar] [CrossRef]
- Portugal, R. ELISpot Assay for the Detection of ASFV-Specific Interferon-Gamma (IFN-gamma)-Producing Cells. Methods Mol. Biol. 2022, 2503, 169–178. [Google Scholar] [CrossRef]
- Jiang, W.; Jiang, D.; Li, L.; Wang, J.; Wang, P.; Shi, X.; Zhao, Q.; Liu, B.; Ji, P.; Zhang, G. Identification of Two Novel Linear B Cell Epitopes on the CD2v Protein of African Swine Fever Virus Using Monoclonal Antibodies. Viruses 2022, 15, 131. [Google Scholar] [CrossRef]
- Sunwoo, S.Y.; Perez-Nunez, D.; Morozov, I.; Sanchez, E.G.; Gaudreault, N.N.; Trujillo, J.D.; Mur, L.; Nogal, M.; Madden, D.; Urbaniak, K.; et al. DNA-Protein Vaccination Strategy Does Not Protect from Challenge with African Swine Fever Virus Armenia 2007 Strain. Vaccines 2019, 7, 12. [Google Scholar] [CrossRef]
- Feng, Z.; Chen, J.; Liang, W.; Chen, W.; Li, Z.; Chen, Q.; Cai, S. The recombinant pseudorabies virus expressing African swine fever virus CD2v protein is safe and effective in mice. Virol. J. 2020, 17, 180. [Google Scholar] [CrossRef]
- Gerritsen, B.; Pandit, A. The memory of a killer T cell: Models of CD8(+) T cell differentiation. Immunol. Cell Biol. 2016, 94, 236–241. [Google Scholar] [CrossRef]
- Reina-Campos, M.; Scharping, N.E.; Goldrath, A.W. CD8(+) T cell metabolism in infection and cancer. Nat. Rev. Immunol. 2021, 21, 718–738. [Google Scholar] [CrossRef]




| Primers | Sequences (5′–3′) |
|---|---|
| pEGFP-N1-230-290-F | gctagcATGTCCCTCCGCAAGCGCAAGAA |
| pEGFP-N1-230-290-R | ggatccGTGCGCATGTAGTAGATGGGGGT |
| pEGFP-N1-270-336-F | gctagcATGCCTCTGCTGCCTAAGCCCTA |
| pEGFP-N1-270-336-R | ggatccGTGGAAGGGAGAGGCTTGGGAG |
| pEGFP-N1-300-360-F | gctagcATGCTCCCCAAGCCTTGCCCTCCTCCTAAGCCTTGCCCCCCTCCCAAGCCCT |
| pEGFP-N1-300-360-R | ggatccGTGATGATGCGATCGACGTGGATGA |
| pEGFP-N1-241-336-F | ctcgagATGATCGAGTCCCCTCCTC |
| pEGFP-N1-251-336-F | ctcgagATGGAGGAGCAATGCCAAC |
| pEGFP-N1-262-336-F | ctcgagATGATCCACGAGCCCTCCCCT |
| pEGFP-N1-230-280-R | gatcggatccGTCTGGTAGCGAGAGTAGGGCTT |
| pEGFP-N1-230-275-R | gatcggatccGTGGGCTTAGGCAGCAGAGGCT |
| Clone | 1F3 |
|---|---|
| IgG1 | 1.5 |
| IgG2a | 0.03 |
| IgG2b | 0.03 |
| IgG2c | 0.02 |
| IgG3 | 0.04 |
| Blank | 0.02 |
| mAb | Heavy Chain | Light Chain |
|---|---|---|
| 1F3 | QLQESGGGLVQPGGSMKLSCAASGFPFSDAWMDWVRQSPERGLEWVAEIRSKANNHATYYAESVKGRFTISRDDSKSRVYLHMNSLRGEDTGIYYCNGPAAGVWGQGTTVTVSS | DIELTQSPVSITASRGEKVTITCRANSSISSNNFHWYLRRPGSSPKLLIYRTSILASGVLDSFSGRGSESSSTLTIHYMQDEVAATYYCQQGSICPPRSEGGPSSRSN |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lu, W.; Bai, Y.; Zhang, S.; Zhao, X.; Jin, J.; Zhu, X.; Wang, R.; Wu, Y.; Zhang, A.; Zhang, G.; et al. An Intracellular Epitope of ASFV CD2v Protein Elicits Humoral and Cellular Immune Responses. Animals 2023, 13, 1967. https://doi.org/10.3390/ani13121967
Lu W, Bai Y, Zhang S, Zhao X, Jin J, Zhu X, Wang R, Wu Y, Zhang A, Zhang G, et al. An Intracellular Epitope of ASFV CD2v Protein Elicits Humoral and Cellular Immune Responses. Animals. 2023; 13(12):1967. https://doi.org/10.3390/ani13121967
Chicago/Turabian StyleLu, Wenlong, Yilin Bai, Shuai Zhang, Xuyang Zhao, Jiaxin Jin, Xiaojing Zhu, Rui Wang, Yanan Wu, Angke Zhang, Gaiping Zhang, and et al. 2023. "An Intracellular Epitope of ASFV CD2v Protein Elicits Humoral and Cellular Immune Responses" Animals 13, no. 12: 1967. https://doi.org/10.3390/ani13121967
APA StyleLu, W., Bai, Y., Zhang, S., Zhao, X., Jin, J., Zhu, X., Wang, R., Wu, Y., Zhang, A., Zhang, G., Zhuang, G., & Sun, A. (2023). An Intracellular Epitope of ASFV CD2v Protein Elicits Humoral and Cellular Immune Responses. Animals, 13(12), 1967. https://doi.org/10.3390/ani13121967





