Heterogeneity of Fish Taxonomic and Functional Diversity Evaluated by eDNA and Gillnet along a Mangrove–Seagrass–Coral Reef Continuum
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
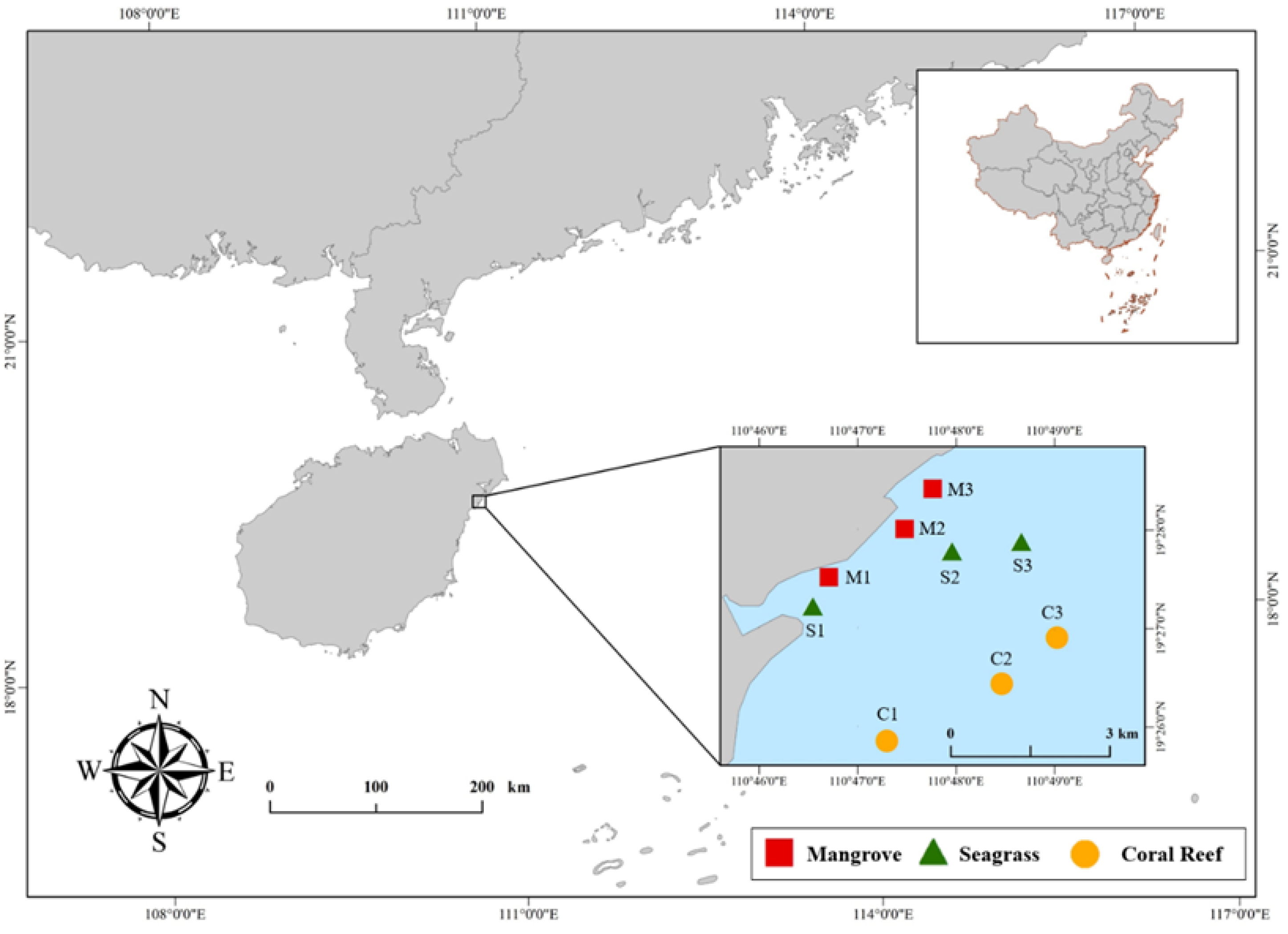
2.1. Study Area
2.2. Environmental DNA Sampling and Library Construction of 12S-rDNA Barcodes
2.3. Gillnet Sample Collection and Species Identification
2.4. Data Analysis
3. Results
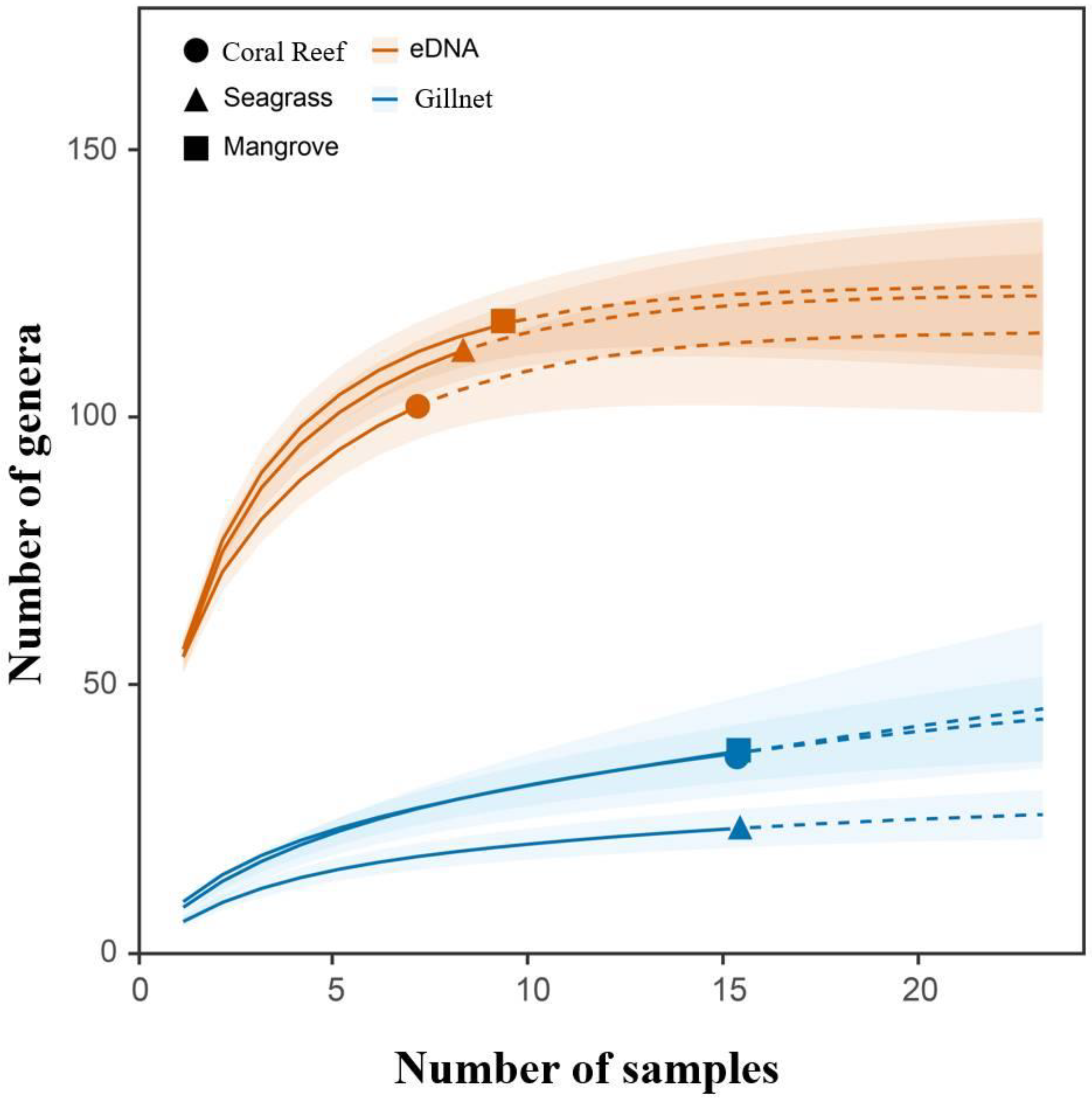
3.1. Species Accumulation Curves
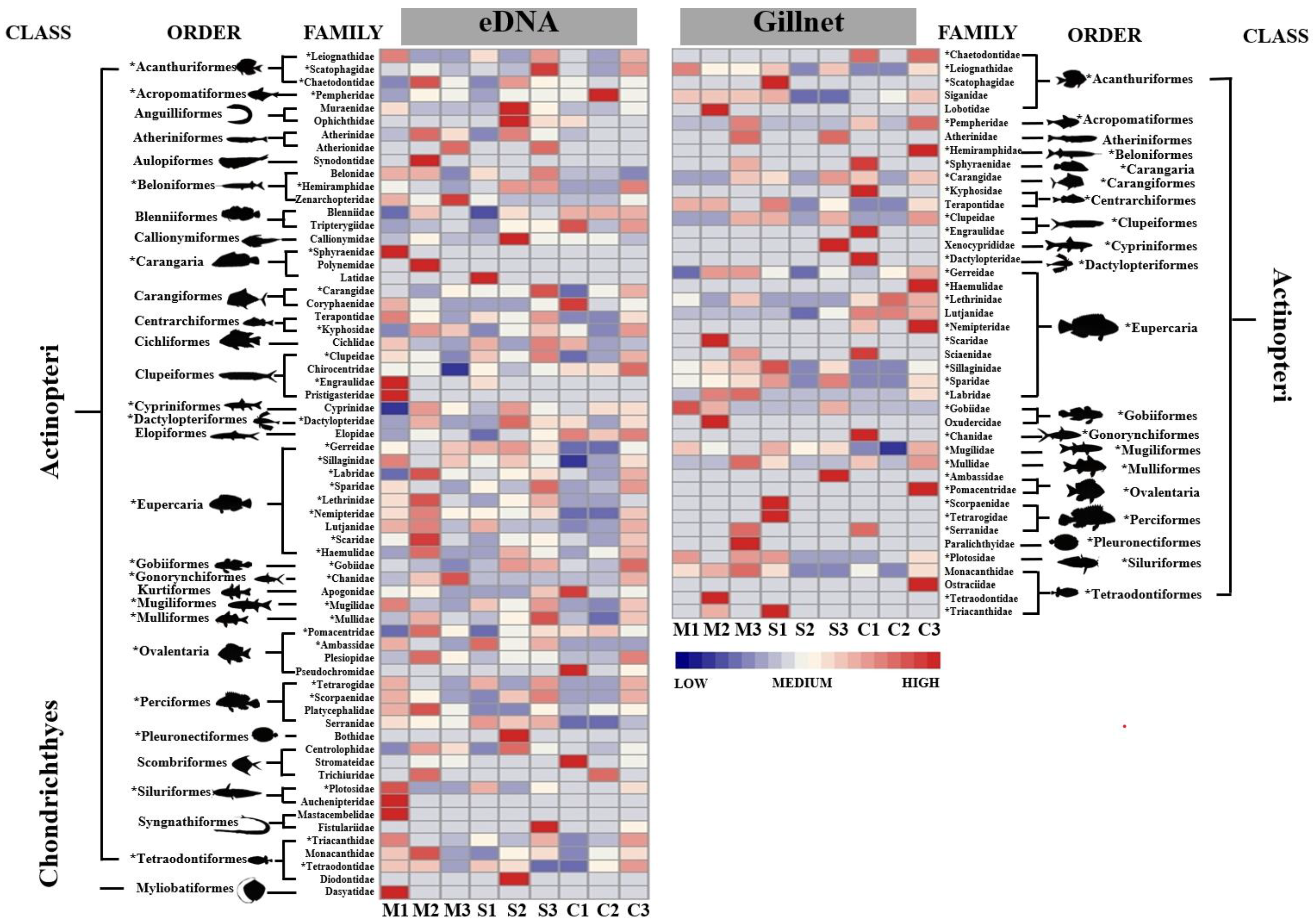
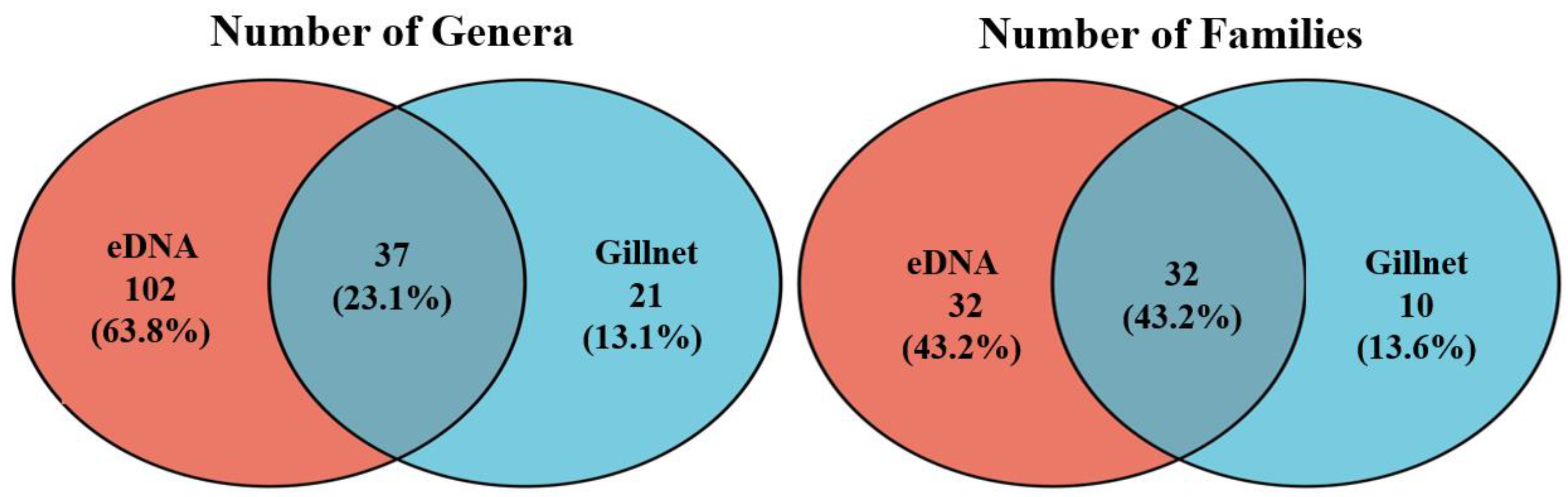
3.2. Fish Taxonomic Information
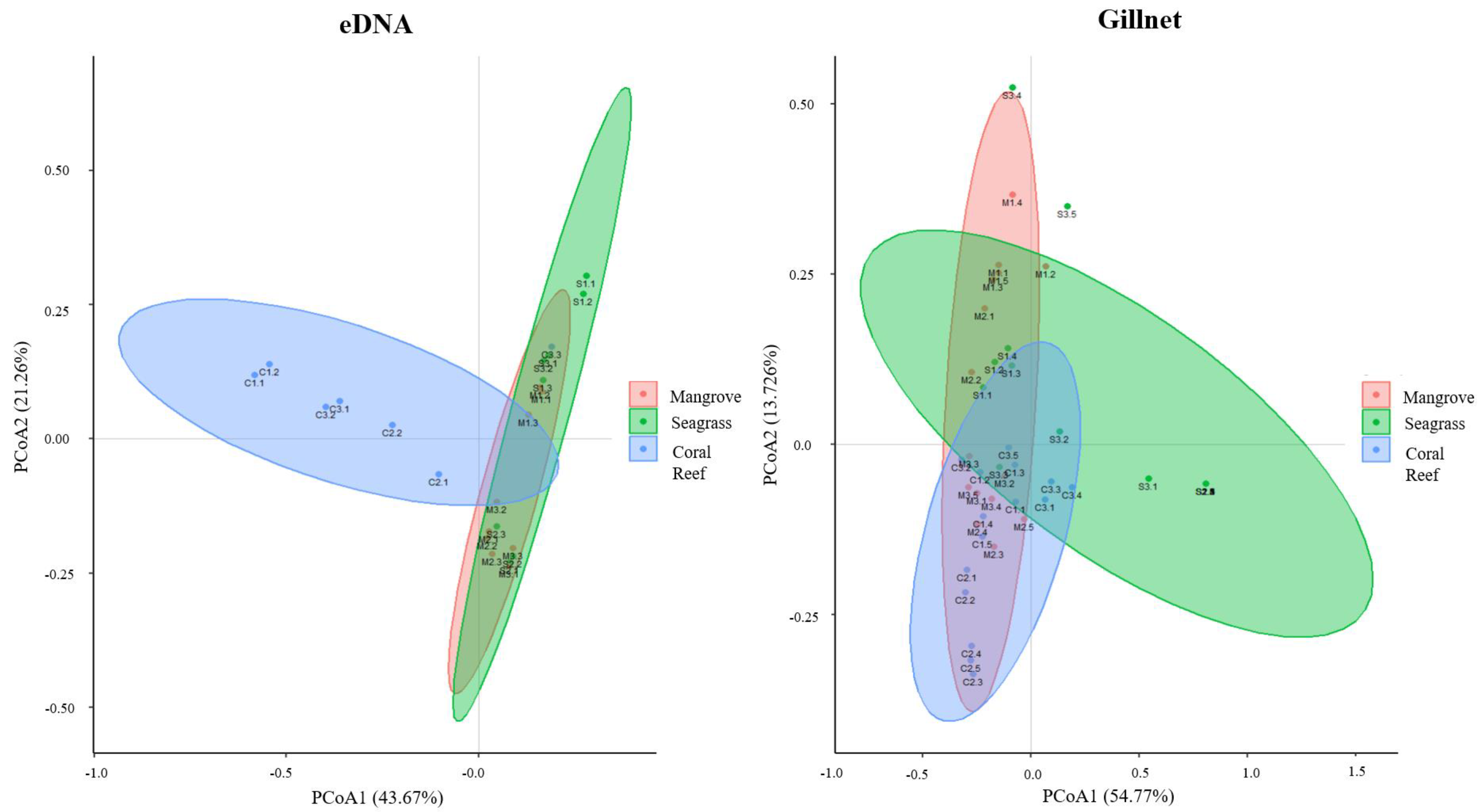
3.3. Fish Communities
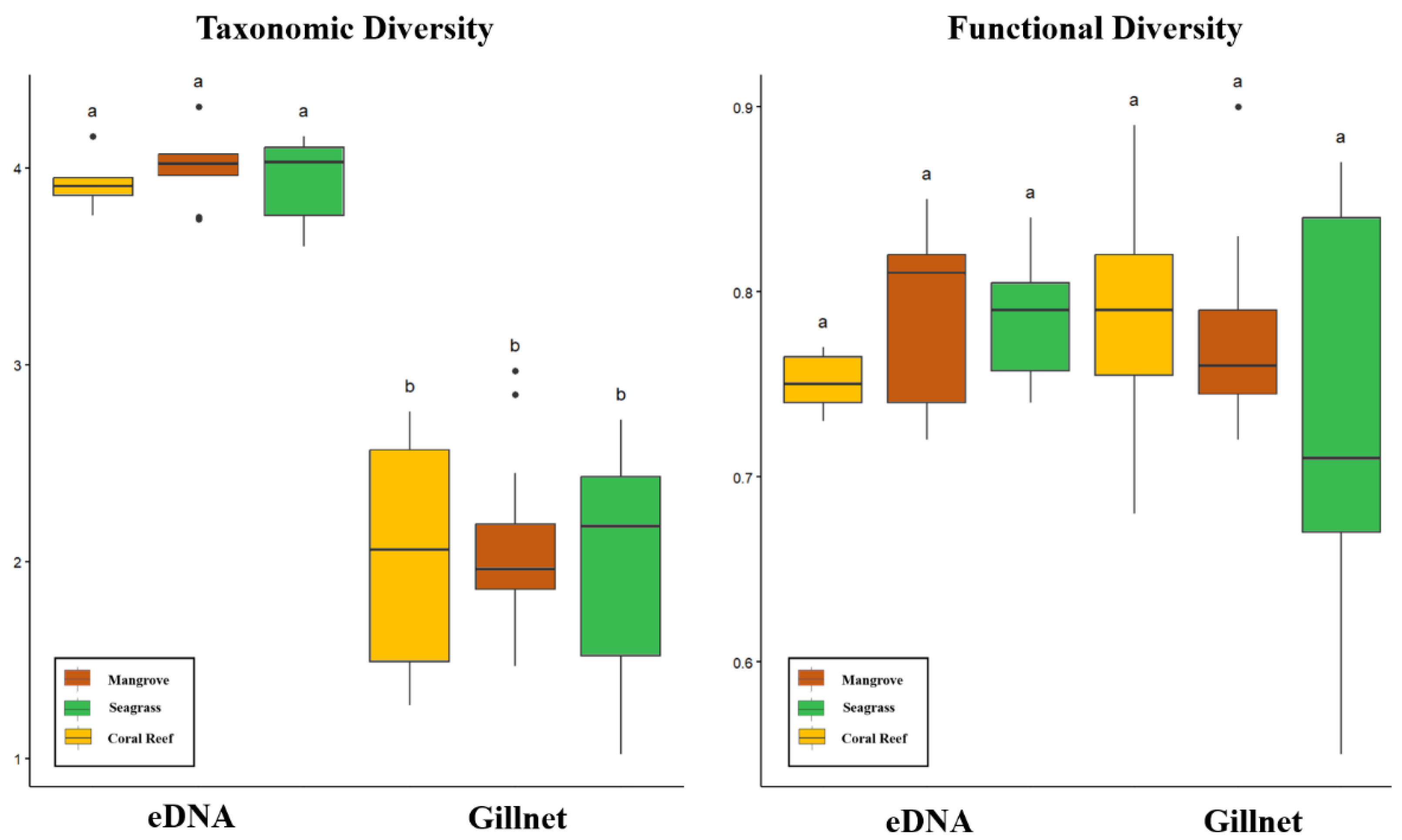
3.4. Taxonomic Diversity and Functional Diversity
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Carlson, R.; Evans, L.; Foo, S.; Grady, B.; Li, J.; Seeley, M.; Xu, Y.; Asner, G. Synergistic benefits of conserving land-sea ecosystems. Glob. Ecol. Conserv. 2021, 28, e01684. [Google Scholar] [CrossRef]
- Berkström, C.; Eggertsen, L.; Goodell, W.; Cordeiro, C.A.M.M.; Lucena, M.B.; Gustafsson, R.; Bandeira, S.; Jiddawi, N.; Ferreira, C.E.L. Thresholds in Seascape Connectivity: The spatial arrangement of nursery habitats structure fish communities on nearby reefs. Ecography 2020, 43, 882–896. [Google Scholar] [CrossRef]
- Alongi, D.M. Present state and future of the world’s mangrove forests. Environ. Conserv. 2002, 29, 331–349. [Google Scholar] [CrossRef]
- Waycott, M.; Duarte, C.M.; Carruthers, T.J.B.; Orth, R.J.; Dennison, W.C.; Olyarnik, S.; Calladine, A.; Fourqurean, J.W.; Heck, K.L.; Hughes, A.R.; et al. Accelerating loss of seagrasses across the globe threatens coastal ecosystems. Proc. Natl. Acad. Sci. USA 2009, 106, 12377–12381. [Google Scholar] [CrossRef] [PubMed]
- McKenzie, L.; Nordlund, L.M.; Jones, B.L.; Cullen-Unsworth, L.C.; Roelfsema, C.M.; Unsworth, R. The global distribution of seagrass meadows. Environ. Res. Lett. 2020, 15, 0740401. [Google Scholar] [CrossRef]
- Cesar, H.; Burke, L.; Pet-Soede, L. The Economics of Worldwide Coral Reef Degradation; International Coral Reef Action Network: Cambridge, UK, 2003. [Google Scholar]
- Guan, Y.; Hohn, S.; Wild, C.; Merico, A. Vulnerability of global coral reef habitat suitability to ocean warming, acidification and eutrophication. Glob. Chang. Biol. 2020, 26, 5646–5660. [Google Scholar] [CrossRef]
- Stat, M.; John, J.; DiBattista, J.D.; Newman, S.J.; Bunce, M.; Harvey, E.S. Combined use of eDNA metabarcoding and video surveillance for the assessment of fish biodiversity. Conserv. Biol. 2018, 33, 196–205. [Google Scholar] [CrossRef]
- Sigsgaard, E.E.; Torquato, F.; Frøslev, T.G.; Moore, A.B.M.; Sørensen, J.M.; Range, P.; Ben-Hamadou, R.; Bach, S.S.; Møller, P.R.; Thomsen, P.F. Using vertebrate Environmental DNA from seawater in biomonitoring of marine habitats. Conserv. Biol. 2019, 34, 697–710. [Google Scholar] [CrossRef]
- Li, C.; Long, H.; Yang, S.; Zhang, Y.; Tang, F.; Jin, W.; Wang, G.; Chang, W.; Pi, Y.; Gao, L.; et al. EDNA Assessment of pelagic fish diversity, distribution, and abundance in the central Pacific Ocean. Reg. Stud. Mar. Sci. 2022, 56, 102661. [Google Scholar] [CrossRef]
- Martinez-Haro, M.; Beiras, R.; Bellas, J.; Capela, R.; Coelho, J.P.; Lopes, I.; Moreira-Santos, M.; Reis-Henriques, A.M.; Ribeiro, R.; Santos, M.M.; et al. A review on the ecological quality status assessment in aquatic systems using community based indicators and ecotoxicological tools: What might be the added value of their combination? Ecol. Indic. 2015, 48, 8–16. [Google Scholar] [CrossRef]
- Sigsgaard, E.E.; Jensen, M.R.; Winkelmann, I.E.; Møller, P.R.; Hansen, M.M.; Thomsen, P.F. Population-level inferences from environmental DNA—Current status and future perspectives. Evol. Appl. 2019, 13, 245–262. [Google Scholar] [CrossRef] [PubMed]
- Watson, D.L.; Harvey, E.S.; Anderson, M.J.; Kendrick, G.A. A comparison of temperate reef fish assemblages recorded by three underwater stereo-video techniques. Mar. Biol. 2005, 148, 415–425. [Google Scholar] [CrossRef]
- Hopkins, G.W.; Freckleton, R.P. Declines in the numbers of amateur and professional taxonomists: Implications for conservation. Anim. Conserv. 2002, 5, 245–249. [Google Scholar] [CrossRef]
- Jeunen, G.; Knapp, M.; Spencer, H.G.; Lamare, M.D.; Taylor, H.R.; Stat, M.; Bunce, M.; Gemmell, N.J. Environmental DNA (eDNA) metabarcoding reveals strong discrimination among diverse marine habitats connected by water movement. Mol. Ecol. Resour. 2019, 19, 426–438. [Google Scholar] [CrossRef] [PubMed]
- Stoeckle, M.Y.; Adolf, J.; Charlop-Powers, Z.; Dunton, K.J.; Hinks, G.; VanMorter, S.M. Trawl and eDNA assessment of marine fish diversity, seasonality, and relative abundance in coastal New Jersey, USA. ICES J. Mar. Sci. 2021, 78, 293–304. [Google Scholar] [CrossRef]
- Kelly, R.P.; Closek, C.J.; O’Donnell, J.L.; Kralj, J.E.; Shelton, A.O.; Samhouri, J.F. Genetic and manual survey methods yield different and complementary views of an ecosystem. Front. Mar. Sci. 2017, 3, 283. [Google Scholar] [CrossRef]
- Momota, K.; Hosokawa, S.; Komuro, T. Small-scale heterogeneity of fish diversity evaluated by environmental DNA analysis in eelgrass beds. Mar. Ecol. Prog. Ser. 2022, 688, 99–112. [Google Scholar] [CrossRef]
- Doxa, A.; Holon, F.; Deter, J.; Villéger, S.; Boissery, P.; Mouquet, N. Mapping biodiversity in three-dimensions challenges marine conservation strategies: The example of coralligenous assemblages in North-Western Mediterranean Sea. Ecol. Indic. 2016, 61, 1042–1054. [Google Scholar] [CrossRef]
- Da Silva, V.E.L.; Dolbeth, M.; Fabré, N.N. Assessing tropical coastal dynamics across habitats and seasons through different dimensions of fish diversity. Mar. Environ. Res. 2021, 171, 105458. [Google Scholar] [CrossRef]
- Loreau, M.; de Mazancourt, C. Biodiversity and ecosystem stability: A synthesis of underlying mechanisms. Ecol. Lett. 2013, 16, 106–115. [Google Scholar] [CrossRef]
- Bellwood, D.R.; Streit, R.P.; Brandl, S.J.; Tebbett, S.B. The meaning of the term “Function” in ecology: A coral reef perspective. Funct. Ecol. 2019, 33, 948–961. [Google Scholar] [CrossRef]
- Streit, R.P.; Cumming, G.S.; Bellwood, D.R. Patchy delivery of functions undermines functional redundancy in a high diversity system. Funct. Ecol. 2019, 33, 1144–1155. [Google Scholar] [CrossRef]
- Vallée, V.; Villanueva, M.; Blanchard, F. Functional richness and turnover patterns reveal assembly rules structuring marine fish communities on the continental shelf of French Guiana. Mar. Ecol. Prog. Ser. 2019, 630, 183–195. [Google Scholar] [CrossRef]
- Navarro-Martínez, Z.; Armenteros, M.; Espinosa, L.; Lake, J.; Apprill, A. Taxonomic and functional assemblage structure of coral reef fishes from Jardines de La Reina (Caribbean Sea, Cuba). Mar. Ecol. Prog. Ser. 2022, 690, 113–132. [Google Scholar] [CrossRef]
- Aglieri, G.; Baillie, C.; Mariani, S.; Cattano, C.; Calò, A.; Turco, G.; Spatafora, D.; Di Franco, A.; Di Lorenzo, M.; Guidetti, P.; et al. Environmental DNA effectively captures functional diversity of coastal fish communities. Mol. Ecol. 2020, 30, 3127–3139. [Google Scholar] [CrossRef]
- Polanco, F.A.; Waldock, C.; Keggin, T.; Marques, V.; Rozanski, R.; Valentini, A.; Dejean, T.; Manel, S.; Vermeij, M.; Albouy, C.; et al. Ecological indices from environmen DNA to contrast coastal reefs under different anthropogenic pressures. Ecol. Evol. 2022, 12, e9212. [Google Scholar] [CrossRef]
- Zhu, B.; Liao, J.; Shen, G. Spatio-temporal simulation of mangrove forests under different scenarios: A case study of mangrove protected areas, Hainan Island, China. Remote Sens. 2021, 13, 4059. [Google Scholar] [CrossRef]
- Xu, B.; Zhang, J.; Lang, S.; Chen, S.; Wu, Z.; Wang, D. Ecological status and degradation factors in seagrass meadows along the coast of Wenchang, Hainan. J. Appl. Oceanogr. 2022, 4, 11. [Google Scholar] [CrossRef]
- Chen, H.; Li, Y. The health status of the coral reef ecosystem and its ecological response to reclamation in Yelin Bay, Wenchang, Hainan Province. Mar. Environ. Sci. 2019, 38, 533–539. [Google Scholar] [CrossRef]
- Du, J.; Xie, M.; Wang, Y.; Chen, Z.; Liu, W.; Liao, J.; Chen, B. Connectivity of fish assemblages along the mangrove-seagrass-coral reef continuum in Wenchang, China. Acta Oceanol. Sin. 2019, 39, 43–52. [Google Scholar] [CrossRef]
- Liao, J.; Zhen, J.; Li, Z.; Metternicht, G. Understanding dynamics of mangrove forest on protected areas of Hainan Island, China: 30 years of evidence from remote sensing. Sustainability 2019, 11, 5356. [Google Scholar] [CrossRef]
- Chen, S.; Wang, D.; Wu, Z.; Zhang, G.; Li, Y.; Tu, Z.; Yao, H.; Cai, Z. Discussion of the change trend of the seagrass meadows in the east coast of Hainan Island in nearly a decade. Mar. Environ. Sci. 2015, 34, 4853. [Google Scholar]
- Yang, C.; Xue, Y.; Su, S.; Wang, X.; Lin, Z.; Chen, C. The survey on plant community characteristics of Bamenwan Mangroves in Wenchang City. Chin. J. Trop. Agric. 2017, 37, 4852. [Google Scholar]
- Tu, Z.; Wu, R.; Zhang, G.; Lan, J.; Chen, X.; Wang, D. Types and characteristics of mangrove plants of Qinglan Harbour in Hainan Island. Chin. J. Trop. Agric. 2015, 35, 21–25. [Google Scholar]
- Froese, R.; Pauly, D. Search FishBase. Available online: https://www.fishbase.de/ (accessed on 1 January 2021).
- Miya, M.; Sato, Y.; Fukunaga, T.; Sado, T.; Poulsen, J.Y.; Sato, K.; Minamoto, T.; Yamamoto, S.; Yamanaka, H.; Araki, H.; et al. MiFish, a set of universal PCR primers for metabarcoding environmental DNA from fishes: Detection of more than 230 subtropical marine species. R. Soc. Open Sci. 2015, 2, 150088. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Zhao, J.; Yao, M. A Comprehensive and comparative evaluation of primers for metabarcoding eDNA from Fish. Methods Ecol. Evol. 2020, 11, 1609–1625. [Google Scholar] [CrossRef]
- Polanco, F.A.; Richards, E.; Flück, B.; Valentini, A.; Altermatt, F.; Brosse, S.; Walser, J.; Eme, D.; Marques, V.; Manel, S.; et al. Comparing the performance of 12S mitochondrial primers for fish environmental DNA across ecosystems. Environ. DNA 2021, 3, 1113–1127. [Google Scholar] [CrossRef]
- Zhang, J.; Kobert, K.; Flouri, T.; Stamatakis, A. PEAR: A fast and accurate Illumina Paired-End reAd mergeR. Bioinformatics 2014, 30, 614–620. [Google Scholar] [CrossRef]
- Edgar, R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010, 26, 2460–2461. [Google Scholar] [CrossRef]
- Du, J.; Liu, X.; Zheng, X.; Hu, W. The Atlas of Common Coral Reef Organisms and Their Distribution in the Xisha Islands; Science Press: Beijing, China, 2022. [Google Scholar]
- Lai, T.; He, B. Marine Bony Fishes in Beibu Gulf of Guangxi; Science Press: Beijing, China, 2016. [Google Scholar]
- Hsieh, T.C.; Ma, K.H.; Chao, A. INEXT: An R Package for rarefaction and extrapolation of species diversity (Hill Numbers). Methods Ecol. Evol. 2016, 7, 1451–1456. [Google Scholar] [CrossRef]
- Clarke, K.R. Non-parametric multivariate analyses of changes in community structure. Aust. J. Ecol. 1993, 18, 117–143. [Google Scholar] [CrossRef]
- Dixon, P. VEGAN, a Package of R functions for community ecology. J. Veg. Sci. 2003, 14, 927–930. [Google Scholar] [CrossRef]
- Magneville, C.; Loiseau, N.; Albouy, C.; Casajus, N.; Claverie, T.; Escalas, A.; Leprieur, F.; Maire, E.; Mouillot, D.; Villéger, S. mFD: An R Package to compute and illustrate the multiple facets of functional diversity. Ecography 2022, 2022. [Google Scholar] [CrossRef]
- Wickham, H. Ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016; Available online: https://ggplot2.tidyverse.org (accessed on 1 January 2021).
- Shao, K. Taiwan Fish Database. 2023. Available online: https://fishdb.sinica.edu.tw/eng/home.php (accessed on 1 January 2021).
- Pierce, C.L.; Corcoran, A.M.; Gronbach, A.N.; Hsia, S.; Mullarkey, B.J.; Schwartzhoff, A.J. Influence of diel period on electrofishing and beach seining assessments of littoral fish assemblages. N. Am. J. Fish. Manag. 2001, 21, 918–926. [Google Scholar] [CrossRef]
- Prchalová, M.; Kubečka, J.; Říha, M.; Litvín, R.; Čech, M.; Frouzová, J.; Hladík, M.; Hohausová, E.; Peterka, J.; Vašek, M. Overestimation of Percid fishes (Percidae) in gillnet sampling. Fish. Res. 2008, 91, 79–87. [Google Scholar] [CrossRef]
- Andres, K.J.; Lambert, T.D.; Lodge, D.M.; Andrés, J.; Jackson, J.R. Combining sampling gear to optimally inventory species highlights the efficiency of eDNA metabarcoding. Environ. DNA 2022, 5, 146–157. [Google Scholar] [CrossRef]
- Beng, K.C.; Corlett, R.T. Applications of environmental DNA (eDNA) in ecology and conservation: Opportunities, challenges and prospects. Biodivers. Conserv. 2020, 29, 2089–2121. [Google Scholar] [CrossRef]
- Kelly, R.P.; Shelton, A.O.; Gallego, R. Understanding PCR processes to draw meaningful conclusions from environmental DNA Studies. Sci. Rep. 2019, 9, 12133. [Google Scholar] [CrossRef]
- Andruszkiewicz, E.A.; Starks, H.A.; Chavez, F.P.; Sassoubre, L.M.; Block, B.A.; Boehm, A.B. Biomonitoring of marine vertebrates in Monterey Bay using eDNA metabarcoding. PLoS ONE 2017, 12, e0176343. [Google Scholar] [CrossRef]
- Sakata, M.K.; Yamamoto, S.; Gotoh, R.O.; Miya, M.; Yamanaka, H.; Minamoto, T. Sedimentary EDNA Provides different information on timescale and fish species composition compared with aqueous eDNA. Environ. DNA 2020, 2, 505–518. [Google Scholar] [CrossRef]
- Honda, K.; Nakamura, Y.; Nakaoka, M.; Uy, W.H.; Fortes, M.D. Habitat use by fishes in coral reefs, seagrass meadows and mangrove habitats in the Philippines. PLoS ONE 2013, 8, e65735. [Google Scholar] [CrossRef] [PubMed]
- Aguilar-Perera, A.; Appeldoorn, R.S. Spatial distribution of marine fishes along a cross-shelf gradient containing a continuum of mangrove–seagrass–coral reefs off southwestern Puerto Rico. Estuar. Coast. Shelf Sci. 2008, 76, 378–394. [Google Scholar] [CrossRef]
- Yamanaka, H.; Minamoto, T. The use of environmental DNA of fishes as an efficient method of determining habitat connectivity. Ecol. Indic. 2016, 62, 147–153. [Google Scholar] [CrossRef]
- Xie, R.; Zhao, G.; Yang, J.; Wang, Z.; Xu, Y.; Zhang, X.; Wang, Z. eDNA metabarcoding revealed differential structures of aquatic communities in a dynamic freshwater ecosystem shaped by habitat heterogeneity. Environ. Res. 2021, 201, 111602. [Google Scholar] [CrossRef]
- Andruszkiewicz, E.A.; Koseff, J.R.; Fringer, O.B.; Ouellette, N.T.; Lowe, A.B.; Edwards, C.A.; Boehm, A.B. Modeling environmental DNA transport in the coastal ocean using Lagrangian particle tracking. Front. Mar. Sci. 2019, 6, 477. [Google Scholar] [CrossRef]
- Xing, Y.; Gao, W.; Shen, Z.; Zhang, Y.; Bai, J.; Cai, X.; Ouyang, J.; Zhao, Y. A Review of environmental DNA Field and laboratory protocols applied in fish ecology and environmental Health. Front. Environ. Sci. 2022, 10, 73. [Google Scholar] [CrossRef]
- Jo, T.; Minamoto, T. Complex interactions between environmental DNA (eDNA) state and water chemistries on eDNA persistence suggested by meta-analyses. Mol. Ecol. Resour. 2021, 21, 1490–1503. [Google Scholar] [CrossRef]
- Yates, M.C.; Derry, A.M.; Cristescu, M.E. Environmental RNA: A revolution in ecological resolution? Trends Ecol. Evol. 2021, 36, 601–609. [Google Scholar] [CrossRef]
- Ruppert, K.M.; Kline, R.J.; Rahman, M.S. Past, Present, and future perspectives of environmental DNA (eDNA) metabarcoding: A systematic review in methods, monitoring, and applications of global eDNA. Glob. Ecol. Conserv. 2019, 17, e00547. [Google Scholar] [CrossRef]
- Sassoubre, L.M.; Yamahara, K.M.; Gardner, L.D.; Block, B.A.; Boehm, A.B. Quantification of environmental DNA (eDNA) shedding and decay rates for three marine fish. Environ. Sci. Technol. 2016, 50, 10456–10464. [Google Scholar] [CrossRef]
- DiBattista, J.D.; Coker, D.J.; Sinclair-Taylor, T.H.; Stat, M.; Berumen, M.L.; Bunce, M. Assessing the utility of eDNA as a tool to survey reef-fish communities in the Red Sea. Coral Reefs 2017, 36, 1245–1252. [Google Scholar] [CrossRef]
- Bista, I.; Carvalho, G.R.; Walsh, K.; Seymour, M.; Hajibabaei, M.; Lallias, D.; Christmas, M.; Creer, S. Annual time-series analysis of aqueous eDNA reveals ecologically relevant dynamics of lake ecosystem biodiversity. Nat. Commun. 2017, 8, 14087. [Google Scholar] [CrossRef] [PubMed]
- Deiner, K.; Bik, H.M.; Mächler, E.; Seymour, M.; Lacoursière-Roussel, A.; Altermatt, F.; Creer, S.; Bista, I.; Lodge, D.M.; Vere, N.; et al. Environmental DNA metabarcoding: Transforming how we survey animal and plant communities. Mol. Ecol. 2017, 26, 5872–5895. [Google Scholar] [CrossRef] [PubMed]
- Porter, T.M.; Hajibabaei, M. Scaling Up: A guide to high-throughput genomic approaches for biodiversity analysis. Mol. Ecol. 2018, 27, 313–338. [Google Scholar] [CrossRef] [PubMed]
- Fediajevaite, J.; Priestley, V.; Arnold, R.; Savolainen, V. Meta-analysis shows that environmental DNA outperforms traditional surveys, but warrants better reporting standards. Ecol. Evol. 2021, 11, 4803–4815. [Google Scholar] [CrossRef] [PubMed]
| Source | Df | Pseudo-F | p |
|---|---|---|---|
| Habitat (eDNA) | 2 | 0.25972 | 0.001 |
| Habitat (gillnet) | 2 | 4.8562 | 0.001 |
| Method | 1 | 0.06979 | 0.001 |
| Method | Habitats | Genera that Contributed the Greatest Dissimilarity | ||
|---|---|---|---|---|
| eDNA & gillnet | Plotosus 70.3% | Triacanthus 67.8% | Leiognathus 65.0% | |
| eDNA | Mangrove & Seagrass | Capoeta 71.7% | Pelates 68.5% | Mugil 64.7% |
| Seagrass & Coral Reef | Plotosus 72.0% | Capoeta 69.5% | Triacanthus 66.6% | |
| Mangrove & Coral Reef | Capoeta 72.2% | Plotosus 69.5% | Terapon 66.6% | |
| gillnet | Mangrove & Seagrass | Terapon 74.4% | Gerres 67.6% | Eubleekeria 58.8% |
| Seagrass & Coral Reef | Terapon 70.1% | Eubleekeria 64.2% | Lutjanus 58.9% | |
| Mangrove & Coral Reef | Plotosus 73.6% | Terapon 69.7% | Pelates 65.1% | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qiu, S.; Ooi, J.L.S.; Chen, W.; Poong, S.-W.; Zhang, H.; He, W.; Su, S.; Luo, H.; Hu, W.; Affendi, Y.A.; et al. Heterogeneity of Fish Taxonomic and Functional Diversity Evaluated by eDNA and Gillnet along a Mangrove–Seagrass–Coral Reef Continuum. Animals 2023, 13, 1777. https://doi.org/10.3390/ani13111777
Qiu S, Ooi JLS, Chen W, Poong S-W, Zhang H, He W, Su S, Luo H, Hu W, Affendi YA, et al. Heterogeneity of Fish Taxonomic and Functional Diversity Evaluated by eDNA and Gillnet along a Mangrove–Seagrass–Coral Reef Continuum. Animals. 2023; 13(11):1777. https://doi.org/10.3390/ani13111777
Chicago/Turabian StyleQiu, Shuting, Jillian Lean Sim Ooi, Weilin Chen, Sze-Wan Poong, Han Zhang, Weiyi He, Shangke Su, Hao Luo, Wenjia Hu, Yang Amri Affendi, and et al. 2023. "Heterogeneity of Fish Taxonomic and Functional Diversity Evaluated by eDNA and Gillnet along a Mangrove–Seagrass–Coral Reef Continuum" Animals 13, no. 11: 1777. https://doi.org/10.3390/ani13111777
APA StyleQiu, S., Ooi, J. L. S., Chen, W., Poong, S.-W., Zhang, H., He, W., Su, S., Luo, H., Hu, W., Affendi, Y. A., Du, J., & Loh, K.-H. (2023). Heterogeneity of Fish Taxonomic and Functional Diversity Evaluated by eDNA and Gillnet along a Mangrove–Seagrass–Coral Reef Continuum. Animals, 13(11), 1777. https://doi.org/10.3390/ani13111777





