Dynamic Profile of the Yak Mammary Transcriptome during the Lactation Cycle
Abstract
Simple Summary
Abstract
1. Introduction
2. Material and Methods
2.1. Milk Yield Measurement
2.2. Animals, Experimental Design, and Diet
2.3. Mammary Gland Collection and Preparation
2.4. Microarray
2.4.1. Hybridization of DNA Microarray and RNA Extraction
2.4.2. RNA Amplification
2.4.3. Labelling and Hybridization
2.5. Validation of Microarray Data with RTqPCR
2.6. Bioinformatics Analyses
2.7. Statistical Analyses
3. Results
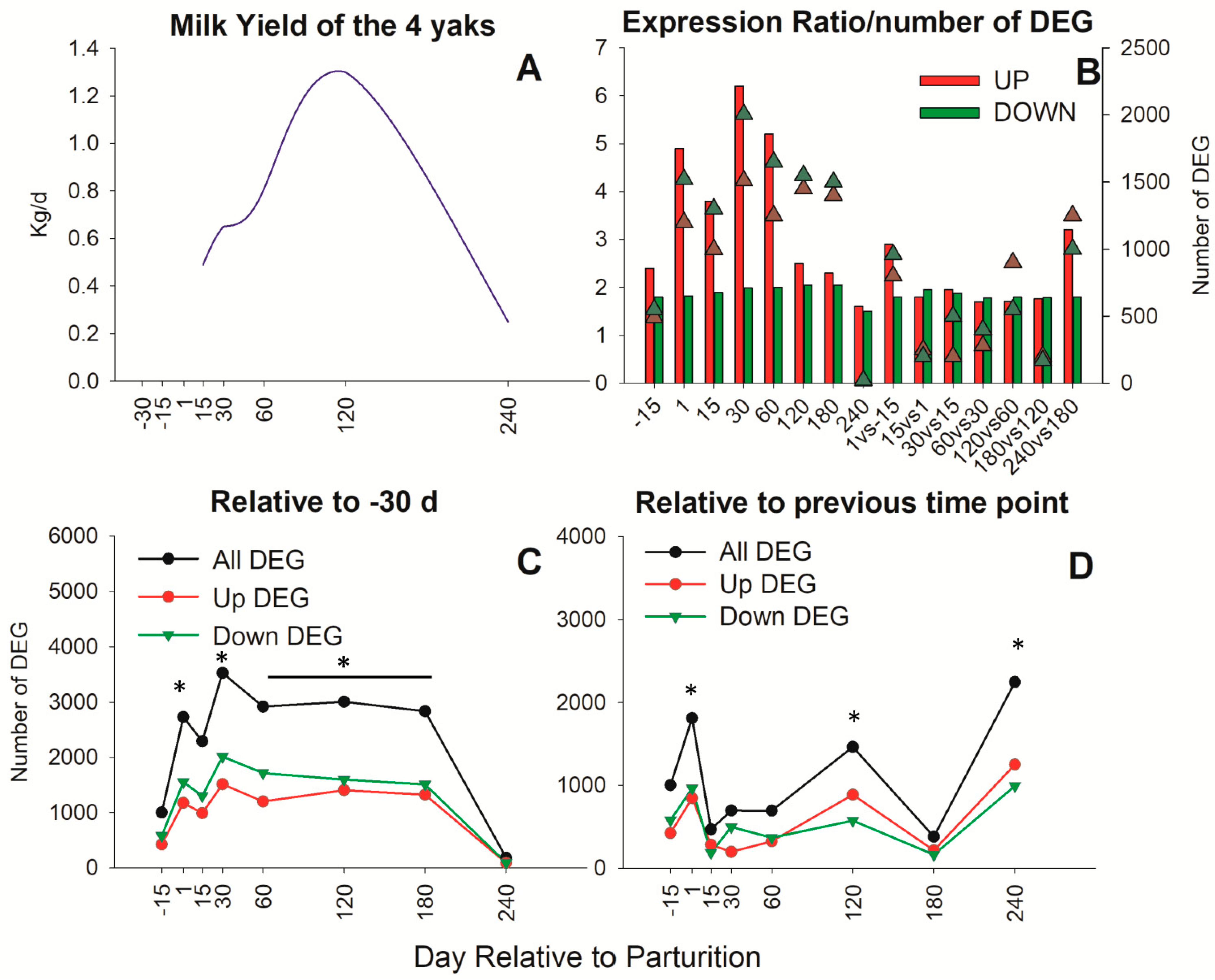
3.1. Overall Transcriptome Expression Profile in Yak Mammary Tissue
3.2. Impact of Stage of Lactation on Chromosomes
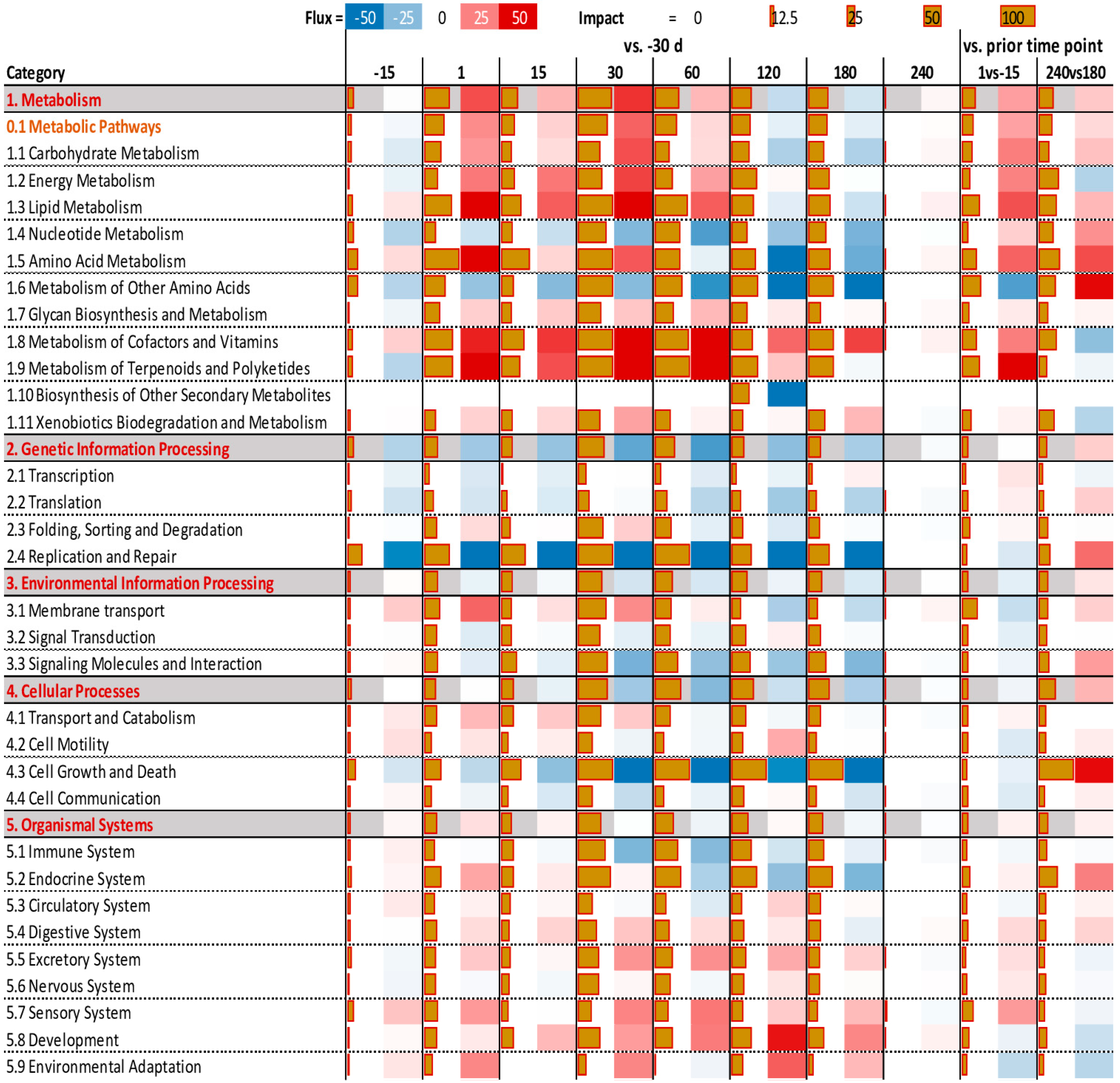
3.3. Functional Analysis of DEG Revealed by DIA
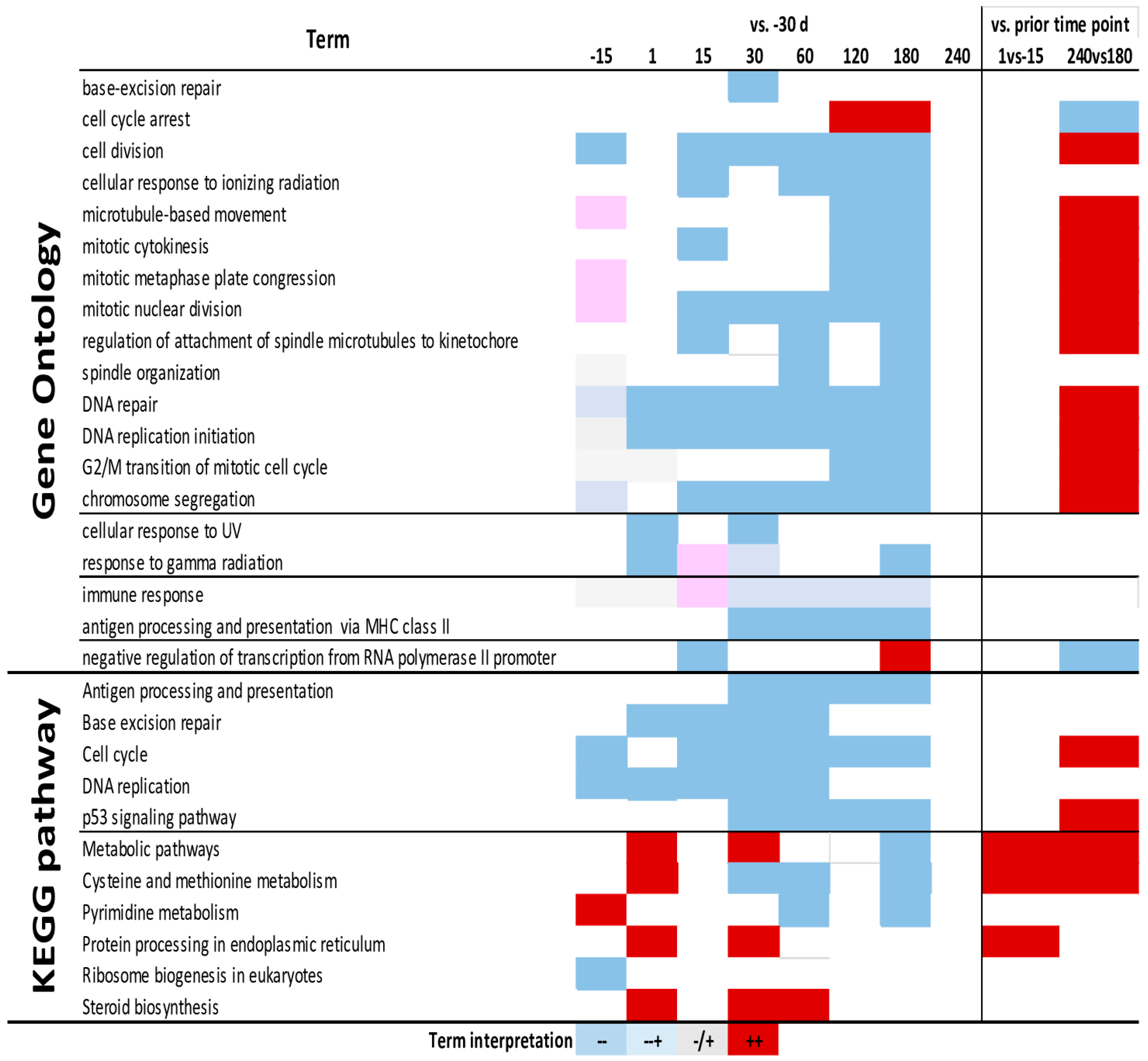
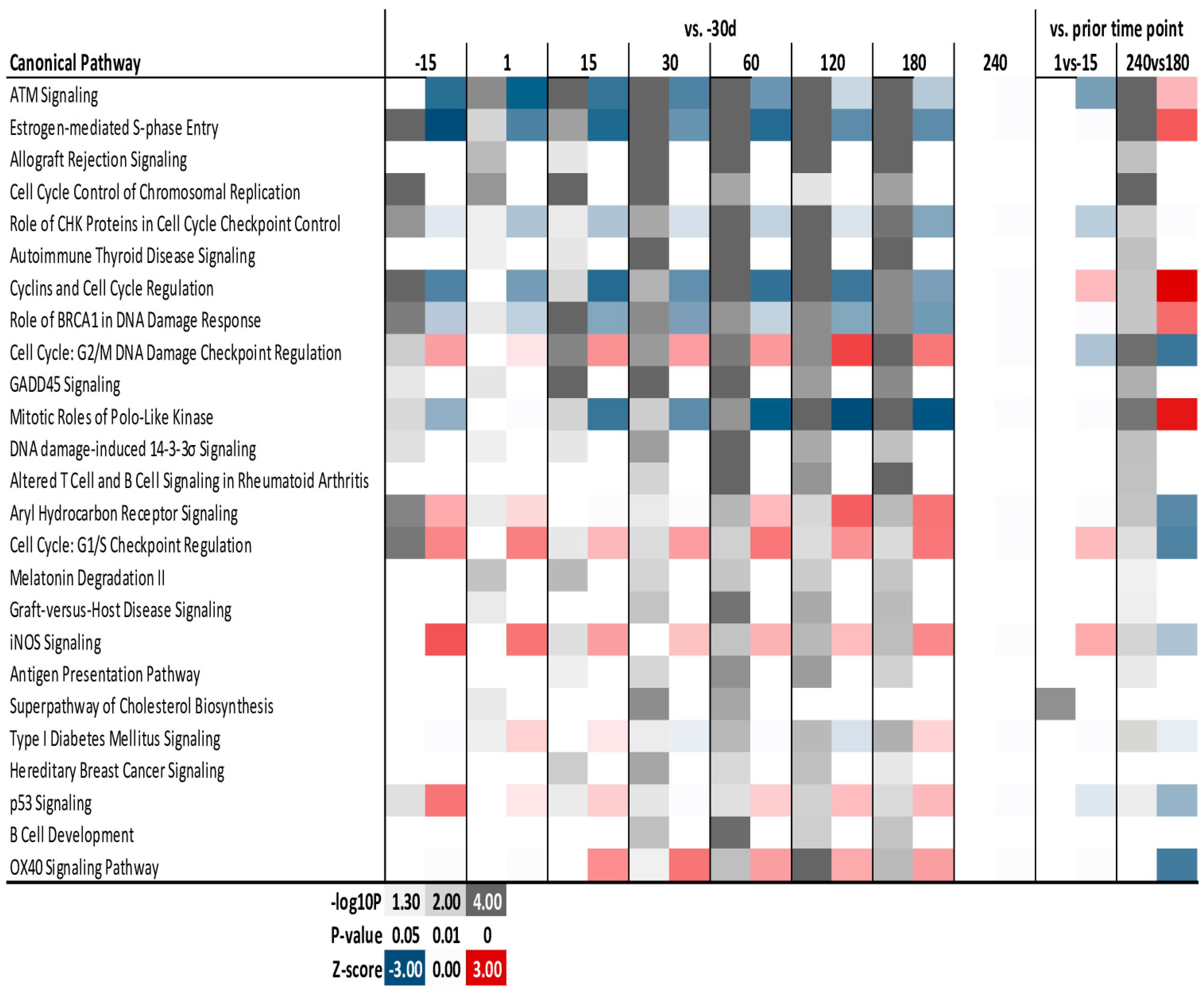
3.4. Enrichment Analysis
4. Discussion
4.1. Impact of Lactation on Yak Chromosome
4.2. The Onset and End of Lactation in Yak Mammary Gland
4.3. Lipid Metabolism Is Induced in Mammary Tissue of Yak during the Lactation Cycle
4.4. Amino Acid Metabolism
4.5. Glycan Biosynthesis and Metabolism
4.6. Cell Cycle
4.7. Immune Response System
4.8. Adaptation to Low Oxygen and High Radiation Environment
4.9. Limitations
5. Summary and Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Harris, R.B.; Pletscher, D.H.; Loggers, C.O.; Miller, D.J. Status and trends of Tibetan plateau mammalian fauna, Yeniugou, China. Biol. Conserv. 1999, 87, 13–19. [Google Scholar] [CrossRef]
- Zhang, J.; Yang, M.; Cai, D.; Hao, Y.; Zhao, X.; Zhu, Y.; Zhu, H.; Yang, Z. Composition, coagulation characteristics, and cheese making capacity of yak milk. J. Dairy Sci. 2020, 103, 1276–1288. [Google Scholar] [CrossRef]
- Bionaz, M.; Periasamy, K.; Rodriguez-Zas, S.L.; Everts, R.E.; Lewin, H.A.; Hurley, W.L.; Loor, J.J. Old and new stories: Revelations from functional analysis of the bovine mammary transcriptome during the lactation cycle. PLoS ONE 2012, 7, e33268. [Google Scholar] [CrossRef]
- Maningat, P.D.; Sen, P.; Rijnkels, M.; Sunehag, A.L.; Hadsell, D.L.; Bray, M.; Haymond, M.W. Gene expression in the human mammary epithelium during lactation: The milk fat globule transcriptome. Physiol. Genom. 2009, 37, 12–22. [Google Scholar] [CrossRef] [PubMed]
- Rudolph, M.C.; McManaman, J.L.; Hunter, L.; Phang, T.; Neville, M.C. Functional development of the mammary gland: Use of expression profiling and trajectory clustering to reveal changes in gene expression during pregnancy, lactation, and involution. J. Mammary Gland. Biol. Neoplasia 2003, 8, 287–307. [Google Scholar] [CrossRef] [PubMed]
- Bauman, D.E.; Mather, I.H.; Wall, R.J.; Lock, A.L. Major advances associated with the biosynthesis of milk. J. Dairy Sci. 2006, 89, 1235–1243. [Google Scholar] [CrossRef]
- Osorio, J.S.; Lohakare, J.; Bionaz, M. Biosynthesis of milk fat, protein, and lactose: Roles of transcriptional and posttranscriptional regulation. Physiol. Genom. 2016, 48, 231–256. [Google Scholar] [CrossRef]
- Loor, J.J.; Cohick, W.S. ASAS centennial paper: Lactation biology for the twenty-first century. J. Anim. Sci. 2009, 87, 813–824. [Google Scholar] [CrossRef]
- Sun, H.; Miao, Z.; Zhang, X.; Chan, U.I.; Su, S.M.; Guo, S.; Wong, C.K.H.; Xu, X.; Deng, C.-X. Single-cell RNA-Seq reveals cell heterogeneity and hierarchy within mouse mammary epithelia. J. Biol. Chem. 2018, 293, 8315–8329. [Google Scholar] [CrossRef]
- Dado-Senn, B.; Skibiel, A.L.; Fabris, T.F.; Zhang, Y.; Dahl, G.E.; Peñagaricano, F.; Laporta, J. RNA-Seq reveals novel genes and pathways involved in bovine mammary involution during the dry period and under environmental heat stress. Sci. Rep. 2018, 8, 11096. [Google Scholar] [CrossRef]
- Wang, Z.; Gerstein, M.; Snyder, M. RNA-Seq: A revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009, 10, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Su, Z.; Fang, H.; Hong, H.; Shi, L.; Zhang, W.; Zhang, W.; Zhang, Y.; Dong, Z.; Lancashire, L.J.; Bessarabova, M. An investigation of biomarkers derived from legacy microarray data for their utility in the RNA-seq era. Genome Biol. 2014, 15, 523. [Google Scholar] [CrossRef] [PubMed]
- Mantione, K.J.; Kream, R.M.; Kuzelova, H.; Ptacek, R.; Raboch, J.; Samuel, J.M.; Stefano, G.B. Comparing bioinformatic gene expression profiling methods: Microarray and RNA-Seq. Med. Sci. Monit. Basic Res. 2014, 20, 138–141. [Google Scholar]
- Fu, M.; Chen, Y.; Xiong, X.; Lan, D.; Li, J. Establishment of mammary gland model in vitro: Culture and evaluation of a yak mammary epithelial cell line. PLoS ONE 2014, 9, e113669. [Google Scholar] [CrossRef] [PubMed]
- Jaswal, S.; Anand, V.; Ali, S.A.; Jena, M.K.; Kumar, S.; Kaushik, J.K.; Mohanty, A.K. TMT based deep proteome analysis of buffalo mammary epithelial cells and identification of novel protein signatures during lactogenic differentiation. FASEB J. 2021, 35, e21621. [Google Scholar] [CrossRef]
- Yuan, M.; Xia, W.; Zhang, X.; Liu, Y.; Jiang, M. Identification and verification of differentially expressed genes in yak mammary tissue during the lactation cycle. J. Dairy Res. 2020, 87, 158–165. [Google Scholar] [CrossRef]
- Xia, W.; Osorio, J.S.; Yang, Y.; Liu, D.; Jiang, M.F. Characterization of gene expression profiles related to yak milk protein synthesis during the lactation cycle. J. Dairy Sci. 2018, 101, 11150–11158. [Google Scholar] [CrossRef]
- Wu, X.; Zhou, X.; Xiong, L.; Pei, J.; Yao, X.; Liang, C.; Bao, P.; Chu, M.; Guo, X.; Yan, P. Transcriptome Analysis Reveals the Potential Role of Long Non-coding RNAs in Mammary Gland of Yak During Lactation and Dry Period. Front. Cell Dev. Biol. 2020, 8, 1452. [Google Scholar] [CrossRef]
- Shi, W.; Yuan, X.; Cui, K.; Li, H.; Fu, P.; Rehman, S.-U.; Shi, D.; Liu, Q.; Li, Z. LC-MS/MS Based Metabolomics Reveal Candidate Biomarkers and Metabolic Changes in Different Buffalo Species. Animals 2021, 11, 560. [Google Scholar] [CrossRef]
- Bionaz, M.; Hurley, W.; Loor, J. Milk protein synthesis in the lactating mammary gland: Insights from transcriptomics analyses. Milk Protein 2012, 11, 285–324. [Google Scholar]
- Lemay, D.G.; Neville, M.C.; Rudolph, M.C.; Pollard, K.S.; German, J.B. Gene regulatory networks in lactation: Identification of global principles using bioinformatics. BMC Syst. Biol. 2007, 1, 56. [Google Scholar] [CrossRef]
- Paul, M.; Poyan Mehr, A.; Kreutz, R. Physiology of local renin-angiotensin systems. Physiol. Rev. 2006, 86, 747–803. [Google Scholar] [CrossRef]
- Jiang, M.; Lee, J.N.; Bionaz, M.; Deng, X.Y.; Wang, Y. Evaluation of suitable internal control genes for RT-qPCR in yak mammary tissue during the lactation cycle. PLoS ONE 2016, 11, e0147705. [Google Scholar] [CrossRef]
- Lee, J.N.; Wang, Y.; Xu, Y.O.; Li, Y.C.; Tian, F.; Jiang, M.F. Characterisation of gene expression related to milk fat synthesis in the mammary tissue of lactating yaks. J. Dairy Res. 2017, 84, 283–288. [Google Scholar] [CrossRef]
- Bionaz, M.; Periasamy, K.; Rodriguez-Zas, S.L.; Hurley, W.L.; Loor, J.J. A Novel Dynamic Impact Approach (DIA) for Functional Analysis of Time-Course Omics Studies: Validation Using the Bovine Mammary Transcriptome. PLoS ONE 2012, 7, e32455. [Google Scholar] [CrossRef]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B (Methodol.) 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Khatkar, M.S.; Thomson, P.C.; Tammen, I.; Raadsma, H.W. Quantitative trait loci mapping in dairy cattle: Review and meta-analysis. Genet. Sel. Evol. 2004, 36, 163–190. [Google Scholar] [CrossRef]
- Tal-Stein, R.; Fontanesi, L.; Dolezal, M.; Scotti, E.; Bagnato, A.; Russo, V.; Canavesi, F.; Friedmann, A.; Soller, M.; Lipkin, E. A genome scan for quantitative trait loci affecting milk somatic cell score in Israeli and Italian Holstein cows by means of selective DNA pooling with single- and multiple-marker mapping. J. Dairy Sci. 2010, 93, 4913–4927. [Google Scholar] [CrossRef] [PubMed]
- Ron, M.; Kliger, D.; Feldmesser, E.; Seroussi, E.; Ezra, E.; Weller, J.I. Multiple quantitative trait locus analysis of bovine chromosome 6 in the Israeli Holstein population by a daughter design. Genetics 2001, 159, 727. [Google Scholar] [CrossRef]
- Freyer, G.; Sorensen, P.; Kuehn, C.; Weikard, R.; Hoeschele, I. Search for pleiotropic QTL on chromosome BTA6 affecting yield traits of milk production. J. Dairy Sci. 2003, 86, 999–1008. [Google Scholar] [CrossRef]
- Silva, A.A.; Azevedo, A.L.S.; Verneque, R.S.; Gasparini, K.; Peixoto, M.G.C.D.; Silva, M.V.G.B.D.; Lopes, P.S.; Guimarães, S.E.F.; Machado, M.A. Quantitative trait loci affecting milk production traits on bovine chromosome 6 in zebuine Gyr breed. J. Dairy Sci. 2011, 94, 971–980. [Google Scholar] [CrossRef] [PubMed]
- Sheehy, P.A.; Riley, L.G.; Raadsma, H.W.; Williamson, P.; Wynn, P.C. A functional genomics approach to evaluate candidate genes located in a QTL interval for milk production traits on BTA6. Anim. Genet. 2010, 40, 492–498. [Google Scholar] [CrossRef]
- Cohen-Zinder, M.; Seroussi, E.; Larkin, D.M.; Loor, J.J.; Everts-Van Der Wind, A.; Lee, J.-H.; Drackley, J.K.; Band, M.R.; Hernandez, A.; Shani, M. Identification of a missense mutation in the bovine ABCG2 gene with a major effect on the QTL on chromosome 6 affecting milk yield and composition in Holstein cattle. Genome Res. 2005, 15, 936–944. [Google Scholar] [CrossRef] [PubMed]
- Coppieters, W.; Riquet, J.; Arranz, J.J.; Berzi, P.; Cambisano, N.; Grisart, B.; Karim, L.; Marcq, F.; Moreau, L.; Nezer, C. A QTL with major effect on milk yield and composition maps to bovine Chromosome 14. Mamm. Genome 1998, 9, 540–544. [Google Scholar] [CrossRef]
- Wibowo, T.A.; Gaskins, C.T.; Newberry, R.C.; Thorgaard, G.H.; Jiang, Z. Genome Assembly Anchored QTL Map of Bovine Chromosome 14. Int. J. Biol. Sci. 2008, 4, 406–414. [Google Scholar] [CrossRef]
- Grisart, B.; Coppieters, W.; Farnir, F.; Karim, L.; Ford, C.; Berzi, P.; Cambisano, N.; Mni, M.; Reid, S.; Simon, P.; et al. Positional Candidate Cloning of a QTL in Dairy Cattle: Identification of a Missense Mutation in the Bovine DGAT1 Gene with Major Effect on Milk Yield and Composition. Genome Res. 2002, 12, 222–231. [Google Scholar] [CrossRef]
- Bionaz, M.; Loor, J.J. Gene networks driving bovine milk fat synthesis during the lactation cycle. BMC Genom. 2008, 9, 366–387. [Google Scholar] [CrossRef]
- Loor, J.J.; Bionaz, M.; Invernizzi, G. Systems biology and animal nutrition: Insights from the dairy cow during growth and the lactation cycle. Syst. Biol. Livest. Sci. 2011, 19, 24. [Google Scholar]
- Watson, C.J. Key stages in mammary gland development-Involution: Apoptosis and tissue remodelling that convert the mammary gland from milk factory to a quiescent organ. Breast Cancer Res. 2006, 8, 203. [Google Scholar] [CrossRef]
- Stein, T.; Salomonis, N.; Gusterson, B.A. Mammary gland involution as a multi-step process. J. Mammary Gland. Biol. Neoplasia 2007, 12, 25–35. [Google Scholar] [CrossRef] [PubMed]
- Wesselink, E.; Koekkoek, W.; Grefte, S.; Witkamp, R.; van Zanten, A. Feeding mitochondria: Potential role of nutritional components to improve critical illness convalescence. Clin. Nutr. 2019, 38, 982–995. [Google Scholar] [CrossRef]
- Murray, A.J.; Horscroft, J.A. Mitochondrial function at extreme high altitude. J. Physiol. 2016, 594, 1137–1149. [Google Scholar] [CrossRef]
- Long, L.; Zhu, Y.; Li, Z.; Zhang, H.; Liu, L.; Bai, J. Differential expression of skeletal muscle mitochondrial proteins in yak, dzo, and cattle: A proteomics-based study. J. Vet. Med. Sci. 2020, 82, 1178–1186. [Google Scholar] [CrossRef]
- An, X.; Song, Y.; Hou, J.; Han, P.; Peng, J.; Zhang, L.; Wang, J.; Cao, B. Mutations in the MTHFR gene and their associations with milk production traits in dairy goats. Small Rumin. Res. 2015, 130, 76–80. [Google Scholar] [CrossRef]
- Yang, J.; Jin, Z.-B.; Chen, J.; Huang, X.-F.; Li, X.-M.; Liang, Y.-B.; Mao, J.-Y.; Chen, X.; Zheng, Z.; Bakshi, A. Genetic signatures of high-altitude adaptation in Tibetans. Proc. Natl. Acad. Sci. USA 2017, 114, 4189–4194. [Google Scholar] [CrossRef]
- Rudolph, M.C.; McManaman, J.L.; Phang, T.; Russell, T.; Kominsky, D.J.; Serkova, N.J.; Stein, T.; Anderson, S.M.; Neville, M.C. Metabolic regulation in the lactating mammary gland: A lipid synthesizing machine. Physiol. Genom. 2007, 28, 323–336. [Google Scholar] [CrossRef] [PubMed]
- Palmquist, D.; Davis, C.; Brown, R.; Sachan, D. Availability and metabolism of various substrates in ruminants. V. Entry rate into the body and incorporation into milk fat of d (−) β-hydroxybutyrate. J. Dairy Sci. 1969, 52, 633–638. [Google Scholar] [CrossRef]
- Kinsella, J. Stearic acid metabolism by mammary cells. J. Dairy Sci. 1970, 53, 1757–1765. [Google Scholar] [CrossRef]
- Yadav, P.; Kumar, P.; Mukesh, M.; Kataria, R.; Yadav, A.; Mohanty, A.; Mishra, B. Kinetics of lipogenic genes expression in milk purified mammary epithelial cells (MEC) across lactation and their correlation with milk and fat yield in buffalo. Res. Vet. Sci. 2015, 99, 129–136. [Google Scholar] [CrossRef]
- Bartley, J.C. Modulation by ketone bodies of the rate of fatty acid synthesis in mammary gland slices from lactating rats. Lipids 1976, 11, 774–777. [Google Scholar] [CrossRef] [PubMed]
- Viturro, E.; Koenning, M.; Kroemer, A.; Schlamberger, G.; Wiedemann, S.; Kaske, M.; Meyer, H.H. Cholesterol synthesis in the lactating cow: Induced expression of candidate genes. J. Steroid Biochem. Mol. Biol. 2009, 115, 62–67. [Google Scholar] [CrossRef] [PubMed]
- Ontsouka, E.C.; Albrecht, C. Cholesterol transport and regulation in the mammary gland. J. Mammary Gland. Biol. Neoplasia 2014, 19, 43–58. [Google Scholar] [CrossRef]
- Renterghem, R.V. The riboflavin (vitamin B2) content of milk. Arch. Belg. 1984, 42, 339–348. [Google Scholar]
- Stincone, A.; Prigione, A.; Cramer, T.; Wamelink, M.M.; Campbell, K.; Cheung, E.; Olin-Sandoval, V.; Grüning, N.M.; Krüger, A.; Tauqeer Alam, M. The return of metabolism: Biochemistry and physiology of the pentose phosphate pathway. Biol. Rev. 2015, 90, 927–963. [Google Scholar] [CrossRef]
- Mashek, D.G.; Coleman, R.A. Cellular fatty acid uptake: The contribution of metabolism. Curr. Opin. Lipidol. 2006, 17, 274–278. [Google Scholar] [CrossRef] [PubMed]
- Khodamoradi, S.; Fatahnia, F.; Taherpour, K.; Pirani, V.; Azarfar, A. Effect of monensin and vitamin E on milk production and composition of lactating dairy cows. J. Anim. Physiol. Anim. Nutr. 2013, 97, 666–674. [Google Scholar] [CrossRef]
- Piamphon, N.; Wachirapakorn, C.; Wanapat, M.; Navanukraw, C. Effects of Protected Conjugated Linoleic Acid Supplementation on Milk Fatty Acid in Dairy Cows. Asian Australas. J. Anim. Sci. 2009, 22, 49–56. [Google Scholar] [CrossRef]
- Khatri, P.; Sirota, M.; Butte, A.J. Ten years of pathway analysis: Current approaches and outstanding challenges. PLoS Comput. Biol. 2012, 8, e1002375. [Google Scholar] [CrossRef] [PubMed]
- Bionaz, M.; Osorio, J.; Loor, J. TRIENNIAL LACTATION SYMPOSIUM: Nutrigenomics in dairy cows: Nutrients, transcription factors, and techniques. J. Anim. Sci. 2015, 93, 5531–5553. [Google Scholar] [CrossRef]
- Takimori, S.; Shimaoka, H.; Furukawa, J.I.; Yamashita, T.; Amano, M.; Fujitani, N.; Takegawa, Y.; Hammarström, L.; Kacskovics, I.; Shinohara, Y. Alteration of the N-glycome of bovine milk glycoproteins during early lactation. Febs J. 2011, 278, 3769–3781. [Google Scholar] [CrossRef]
- Tao, N.; Depeters, E.J.; Freeman, S.; German, J.B.; Grimm, R.; Lebrilla, C.B. Bovine Milk Glycome. J. Dairy Sci. 2008, 91, 3768–3778. [Google Scholar] [CrossRef]
- Robert, K.Y.; Schengrund, C.-L. Glycobiology of the Nervous System; Springer: Berlin/Heidelberg, Germany, 2014; Volume 9. [Google Scholar]
- Spiro, R.G. Protein glycosylation: Nature, distribution, enzymatic formation, and disease implications of glycopeptide bonds. Glycobiology 2002, 12, 43R–56R. [Google Scholar] [CrossRef] [PubMed]
- Merlo, G.R.; Basolo, F.; Fiore, L.; Hynes, D.N.E. p53-Dependent and p53-Independent Activation of Apoptosis in Mammary Epithelial Cells Reveals a Survival Function of EGF and Insulin. J. Cell Biol. 1995, 128, 1185–1196. [Google Scholar] [CrossRef] [PubMed]
- Brew, C.T.; Aronchik, I.; Hsu, J.C.; Sheen, J.H.; Dickson, R.B.; Bjeldanes, L.F.; Firestone, G.L. Indole-3-carbinol activates the ATM signaling pathway independent of DNA damage to stabilize p53 and induce G1 arrest of human mammary epithelial cells. Int. J. Cancer 2010, 118, 857–868. [Google Scholar] [CrossRef] [PubMed]
- Capuco, A.V.; Wood, D.L.; Baldwin, R. Mammary cell number, proliferation, and apoptosis during a bovine lactation: Relation to milk production and effect of bST. J. Dairy Sci. 2001, 84, 2177–2187. [Google Scholar] [CrossRef] [PubMed]
- Rus, H.; Cudrici, C.; Niculescu, F. The role of the complement system in innate immunity. Immunol. Res. 2005, 33, 103–112. [Google Scholar] [CrossRef]
- Kitchens, R.L. Role of CD14 in Cellular Recognition of Bacterial Lipopolysaccharides. Chem. Immunol. 2000, 74, 61–82. [Google Scholar] [PubMed]
- Qiu, Q.; Zhang, G.; Ma, T.; Qian, W.; Wang, J.; Ye, Z.; Cao, C.; Hu, Q.; Kim, J.; Larkin, D.M. The yak genome and adaptation to life at high altitude. Nat. Genet. 2012, 44, 946–949. [Google Scholar] [CrossRef]
- Pepper, M.S.; Baetens, D.; Mandriota, S.J.; Di Sanza, C.; Oikemus, S.; Lane, T.F.; Soriano, J.V.; Montesano, R.; Iruela-Arispe, M.L. Regulation of VEGF and VEGF receptor expression in the rodent mammary gland during pregnancy, lactation, and involution. Dev. Dyn. 2000, 218, 507–524. [Google Scholar] [CrossRef]
- Rossiter, H.; Barresi, C.; Ghannadan, M.; Gruber, F.; Mildner, M.; Födinger, D.; Tschachler, E. Inactivation of VEGF in mammary gland epithelium severely compromises mammary gland development and function. FASEB J. 2007, 21, 3994–4004. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.-C.; Norsang, G.; Pingcuo, N.; Dahlback, A.; Frette, Y.; Kjeldstad, B.; Hamre, B.; Stamnes, K.; Stamnes, J.J. Solar UV radiation measurements across the Tibetan Plateau. In AIP Conference Proceedings; American Institute of Physics: College Park, MD, USA, 2013; pp. 848–851. [Google Scholar]
- Ayalew, W.; Chu, M.; Liang, C.; Wu, X.; Yan, P. Adaptation Mechanisms of Yak (Bos grunniens) to High-Altitude Environmental Stress. Animals 2021, 11, 2344. [Google Scholar] [CrossRef] [PubMed]
- Ning, Z. Analysis on the Relationship Between Ultraviolet Radiation Characteristics and Asphalt Pavement Disease in Tibet Plateau. Am. J. Civ. Eng. 2016, 4, 367–371. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xia, W.; Liu, Y.; Loor, J.J.; Bionaz, M.; Jiang, M. Dynamic Profile of the Yak Mammary Transcriptome during the Lactation Cycle. Animals 2023, 13, 1710. https://doi.org/10.3390/ani13101710
Xia W, Liu Y, Loor JJ, Bionaz M, Jiang M. Dynamic Profile of the Yak Mammary Transcriptome during the Lactation Cycle. Animals. 2023; 13(10):1710. https://doi.org/10.3390/ani13101710
Chicago/Turabian StyleXia, Wei, Yili Liu, Juan J. Loor, Massimo Bionaz, and Mingfeng Jiang. 2023. "Dynamic Profile of the Yak Mammary Transcriptome during the Lactation Cycle" Animals 13, no. 10: 1710. https://doi.org/10.3390/ani13101710
APA StyleXia, W., Liu, Y., Loor, J. J., Bionaz, M., & Jiang, M. (2023). Dynamic Profile of the Yak Mammary Transcriptome during the Lactation Cycle. Animals, 13(10), 1710. https://doi.org/10.3390/ani13101710






