Identification of Candidate Genes Regulating Carcass Depth and Hind Leg Circumference in Simmental Beef Cattle Using Illumina Bovine Beadchip and Next-Generation Sequencing Analyses
Abstract
Simple Summary
Abstract
1. Introduction
2. Material and Methods
2.1. Ethics Statement
2.2. Animal Resources and Phenotype Data
2.3. Genotype Examination and Quality Control
2.4. Resequencing
2.5. Variant Imputation
2.6. Statistical Analysis
2.7. Heritability Estimation
3. Results
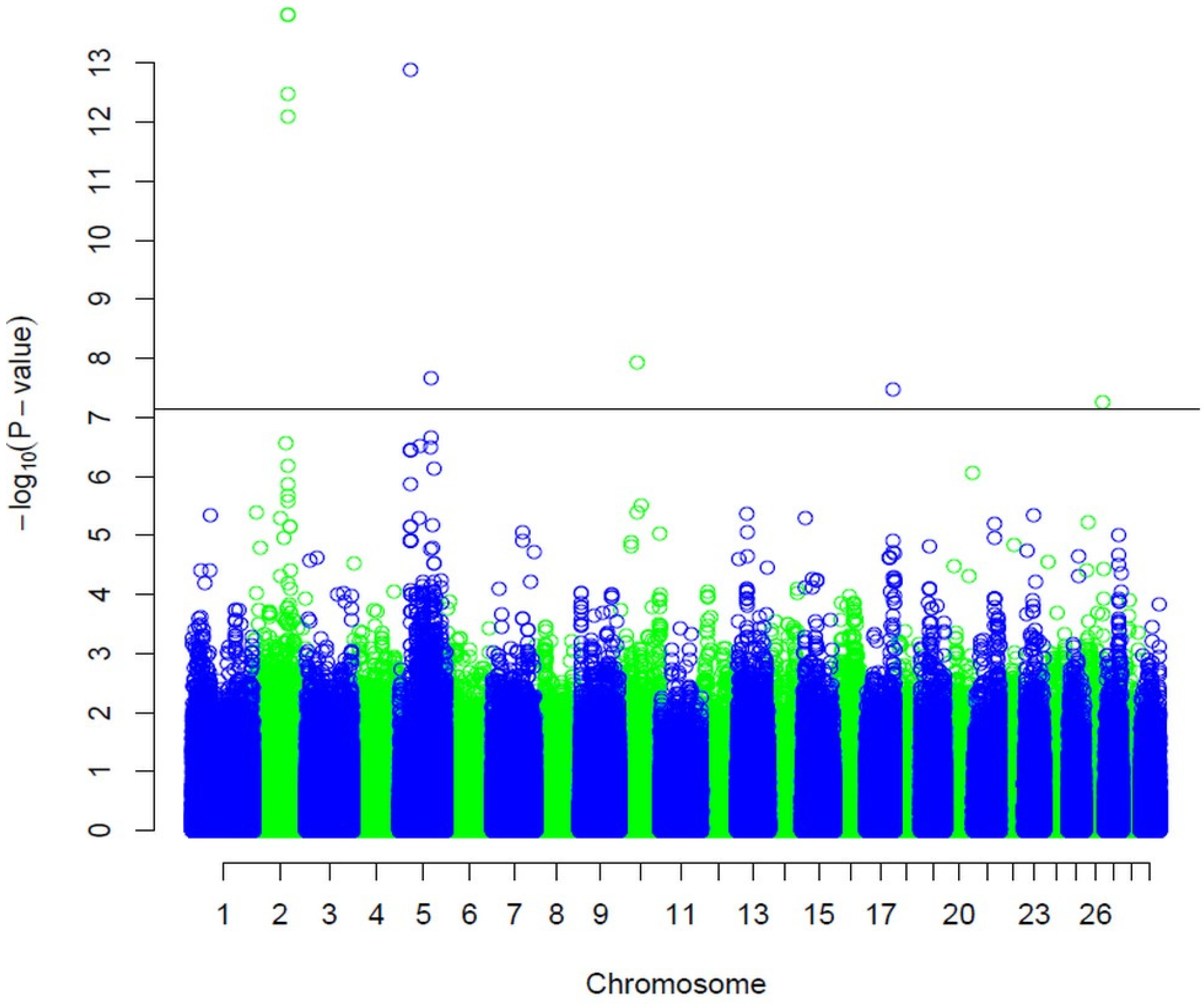
3.1. SNPs Associated with Carcass Depth
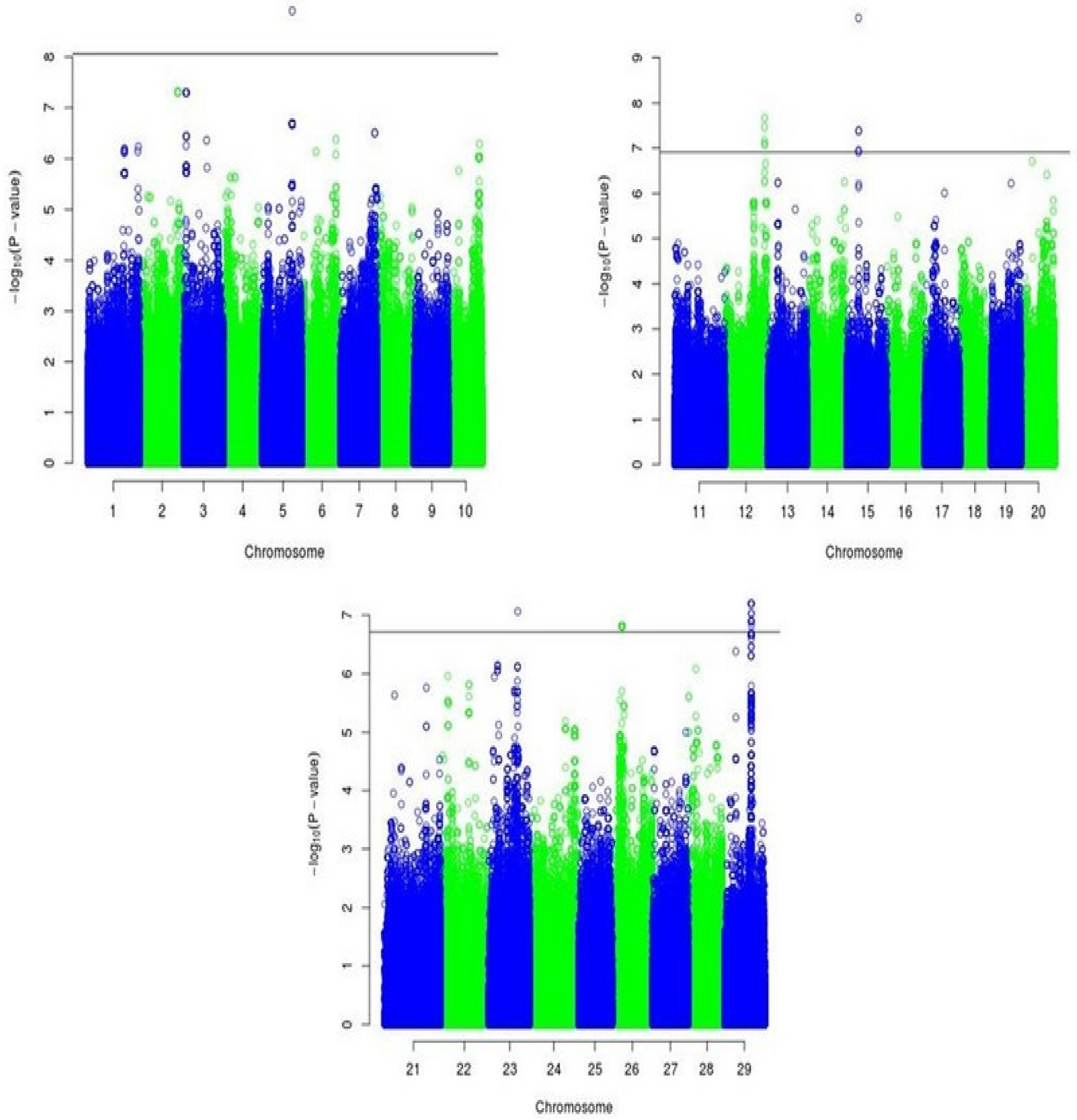
3.2. SNPS Associated with Hind Leg Circumference
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Seré, C.; Steinfeld, H. World Livestock Production Systems; Animal Production and Health Paper 127; FAO: Rome, Italy, 1996; Available online: http://www.fao.org/3/a-w0027e.pdf (accessed on 7 July 2021).
- Harris, D.L. Breeding for Efficiency in Livestock Production: Defining the Economic Objectives. J. Anim. Sci. 1970, 30, 860–865. [Google Scholar] [CrossRef]
- Yuan, Z.; Li, J.; Li, J.; Gao, X.; Gao, H.; Xu, S. Effects of DGAT1 gene on meat and carcass fatness quality in Chinese commercial cattle. Mol. Biol. Rep. 2012, 40, 1947–1954. [Google Scholar] [CrossRef] [PubMed]
- Albertí, P.; Panea, B.; Sañudo, C.; Olleta, J.L.; Ripoll, G.; Ertbjerg, P.; Christensen, S.; Gigli, S.; Failla, S.; Concetti, J.F.; et al. Live weight, body size and carcass characteristics of young bulls of fifteen European breeds. Livest. Sci. 2008, 114, 19–30. [Google Scholar] [CrossRef]
- Chambaz, A.; Scheeder, M.R.; Kreuzer, M.; Dufey, P.A. Meat quality of Angus, Simmental, Charolais and Limousin steers compared at the same intramuscular fat content. Meat Sci. 2003, 63, 491–500. [Google Scholar] [CrossRef]
- Cuvelier, C.; Cabaraux, J.F.; Dufrasne, I.; Clinquart, A.; Hocquette, J.F.; Istasse, L.; Hornick, J.L. Performance, slaughter characteristics and meat quality of young bulls from Belgian Blue, Limousin and Aberdeen Angus breeds fattened with a sugar-beet pulp or a cereal-based diet. Anim. Sci. 2006, 82, 125–132. [Google Scholar] [CrossRef]
- Vieira, C.; Cerdeño, A.; Serrano, E.; Lavín, P.; Mantecón, A.R. Breed and ageing extent on carcass and meat quality of beef from adult steers (oxen). Livest. Sci. 2007, 107, 62–69. [Google Scholar] [CrossRef]
- May, M.L.; Dikeman, M.E.; Schalles, R. Longissimus Muscle Histological Characteristics of Simmental × Angus, Hereford × Angus and Limousin × Angus Crossbred Steers as Related to Carcass Composition and Meat Palatability Traits. J. Anim. Sci. 1977, 44, 571–580. [Google Scholar] [CrossRef]
- Ozlütürk, A.; Tüzemen, N.; Yanar, M.; Esenbuga, N.; Dursun, E. Fattening performance, carcass traits and meat quality characteristics of calves sired by Charolais, Simmental and Eastern Anatolian Red sires mated to Eastern Anatolian Red dams. Meat Sci. 2004, 67, 463–470. [Google Scholar] [CrossRef]
- Sami, A.S.; Augustini, C.; Schwarz, F.J. Effects of feeding intensity and time on feed on performance, carcass characteristics and meat quality of Simmental bulls. Meat Sci. 2004, 67, 195–201. [Google Scholar] [CrossRef]
- Kebede, A.; Komlosi, I. Evaluation of genetic parameters and growth traits of Hungarian Simmental cattle breed. Livest. Res. 2015, 27, Article #172. Available online: http://www.lrrd.org/lrrd27/9/dami27172.html (accessed on 8 July 2021).
- Tabor, H.K.; Risch, N.J.; Myers, R.M. Candidate-gene approaches for studying complex genetic traits: Practical considerations. Nat. Rev. Genet. 2002, 3, 391–397. [Google Scholar] [CrossRef] [PubMed]
- Zhu, M.; Zhao, S. Candidate Gene Identification Approach: Progress and Challenges. Int. J. Biol. Sci. 2007, 3, 420–427. [Google Scholar] [CrossRef] [PubMed]
- Xia, J.; Qi, X.; Wu, Y.; Zhu, B.; Xu, L.; Zhang, L.; Gao, X.; Chen, Y.; Li, J.; Gao, H. Genome-wide association study identifies loci and candidate genes for meat quality traits in Simmental beef cattle. Mamm. Genome 2016, 27, 246–255. [Google Scholar] [CrossRef] [PubMed]
- Chang, T.; Xia, J.; Xu, L.; Wang, X.; Zhu, B.; Zhang, L.; Gao, X.; Chen, Y.; Li, J.; Gao, H. A genome-wide association study suggests several novel candidate genes for carcass traits in Chinese Simmental beef cattle. Anim Genet 2018, 49, 312–316. [Google Scholar] [CrossRef]
- Du, L.; Duan, X.; An, B.; Chang, T.; Liang, M.; Xu, L.; Zhang, L.; Li, J.; E, G.; Gao, H. Genome-Wide Association Study Based on Random Regression Model Reveals Candidate Genes Associated with Longitudinal Data in Chinese Simmental Beef Cattle. Animals 2021, 11, 2524. [Google Scholar] [CrossRef]
- Hirschhorn, J.N.; Daly, M.J. Genome-wide association studies for common diseases and complex traits. Nat. Rev. Genet. 2005, 6, 95–108. [Google Scholar] [CrossRef]
- Coon, K.D.; Myers, A.J.; Craig, D.W.; Webster, J.A.; Pearson, J.V.; Lince, D.H.; Zismann, V.L.; Beach, T.G.; Leung, D.; Bryden, L.; et al. A high-density whole-genome association study reveals that APOE is the major susceptibility gene for sporadic late-onset Alzheimer’s disease. J. Clin. Psychiatry 2007, 68, 613–618. [Google Scholar] [CrossRef]
- Gudmundsson, J.; Sulem, P.; Gudbjartsson, D.F.; Blondal, T.; Gylfason, A.; Agnarsson, B.A.; Benediktsdottir, K.R.; Magnusdottir, D.N.; Orlygsdottir, G.; Jakobsdottir, M.; et al. Genome-wide association and replication studies identify four variants associated with prostate cancer susceptibility. Nat. Genet. 2009, 41, 1122–1126. [Google Scholar] [CrossRef]
- Visscher, P.M.; Wray, N.R.; Zhang, Q.; Sklar, P.; McCarthy, M.I.; Brown, M.A.; Yang, J. 10 years of GWAS discovery: Biology, function, and Translation. Am. J. Hum. Genet. 2017, 101, 5–22. [Google Scholar] [CrossRef]
- Sharma, A.; Cho, Y.; Choi, B.H.; Chai, H.H.; Park, J.E.; Lim, D. Limited representation of OMIA causative mutations for cattle in SNP databases. Anim. Genet. 2017, 48, 369–370. [Google Scholar] [CrossRef]
- Shin, D.H.; Lee, H.J.; Cho, S.; Kim, H.J.; Hwang, J.Y.; Lee, C.K.; Jeong, J.; Yoon, D.; Kim, H. Deleted copy number variation of Hanwoo and Holstein using next generation sequencing at the population level. BMC Genom. 2014, 15, 240. [Google Scholar] [CrossRef] [PubMed]
- Bordbar, F.; Jensen, J.; Zhu, B.; Wang, Z.; Xu, L.; Chang, T.; Xu, L.; Du, M.; Zhang, L.; Gao, H.; et al. Identification of muscle-specific candidate genes in Simmental beef cattle using imputed next generation sequencing. PLoS ONE 2019, 14, e0223671. [Google Scholar] [CrossRef] [PubMed]
- Bordbar, F.; Jensen, J.; Du, M.; Abied, A.; Guo, W.; Xu, L.; Gao, H.; Zhang, L.; Li, J. Identification and validation of a novel candidate gene regulating net meat weight in Simmental beef cattle based on imputed next-generation sequencing. Cell Prolif. 2020, 53, e12870. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.W.; Chung, W.H.; Lee, K.T.; Cho, E.S.; Lee, S.W.; Choi, B.H.; Lee, S.H.; Lim, W.; Lim, D.; Lee, Y.G.; et al. Whole-genome resequencing analyses of five pig breeds, including Korean wild and native, and three European origin breeds. DNA Res. 2015, 22, 259–267. [Google Scholar] [CrossRef]
- Piedrafita, J.; Quintanilla, R.; Sanudo, C.; Olleta, J.L.; Campo, M.M.; Panea, B.; Renand, G.; Turin, F.; Jabet, S.; Osoroe, K.; et al. Carcass quality of 10 beef cattle breeds of the Southwest of Europe in their typical production systems. Livest. Prod. Sci. 2003, 82, 1–13. [Google Scholar] [CrossRef]
- Han, R.; Zan, L.; Yang, D.; Hao, R. [SNPs detection of IGF2 gene and its relationship with carcass and meat quality traits in Qinchuan cattle]. Yi Chuan 2008, 30, 1579–1584. [Google Scholar] [CrossRef]
- Mohammadabadi, M.; Bordbar, F.; Jensen, J.; Du, M.; Guo, W. Key Genes Regulating Skeletal Muscle Development and Growth in Farm Animals. Animals 2021, 11, 835. [Google Scholar] [CrossRef]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.R.; Bender, D.; Maller, J.; Sklar, P.; Bakker, P.; Daly, M.J.; et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef] [PubMed]
- Li, H. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 2011, 27, 2987–2993. [Google Scholar] [CrossRef] [PubMed]
- Browning, B.L.; Browning, S.R. Genotype imputation with millions of reference samples. Am. J. Hum. Genet. 2016, 98, 116–126. [Google Scholar] [CrossRef] [PubMed]
- Fuchsberger, C.; Abecasis, G.R.; Hinds, D.A. Minimac2: Faster genotype imputation. Bioinformatics 2014, 31, 782–784. [Google Scholar] [CrossRef] [PubMed]
- Aulchenko, Y.S.; Ripke, S.; Isaacs, A.; Duijn, C. GenABEL: An R library for genome-wide association analysis. Bioinformatics 2007, 23, 1294–1296. [Google Scholar] [CrossRef] [PubMed]
- Nakagawa, S. A farewell to Bonferroni: The problems of low statistical power and publication bias. Behav. Ecol. 2004, 15, 1044–1045. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Takasuga, A.; Watanabe, T.; Mizoguchi, Y.; Hirano, T.; Ihara, N.; Takano, A.; Yokouchi, K.; Fujikawa, A.; Chiba, K.; Kobayashi, N.; et al. Identification of bovine QTL for growth and carcass traits in Japanese Black cattle by replication and identical-by-descent mapping. Mamm. Genome 2017, 18, 125–136. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Chen, B.; Xu, K.; Wu, J.; Song, W.; Bancroft, I.; Harper, A.L.; Trick, M.; Liu, G.; Gao, G.; et al. Genome-wide association study dissects the genetic architecture of seed weight and seed quality in rapeseed (Brassica napus L.). DNA Res. 2014, 21, 355–367. [Google Scholar] [CrossRef] [PubMed]
- Cole, J.B.; Wiggans, G.R.; Ma, L.; Sonstegard, T.S.; Lawlor, T.J.; Crooker, B.A.; Van Tassell, C.P.; Yang, J.; Wang, S.; Lakshmi, K.; et al. Genome-wide association analysis of thirty one production, health, reproduction and body conformation traits in contemporary U.S. Holstein cows. BMC Genom. 2011, 12, 408. [Google Scholar] [CrossRef] [PubMed]
- Bolormaa, S.; Hayes, B.J.; Savin, K.; Hawken, R.; Barendse, W.; Arthur, P.F.; Herd, R.M.; Goddard, M.E. Genome-wide association studies for feedlot and growth traits in cattle. J. Anim. Sci. 2011, 89, 1684–1697. [Google Scholar] [CrossRef]
- Bolormaa, S.; Pryce, J.E.; Reverter, A.; Zhang, Y.; Barendse, W.; Kemper, K.; Tier, B.; Savin, K.; Hayes, B.; Goddard, M. A multi-trait, meta-analysis for detecting pleiotropic polymorphisms for stature, fatness and reproduction in beef cattle. PLoS Genet. 2014, 10, e1004198. [Google Scholar] [CrossRef]
- Wu, X.; Fang, M.; Liu, L.; Wang, S.; Liu, J.; Ding, X.; Zhang, S.; Zhang, Q.; Zhang, Y.; Qiao, L.; et al. Genome wide association studies for body conformation traits in the Chinese Holstein cattle population. BMC Genom. 2013, 14, 897. [Google Scholar] [CrossRef]
- Abo-Ismail, M.K.; Vander Voort, G.; Squires, J.J.; Swanson, K.C.; Mandell, I.B.; Liao, X.; Stothard, P.; Moore, S.; Plastow, G.; Miller, S. Single nucleotide polymorphisms for feed efficiency and performance in crossbred beef cattle. BMC Genet. 2014, 15, 14. [Google Scholar] [CrossRef] [PubMed]
- Feugang, J.M.; Kaya, A.; Page, G.P.; Chen, L.; Mehta, T.; Hirani, K.; Nazareth, L.; Topper, E.; Gibbs, R.; Memili, E. Two-stage genome-wide association study identifies integrin beta 5 as having potential role in bull fertility. BMC Genom. 2009, 10, 176. [Google Scholar] [CrossRef] [PubMed]
- Duijvesteijn, N.; Knol, E.F.; Merks, J.W.; Crooijmans, R.P.; Groenen, M.A.; Bovenhuis, H.; Harlizius, B. A genome-wide association study on androstenone levels in pigs reveals a cluster of candidate genes on chromosome 6. BMC Genet. 2010, 11, 42. [Google Scholar] [CrossRef] [PubMed]
- Barendse, W.; Reverter, A.; Bunch, R.J.; Harrison, B.E.; Barris, W.; Thomas, M.B. A validated whole-genome association study of efficient food conversion in cattle. Genetics 2007, 176, 1893–1905. [Google Scholar] [CrossRef]
- Kolbehdari, D.; Wang, Z.; Grant, J.R.; Murdoch, B.; Prasad, A.; Xiu, Z.; Marques, E.; Stothard, P.; Moore, S. A whole genome scan to map QTL for milk production traits and somatic cell score in Canadian Holstein bulls. J. Ani. Breed. Genet. 2009, 126, 216–227. [Google Scholar] [CrossRef]
- Settles, M.; Zanella, R.; McKay, S.D.; Schnabel, R.D.; Taylor, J.F.; Whitlock, R.; Schukken, Y.; Van Kessel, J.S.; Smith, J.M.; Neibergs, H. A Whole Genome Association analysis identifies loci associated with Mycobacterium avium subsp. paratuberculosis infection status in US Holstein Cattle. Anim. Genet. 2009, 40, 655–662. [Google Scholar] [CrossRef]
- Reis-Filho, J.S. Next-generation sequencing. Breast Cancer Res. 2009, 11, S12. [Google Scholar] [CrossRef]
- Doherty, S.; Maurer, J. CLK1 and Its Effects on Skin Stem Cell Differentiation. Bachelor’s Thesis, Worcester Polytechnic Institute, Worcester, MA, USA, 29 April 2010. [Google Scholar]
- Menegay, H.J.; Myers, M.P.; Moeslein, F.M.; Landreth, G.E. Biochemical Characterization and Localization of the Dual Specificity Kinase CLK1. J. Cell Sci. 2000, 113, 3241–3253. [Google Scholar] [CrossRef]
- Sako, Y.; Ninomiya, K.; Okuno, Y.; Toyomoto, M.; Nishida, A.; Koike, Y.; Ohe, K.; Kii, I.; Yoshida, S.; Hashimoto, N.; et al. Development of an orally available inhibitor of CLK1 for skipping a mutated dystrophin exon in Duchenne muscular dystrophy. Sci. Rep. 2017, 7, 46126. [Google Scholar] [CrossRef]
- Xie, S.H.; Li, J.Q.; Chen, Y.S.; Gao, P.; Zhang, H.; Li, Z.Y. Molecular Cloning, Expression, and Chromosomal Mapping of the Porcine CDC-2-Like Kinase 1 (CLK1) Gene. Biochem. Genet. 2009, 47, 266–275. [Google Scholar] [CrossRef]
- Lee, S.; Gondro, G.; van der Werf, J.; Kim, N.; Lim, D.; Park, E.; Oh, S.; Gibson, J.P.; Thompson, J.M. Use of a bovine genome array to identify new biological pathways for beef marbling in Hanwoo (Korean Cattle). BMC Genom. 2010, 11, 623. [Google Scholar] [CrossRef] [PubMed]
- Lim, D.; Lee, S.; Kim, N.; Cho, Y.; Chai, H.; Seong, H.; Kim, H. Gene Co-expression Analysis to Characterize Genes Related to Marbling Trait in Hanwoo (Korean) Cattle. Asian-Aust. J. Anim. Sci. 2013, 26, 19–29. [Google Scholar] [CrossRef]
- Li, S.; Chai, Z.; Li, Y.; Liu, D.; Bai, Z.; Li, Y.; Li, Y.; Situ, Z. BZW1, a novel proliferation regulator that promotes growth of salivary muocepodermoid carcinoma. Cancer Lett. 2009, 284, 86–94. [Google Scholar] [CrossRef] [PubMed]
- Chiou, J.; Chang, Y.; Jan, Y.; Tsai, H.; Yang, C.; Huang, M.; Yu, Y.; Hsiao, M. Overexpression of BZW1 is an independent poor prognosis marker and its down-regulation suppresses lung adenocarcinoma metastasis. Sci. Rep. 2019, 9, 14624. [Google Scholar] [CrossRef] [PubMed]
- Shi, Z.; Xiao, C.; Lin, T.; Wu, J.; Li, K. BZW1 promotes cell proliferation in prostate cancer by regulating TGF-β1/Smad pathway. Cell Cycle 2021, 20, 894–902. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Mandel, J.; Bouaziz, B.; Commenges, D.; Nabirotchkine, S.; Chumakov, I.; Cohen, D.; Guedj, M.; the Alzheimer’s Disease Neuroimaging Initiative. A Multi-Marker Genetic Association Test Based on the Rasch Model Applied to Alzheimer’s Disease. PLoS ONE 2015, 10, e0138223. [Google Scholar] [CrossRef] [PubMed]
- Hoelker, M.; Salilew-Wondim, D.; Drillich, M.; Christine, G.B.; Ghanem, N.; Goetze, L.; Tesfaye, D.; Schellander, K.; Heuwieser, W. Transcriptional response of the bovine endometrium and embryo to endometrial polymorphonuclear neutrophil infiltration as an indicator of subclinical inflammation of the uterine environment. Reprod. Fertil. Dev. 2012, 24, 778–793. [Google Scholar] [CrossRef]
- Sarin, K.Y.; Lin, Y.; Daneshjou, R.; Ziyatdinov, A.; Thorleifsson, G.; Rubin, A.; Pardo, L.M.; Wu, W.; Khavari, P.A.; Uitterlinden, A.; et al. Genome-wide meta-analysis identifies eight new susceptibility loci for cutaneous squamous cell carcinoma. Nat. Commun. 2020, 11, 820. [Google Scholar] [CrossRef]
- Nayeri, S.; Stothard, P. Tissues, Metabolic Pathways and Genes of Key Importance in Lactating Dairy Cattle. Springer Sci. Rev. 2016, 4, 49–77. [Google Scholar] [CrossRef]
- Oliveira, P.S.N.; Cesar, A.S.M.; Oliveira, G.B.; Tizioto, P.C.; Poleti, M.D.; Diniz, W.J.S.; Lima, A.O.D.; Reecy, J.M.; Coutinho, L.L.; Regitano, L.C.A. miRNAs related to fatty acids composition in Nellore cattle. J. Anim. Sci. 2016, 94, 164. [Google Scholar] [CrossRef]
- Seetharaman, S.; Flemyng, E.; Shen, J.; Conte, M.R.; Ridley, A.J. The RNA-binding protein LARP4 regulates cancer cell migration and invasion. Cytoskeleton 2016, 73, 680–690. [Google Scholar] [CrossRef] [PubMed]
- Egiz, M.; Usui, T.; Ishibashi, M.; Zhang, X.; Shigeta, S.; Toyoshima, M.; Kitatani, K.; Yaegashi, N. La-Related Protein 4 as a Suppressor for Motility of Ovarian Cancer Cells. Tohoku J. Exp. Med. 2019, 247, 59–67. [Google Scholar] [CrossRef] [PubMed]
- Blagden, S.P.; Gatt, M.K.; Archambault, V.; Lada, K.; Ichihara, K.; Lilley, K.S.; Inoue, Y.H.; Glover, D.M. Drosophila Larp associates with poly(A)-binding protein and is required for male fertility and syncytial embryo development. Dev. Biol. 2009, 334, 186–197. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.; Li, C.; Cai, W.; Liu, S.; Yin, H.; Shi, S.; Zhang, Q.; Zhang, S. Genome-Wide Association Study for Milk Protein Composition Traits in a Chinese Holstein Population Using a Single-Step Approach. Front. genet. 2019, 10, 72. [Google Scholar] [CrossRef]
- Suliman, B.A.; Xu, D.; Williams, B.R. The promyelocytic leukemia zinc finger protein: Two decades of molecular oncology. Front. Oncol. 2012, 2, 74. [Google Scholar] [CrossRef]
- Uhlen, M.; Bjorling, E.; Agaton, C.; Szigyarto, C.A.; Amini, B.; Andersen, E.; Angelidou, P.; Asplund, A.; Asplund, C.; Berglund, L.; et al. A human protein atlas for normal and cancer tissues based on antibody proteomics. Mol. Cell Proteomic. 2005, 4, 1920–1932. [Google Scholar] [CrossRef]
- Kolesnichenko, M.; Vogt, P.K. Understanding PLZF: Two transcriptional targets, REDD1 and smooth muscle alpha-actin, define new questions in growth control, senescence, self-renewal and tumor suppression. Cell Cycle 2011, 10, 771–775. [Google Scholar] [CrossRef][Green Version]
- Cheung, M.; Pei, J.; Pei, Y.; Jhanwar, S.C.; Pass, H.I.; Testa, J.R. The promyelocytic leukemia zinc-finger gene, PLZF, is frequently downregulated in malignant mesothelioma cells and contributes to cell survival. Oncogene 2010, 29, 1633–1640. [Google Scholar] [CrossRef]
- Barna, M.; Hawe, N.; Niswander, L.; Pandolfi, P.P. Plzf regulates limb and axial skeletal patterning. Nat. Genet. 2000, 25, 166–172. [Google Scholar] [CrossRef]
- Costoya, J.A.; Hobbs, R.M.; Barna, M.; Cattoretti, G.; Manova, K.; Sukhwani, M.; Orwig, K.E.; Wolgemuth, D.J.; Pandolfi, P.P. Essential role of Plzf in maintenance of spermatogonial stem cells. Nat. Genet. 2004, 36, 653–659. [Google Scholar] [CrossRef]
- Felicetti, F.; Bottero, L.; Felli, N.; Mattia, G.; Labbaye, C.; Alvino, E.; Peschle, C.; Colombo, M.P.; Care, A. Role of PLZF in melanoma progression. Oncogene 2004, 23, 4567–4576. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Qian, J.; Shi, X.; Gao, T.; Liang, T.; Liu, C. Control of hepatic gluconeogenesis by the promyelocytic leukemia zinc finger protein. Mol. Endocrinol. 2014, 28, 1987–1998. [Google Scholar] [CrossRef] [PubMed]
- Marofi, F.; Vahedi, G.; Solali, S.; Alivand, M.; Salarinasab, S.; Heydarabad, M.Z.; Farshdousti Hagh, M. Gene expression of TWIST1 and ZBTB16 is regulated by methylation modifications during the osteoblastic differentiation of mesenchymal stem cells. J. Cell Physiol. 2019, 234, 6230–6234. [Google Scholar] [CrossRef] [PubMed]
- Vincent-Fabert, C.; Platet, N.; Vandevelde, A.; Poplineau, M.; Koubi, M.; Finetti, P.; Tiberi, G.; Imbert, A.M.; Bertucci, F.; Duprez, E. PLZF mutation alters mouse hematopoietic stem cell function and cell cycle progression. Blood 2016, 127, 1881–1885. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Mikkelsen, T.S.; Xu, Z.; Zhang, X.; Wang, L.; Gimble, J.M.; Lander, E.S.; Rosen, E.D. Comparative epigenomic analysis of murine and human adipogenesis. Cell 2010, 143, 156–169. [Google Scholar] [CrossRef]
- Ambele, M.A.; Dessels, C.; Durandt, C.; Pepper, M.S. Genome-wide analysis of gene expression during adipogenesis in human adipose-derived stromal cells reveals novel patterns of gene expression during adipocyte differentiation. Stem Cell Res. 2016, 16, 725–734. [Google Scholar] [CrossRef]
- Plaisier, C.L.; Bennett, B.J.; He, A.; Guan, B.; Lusis, A.J.; Reue, K.; Vergnes, L. Zbtb16 has a role in brown adipocyte bioenergetics. Nutr. Diabetes 2012, 2, e46. [Google Scholar] [CrossRef]
- Wei, S.; Zhang, M.; Zheng, Y.; Yan, P. ZBTB16 Overexpression Enhances White Adipogenesis and Induces Brown-Like Adipocyte Formation of Bovine White Intramuscular Preadipocytes. Cell Physiol. Biochem. 2018, 48, 2528–2538. [Google Scholar] [CrossRef]
- Liu, Y.; Hu, Y.; Zeng, Y.; Li, Z.; Zhang, H.; Deng, J.; Wang, G. Neurexophilin and PC-esterase domain family member 4 (NXPE4) and prostate androgen-regulated mucin-like protein 1 (PARM1) as prognostic biomarkers for colorectal cancer. J. Cell Biochem. 2019, 120, 18041–18052. [Google Scholar] [CrossRef]
- Sarghale, A.J.; Shahrebabak, M.M.; Shahrebabak, H.M.; Javaremi, A.N.; Saatchi, M.; Khansefid, M.; Miar, Y. Genome-wide association studies for methane emission and ruminal volatile fatty acids using Holstein cattle sequence data. BMC Genet. 2020, 21, 129. [Google Scholar]
- Mokhber, M. Genome-wide association study for milk production in Iranian buffalo. In Proceedings of the Conference on organic vs. Conventional agriculture, Ardabil, Iran, 16–17 August 2017. [Google Scholar]
- Steinthorsdottir, V.; Thorleifsson, G.; Reynisdottir, I.; Benediktsson, R.; Jonsdottir, T.; Walters, G.B.; Styrkarsdottir, U.; Gretarsdottir, S.; Emilsson, V.; Ghosh, S.; et al. A variant in CDKAL1 influences insulin response and risk of type 2 diabetes. Nat. Genet. 2007, 39, 770–775. [Google Scholar] [CrossRef] [PubMed]
- Zeggini, E.; Weedon, M.N.; Lindgren, C.M.; Frayling, T.M.; Elliott, K.S.; Lango, H.; Timpson, N.J.; Perry, J.R.B.; Rayner, N.W.; Freathy, R.M.; et al. Replication of genome-wide association signals in UK samples reveals risk loci for type 2 diabetes. Science 2007, 316, 1336–1341. [Google Scholar] [CrossRef] [PubMed]
- Scott, L.J.; Mohlke, K.L.; Bonnycastle, L.L.; Willer, C.J.; Li, Y.; Duren, W.L.; Erdos, M.R.; Stringham, H.M.; Chines, P.S.; Jackson, A.U.; et al. A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science 2007, 316, 1341–1345. [Google Scholar] [CrossRef] [PubMed]
- Lew, J.; Huang, Q.Q.; Qi, Z.; Winkfein, R.J.; Aebersold, R.; Hunt, T.; Wang, J.H. A brain-specific activator of cyclin-dependent kinase 5. Nature 1994, 371, 423–426. [Google Scholar] [CrossRef] [PubMed]
- Knight, J.D.R.; Kothary, R. The myogenic kinome: Protein kinases critical to mammalian skeletal myogenesis. Skelet. Muscle 2011, 1, 29. [Google Scholar] [CrossRef]
- Palmer, C.J.; Bruckner, R.J.; Paulo, J.A.; Kazak, L.; Long, J.Z.; Mina, A.I.; Deng, Z.; LeClair, K.B.; Hall, J.A.; Hong, S.; et al. Cdkal1, a type 2 diabetes susceptibility gene, regulates mitochondrial function in adipose tissue. Mol. Metab. 2017, 6, 1212–1225. [Google Scholar] [CrossRef]
- Asp, P.; Acosta-Alvear, D.; Tsikitis, M.; van Oevelen, C.; Dynlacht, B.D. E2f3b plays an essential role in myogenic differentiation through isoform-specific gene regulation. Gen. Dev. 2009, 23, 37–53. [Google Scholar] [CrossRef]
- Ma, M.; Wang, X.; Chen, X.; Cai, R.; Chen, F.; Dong, W.; Yang, G.; Pang, W. MicroRNA-432 targeting E2F3 and P55PIK inhibits myogenesis through PI3K/AKT/mTOR signaling pathway. RNA Biol. 2017, 14, 347–360. [Google Scholar] [CrossRef]
- Foster, C.S.; Falconer, A.; Dodson, A.R.; Norman, A.R.; Dennis, N.; Fletcher, A.; Southgate, C.; Dowe, A.; Dearnaley, D.; Jhavar, S.; et al. Transcription factor E2F3 overexpressed in prostate cancer independently predicts clinical outcome. Oncogene 2004, 23, 5871–5879. [Google Scholar] [CrossRef]
- Feber, A.; Clark, J.; Goodwin, G.; Dodson, A.R.; Smith, P.H.; Fletcher, A.; Edwards, S.; Flohr, P.; Falconer, A.; Roe, T.; et al. Amplification and overexpression of E2F3 in human bladder cancer. Oncogene 2004, 23, 1627–1630. [Google Scholar] [CrossRef]
- Vimala, K.; Sundarraj, S.; Sujitha, M.V.; Kannan, S. Curtailing overexpression of E2F3 in breast cancer using siRNA (E2F3)-based gene silencing. Arch. Med. Res. 2012, 43, 415–422. [Google Scholar] [CrossRef] [PubMed]
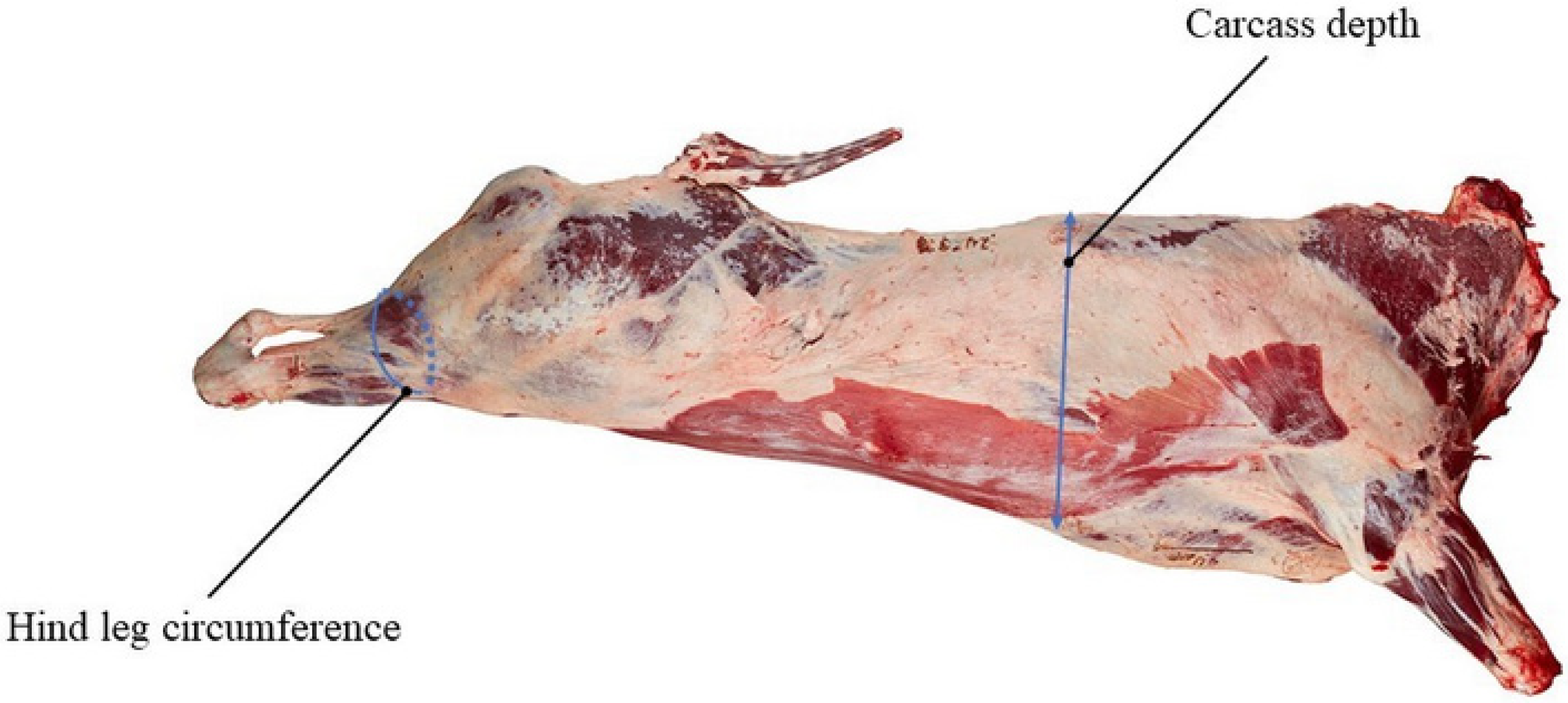
| Trait | Mean (cm) | Standard Deviation (cm) | Maximum (cm) | Minimum (cm) |
|---|---|---|---|---|
| Carcass depth Circumference of hind leg | 63.88 68.01 | 4.62 16.58 | 82 110 | 13 22 |
| SNP | Chromosome | Position | p-Value |
|---|---|---|---|
| BovineHD0200025561 BovineHD0200025562 BovineHD0200025553 BovineHD0200025550 BovineHD0500008673 BovineHD0500023139 BovineHD1000031388 BovineHD1700020542 BovineHD2600013096 | 2 2 2 2 5 5 10 17 26 | 89923045 89926675 89896059 89887085 29582709 81891942 44213755 70557785 46267236 | 1.49 × 10−14 1.49 × 10−14 3.4 × 10−13 8.3 × 10−13 1.3 × 10−13 2.17 × 10−8 1.17 × 10−8 3.47 × 10−8 5.58 × 10−8 |
| SNP | Chromosome | Position | p-Value |
|---|---|---|---|
| rs84436800 rs84450386 rs84441028 rs84429941 rs84428499 rs25168454 rs25439719 rs25457101 rs25237349 rs25224780 | 12 12 12 12 12 15 15 15 15 15 | 84436800 84450386 84441028 84429941 84428499 25168454 25439719 25457101 25237349 25224780 | 2.17 × 10−8 3.45 × 10−8 6.93 × 10−8 8.13 × 10−8 8.24 × 10−8 1.31 × 10−10 4.14 × 10−8 4.14 × 10−8 1.12 × 10−7 1.22 × 10−7 |
| SNP | Chromosome | Position | p-Value |
|---|---|---|---|
| rs37207517 rs12211246 rs12211363 rs12211411 rs34314488 rs34314780 rs34329034 rs34330397 rs34417763 rs34423556 rs34376791 rs34381008 rs34381454 rs34381516 rs34416372 | 23 26 26 26 29 29 29 29 29 29 29 29 29 29 29 | 37207517 12211246 12211363 12211411 34314488 34314780 34329034 34330397 34417763 34423556 34376791 34381008 34381454 34381516 34416372 | 8.61 × 10−8 1.54 × 10−7 1.54 × 10−7 1.54 × 10−7 6.32 × 10−8 6.32 × 10−8 6.32 × 10−8 6.32 × 10−8 9.3 × 10−8 9.3 × 10−8 1.27 × 10−7 1.27 × 10−7 1.27 × 10−7 1.27 × 10−7 1.5 × 10−7 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bordbar, F.; Mohammadabadi, M.; Jensen, J.; Xu, L.; Li, J.; Zhang, L. Identification of Candidate Genes Regulating Carcass Depth and Hind Leg Circumference in Simmental Beef Cattle Using Illumina Bovine Beadchip and Next-Generation Sequencing Analyses. Animals 2022, 12, 1103. https://doi.org/10.3390/ani12091103
Bordbar F, Mohammadabadi M, Jensen J, Xu L, Li J, Zhang L. Identification of Candidate Genes Regulating Carcass Depth and Hind Leg Circumference in Simmental Beef Cattle Using Illumina Bovine Beadchip and Next-Generation Sequencing Analyses. Animals. 2022; 12(9):1103. https://doi.org/10.3390/ani12091103
Chicago/Turabian StyleBordbar, Farhad, Mohammadreza Mohammadabadi, Just Jensen, Lingyang Xu, Junya Li, and Lupei Zhang. 2022. "Identification of Candidate Genes Regulating Carcass Depth and Hind Leg Circumference in Simmental Beef Cattle Using Illumina Bovine Beadchip and Next-Generation Sequencing Analyses" Animals 12, no. 9: 1103. https://doi.org/10.3390/ani12091103
APA StyleBordbar, F., Mohammadabadi, M., Jensen, J., Xu, L., Li, J., & Zhang, L. (2022). Identification of Candidate Genes Regulating Carcass Depth and Hind Leg Circumference in Simmental Beef Cattle Using Illumina Bovine Beadchip and Next-Generation Sequencing Analyses. Animals, 12(9), 1103. https://doi.org/10.3390/ani12091103








