Characterization of Salmonella spp. and E. coli Strains Isolated from Wild Carnivores in Janos Biosphere Reserve, Mexico
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Area
2.2. Experimental Design and Animal Management
2.3. Isolation and Identification of Salmonella spp. from feces
2.4. Isolation and Identification of E. coli from Feces
2.5. E. coli and Salmonella spp. Characterization by PCR
2.6. E. coli Serotyping and Genotyping
2.7. Salmonella spp. Intracellular Survival Assays
2.8. E. coli Adhesion Assays
2.9. Statistical Analysis
3. Results
3.1. Animals Captured
3.2. Isolation and Identification of E. coli and Salmonella spp. from Feces
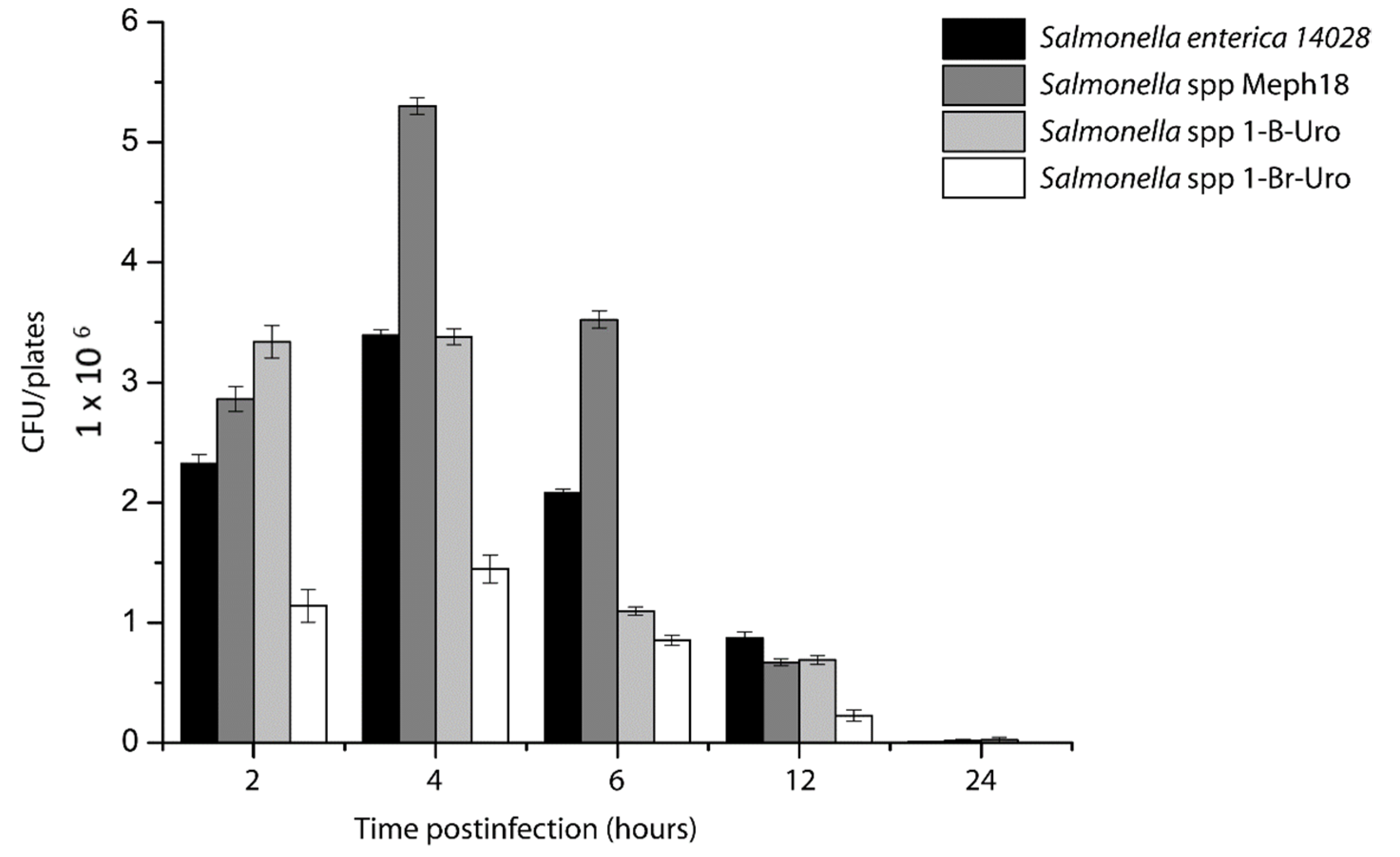
3.3. Salmonella spp. Intracellular Survival Assays
3.4. E. coli Adhesion Assays
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Daszak, P.; Cunningham, A.; Hyatt, A. Anthropogenic environmental change and the emergence of infectious diseases in wildlife. Acta Trop. 2001, 78, 103–116. [Google Scholar] [CrossRef]
- Jones, K.E.; Patel, N.G.; Levy, M.A.; Storeygard, A.; Balk, D.; Gittleman, J.L.; Daszak, P. Global trends in emerging infectious diseases. Nature 2008, 451, 990–993. [Google Scholar] [CrossRef] [PubMed]
- Allen, T.; Murray, K.; Zambrana-Torrelio, C.; Morse, S.S.; Rondinini, C.; Di Marco, M.; Breit, N.; Olival, K.; Daszak, P. Global hotspots and correlates of emerging zoonotic diseases. Nat. Commun. 2017, 8, 1124. [Google Scholar] [CrossRef] [PubMed]
- Wolf, M.K. Bacterial infections of the small intestine and colon. Curr. Opin. Gastroenterol. 2000, 16, 4–11. [Google Scholar] [CrossRef] [PubMed]
- Aboutaleb, N.; Kuijper, E.J.; van Dissel, J.T. Emerging infectious colitis. Curr. Opin. Gastroenterol. 2014, 30, 106–115. [Google Scholar] [CrossRef] [PubMed]
- Murphy, R.; Palm, M.; Mustonen, V.; Warringer, J.; Farewell, A.; Parts, L.; Moradigaravand, D. Genomic Epidemiology and Evolution of Escherichia coli in Wild Animals in Mexico. mSphere 2021, 6, e00738-20. [Google Scholar] [CrossRef]
- García, A.; Fox, J.G.; Besser, T.E. Zoonotic Enterohemorrhagic Escherichia coli: A One Health Perspective. ILAR J. 2010, 51, 221–232. [Google Scholar] [CrossRef]
- Tenaillon, O.; Skurnik, D.; Picard, B.; Denamur, E. The population genetics of commensal Escherichia coli. Nat. Rev. Genet. 2010, 8, 207–217. [Google Scholar] [CrossRef]
- Forshell, L.P.; Wierup, M. Salmonella contamination: A significant challenge to the global marketing of animal food products. Rev. Sci. Tech. 2006, 25, 541–554. [Google Scholar]
- Hilbert, F.; Smulders, F.J.M.; Chopra-Dewasthaly, R.; Paulsen, P. Salmonella in the wildlife-human interface. Food Res. Int. 2012, 45, 603–608. [Google Scholar] [CrossRef]
- Ashbolt, R.; Kirk, M.D. Salmonella Mississippi infections in Tasmania: The role of native Australian animals and untreated drinking water. Epidemiol. Infect. 2006, 134, 1257–1265. [Google Scholar] [CrossRef] [PubMed]
- Kreeger, T.J.; Arnemo, J.M.; Jacobus, P.R. Handbook of Wildlife Chemical Immobilization; Wildlife Pharmaceuticals: Laramie, WY, USA, 2002. [Google Scholar]
- Sambrook, J.; Fritsch, E.F.; Maniatis, T. Molecular Cloning: A Laboratory Manual; Cold Spring Laboratory, Cold Spring Harbor: New York, NY, USA, 1989; Volume 2. [Google Scholar]
- Orskov, F.; Orskov, I. Serotyping of Escherichia coli; Bergan, T., Ed.; Academic Press Ltd.: London, UK, 1984; pp. 43–112. [Google Scholar]
- Clermont, O.; Christenson, J.; Denamour, E.; Gordon, D. The Clermont Escherichia coli phylotyping method revisited: Improvement of specificity and detection of new phylogroups. Environ. Microbiol. Rep. 2013, 5, 58–65. [Google Scholar] [CrossRef] [PubMed]
- Santos, F.F.; Yamamoto, D.; Abe, C.M.; Bryant, J.A.; Hernandes, R.T.; Kitamura, F.C.; Castro, F.S.; Valiatti, T.B.; Piazza, R.M.F.; Elias, W.P.; et al. The Type III Secretion System (T3SS)-Translocon of Atypical Enteropathogenic Escherichia coli (aEPEC) Can Mediate Adherence. Front. Microbiol. 2019, 10, 1527. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Galan, J.E.; Ginocchio, C.; Costeas, P. Molecular and functional characterization of the Salmonella invasion gene invA: Homology of InvA to members of a new protein family. J. Bacteriol. 1992, 174, 4338–4349. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deleu, S.; Choi, K.; Reece, J.M.; Shears, S.B. Pathogenicity of Salmonella: SopE-mediated membrane ruffling is independent of inositol phosphate signals. FEBS Lett. 2006, 580, 1709–1715. [Google Scholar] [CrossRef] [Green Version]
- Ashida, H.; Ogawa, M.; Kim, M.; Mimuro, H.; Sasakawa, C. Bacteria and host interactions in the gut epithelial barrier. Nat. Chem. Biol. 2011, 8, 36–45. [Google Scholar] [CrossRef]
- LaRock, D.; Chaudhary, A.; Miller, S.I. Salmonellae interactions with host processes. Nat. Rev. Genet. 2015, 13, 191–205. [Google Scholar] [CrossRef]
- Pacheco, J.; Ceballos, G.; List, R. Los mamíferos de la región de Janos-Casas Grandes, Chihuahua, México. Rev. Mex. Mastozoología. 2000, 4, 69–83. [Google Scholar]
- Millan, J.; Aduriz, G.; Moreno, B.; Juste, R.; Barral, M. Salmonella isolates from wild birds and mammals in the Basque Country (Spain). Rev. Sci. Tech. 2004, 23, 905–911. [Google Scholar] [CrossRef]
- Miller, M.A.; Byrne, B.A.; Jang, S.S.; Dodd, E.M.; Dorfmeier, E.; Harris, M.D.; Ames, J.; Paradies, D.; Worcester, K.; Jessup, D.A.; et al. Enteric bacterial pathogen detection in southern sea otters (Enhydra lutris nereis) is associated with coastal urbanization and freshwater runoff. Vet. Res. 2009, 41, 1. [Google Scholar] [CrossRef] [Green Version]
- Oliveira, M.; Pedroso, N.M.; Sales-Luís, T.; Santos-Reis, M.; Tavares, L.; Vilela, C.L. Antimicrobial-Resistant Salmonella Isolated from Eurasian Otters (Lutra lutra Linnaeus, 1758) in Portugal. J. Wildl. Dis. 2010, 46, 1257–1261. [Google Scholar] [CrossRef] [Green Version]
- Jardine, C.; Reid-Smith, R.J.; Janecko, N.; Allan, M.; McEwen, S.A. Salmonella in Raccoons (Procyon Lotor) in Southern Ontario, Canada. J. Wildl. Dis. 2011, 47, 344–351. [Google Scholar] [CrossRef] [Green Version]
- Compton, J.A.; Baney, J.A.; Donaldson, S.C.; Houser, B.A.; San Julian, G.J.; Yahner, R.H.; Chmielecki, W.; Reynolds, S.; Jayarao, B.M. Salmonella infections in the common raccoon (Procyon lotor) in western Pennsylvania. J. Clin. Microbiol. 2008, 46, 3084–3086. [Google Scholar] [CrossRef] [Green Version]
- Lee, K.; Iwata, T.; Nakadai, A.; Kato, T.; Hayama, S.; Taniguchi, T.; Hayashidani, H. Prevalence of Salmonella, Yersinia and Campylobacter spp. in Feral Raccoons (Procyon lotor) and Masked Palm Civets (Paguma larvata) in Japan. Zoonoses Public Health 2011, 58, 424–431. [Google Scholar] [CrossRef]
- Baldi, M.; Calvo, E.B.; Hutter, S.E.; Walzer, C. Salmonellosis detection and evidence of antibiotic resistance in an urban raccoon population in a highly populated area, Costa Rica. Zoonoses Public Health 2019, 66, 852–860. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Handeland, K.; Nesse, L.L.; Lillehaug, A.; Vikøren, T.; Djønne, B.; Bergsjø, B. Natural and experimental Salmonella Typhimurium infections in foxes (Vulpes vulpes). Vet. Microbiol. 2008, 132, 129–134. [Google Scholar] [CrossRef] [PubMed]
- Jay-Russell, M.T.; Hake, A.F.; Bengson, Y.; Thiptara, A.; Nguyen, T. Prevalence and Characterization of Escherichia coli and Salmonella Strains Isolated from Stray Dog and Coyote Feces in a Major Leafy Greens Production Region at the United States-Mexico Border. PLoS ONE 2014, 9, e113433. [Google Scholar] [CrossRef]
- Very, K.J.; Kirchner, M.K.; Shariat, N.; Cottrell, W.; Sandt, C.H.; Dudley, E.G.; Kariyawasam, S.; Jayarao, B.M. Prevalence and Spatial Distribution of Salmonella Infections in the Pennsylvania Raccoon (Procyon lotor). Zoonoses Public Health 2015, 63, 223–233. [Google Scholar] [CrossRef] [PubMed]
- Douce, G.R.; Amin, I.I.; Stephen, J. Invasion of HEp-2 cells by strains of Salmonella Typhimurium of different virulence in relation to gastroenteritis. J. Med. Microbiol. 1991, 35, 349–357. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bäumler, A.J.; Tsolis, R.M.; Heffron, F. The lpf fimbrial operon mediates adhesion of Salmonella typhimurium to murine Peyer’s patches. Proc. Natl. Acad. Sci. USA 1996, 93, 279–283. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mokracka, J.; Krzymińska, S.; Ałtunin, D.; Wasyl, D.; Koczura, R.; Dudek, K.; Dudek, M.; Chyleńska, Z.A.; Ekner-Grzyb, A. In Vitro virulence characteristics of rare serovars of Salmonella enterica isolated from sand lizards (Lacerta agilis L.). Int. J. Gen. Mol. Microbiol. 2018, 111, 1863–1870. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McWhorter, A.; Owens, J.; Valcanis, M.; Olds, L.; Myers, C.; Smith, I.; Trott, D.; McLelland, D. In vitro invasiveness and antimicrobial resistance of Salmonella enterica subspecies isolated from wild and captive reptiles. Zoonoses Public Health 2021, 68, 402–412. [Google Scholar] [CrossRef] [PubMed]
- Gese, E.M.; Schultz, R.D.; Johnson, M.R.; Williams, E.S.; Crabtree, R.L.; Ruff, R.L. Serological survey for disease in free-ranging coyotes (Canis latrans) in Yellowstone National Park, Wyoming. J. Wildl. Dis. 1997, 33, 47–56. [Google Scholar] [CrossRef] [PubMed]
- Wilson, J.S.; Hazel, S.M.; Williams, N.J.; Phiri, A.; French, N.; Hart, C.A. Nontyphoidal Salmonellae in United Kingdom Badgers: Prevalence and Spatial Distribution. Appl. Environ. Microbiol. 2003, 69, 4312–4315. [Google Scholar] [CrossRef] [Green Version]
- Chiari, M.; Ferrari, N.; Giardiello, D.; Lanfranchi, P.; Zanoni, M.; Lavazza, A.; Alborali, L.G. Isolation and identification of Salmonella spp. from red foxes (Vulpes vulpes) and badgers (Meles meles) in northern Italy. Acta Vet Scand 2014, 56, 1–4. [Google Scholar] [CrossRef] [Green Version]
- Miranda, J.M.; Mondragón, A.C.; Martinez, B.; Guarddon, M.; A Rodriguez, J. Prevalence and antimicrobial resistance patterns of Salmonella from different raw foods in Mexico. J. Food Prot. 2009, 72, 966–971. [Google Scholar] [CrossRef]
- Glawischnig, W.; Wallner, A.; Lazar, J.; Kornschober, C. Cattle-derived Salmonella enterica serovar Dublin infections in red foxes (Vulpes vulpes) in Tyrol, Austria. J. Wildl. Dis. 2017, 53, 361–363. [Google Scholar] [CrossRef]
- Krause, G.; Zimmermann, S.; Beutin, L. Investigation of domestic animals and pets as a reservoir for intimin- (eae) gene positive Escherichia coli types. Vet. Microbiol. 2005, 106, 87–95. [Google Scholar] [CrossRef]
- Manges, A.R.; Tabor, H.; Tellis, P.; Vincent, C.; Tellier, P.P. Endemic and epidemic lineages of Escherichia coli that cause urinary tract infections. Emerg. Infect. Dis. 2008, 10, 1575–1583. [Google Scholar] [CrossRef]
- Clermont, O.; Olier, M.; Hoede, C.; Diancourt, L.; Brisse, S.; Keroudean, M.; Glodt, J.; Picard, B.; Oswald, E.; Denamur, E. Animal and human pathogenic Escherichia coli strains share common genetic backgrounds. Infect Genet Evol. 2011, 11, 654–662. [Google Scholar] [CrossRef]
- Chiu, T.H.; Chen, T.R.; Hwang, W.Z.; Tsen, H.Y. Sequencing of an internal transcribed spacer region of 16S-23S rRNA gene and designing of PCR primers for the detection of Salmonella spp. in food. Int. J. Food Microbiol. 2005, 97, 259–265. [Google Scholar] [CrossRef] [PubMed]
- Müller, D.; Hagedorn, P.; Brast, S.; Heusipp, G.; Bielaszewska, M.; Friedrich, A.W.; Karch, H.; Schmidt, M.A. Rapid identification and differentiation of clinical isolates of enteropathogenic Escherichia coli (EPEC), atypical EPEC, and Shiga toxin-producing Escherichia coli by a one-step multiplex PCR method. J. Clin. Microbiol. 2006, 44, 2626–2629. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Species | Number of Animals Captured | Salmonella spp. Isolation | E. coli Isolation * |
|---|---|---|---|
| Coyote (Canis latrans) | 15 | 1/15 | 3/15 |
| Gray fox (Urocyon cinereoargenteus) | 6 | 2/6 | 0/6 |
| Desert fox (Vulpes macrotis) | 14 | 0/14 | 2/14 |
| Striped skunk (Mephitis mephitis) | 2 | 1/2 | 0/2 |
| Hooded skunk (Mephitis macroura) | 3 | 0/3 | 2/3 |
| Lynx (Linx rufus) | 5 | 1/5 | 0/5 |
| Raccoon (Procyon lotor) | 4 | 0/4 | 0/4 |
| Badger (Taxidea taxus) | 6 | 0/6 | 0/6 |
| Total animals | 55 | 5/55 (9.09%) | 7/55 (12.72%) |
| Species | Genotyping |
|---|---|
| Gray fox (Urocyon cinereoargenteus) | 2 Salmonella spp. (invA+, sopE+, cdtB+ pegA+, stdA+, lpfA+, stfA+, bcfA+, stiA+, stbA+, steA+, fimA+, sefA+) |
| Striped skunk (Mephitis mephitis) | 1 Salmonella spp. (invA+, sopE+, cdtB+ pegA+, stdA+, lpfA+, stfA+, bcfA+, stiA+, stbA+, steA+, fimA+, sefA+) |
| Coyote (Canis latrans) | 1 Salmonella spp. (invA−, sopE−, cdtB−) |
| Lynx (Linx rufus) | 1 Salmonella spp. (invA−, sopE−, cdtB−) |
| Species | Serotype | Genotyping |
|---|---|---|
| Desert fox (Vulpes macrotis) | 8 E. coli O153H21 9 E. coli O153H7 1 E. coli O175H16 4 E. coli O139H21 1 E. coli O139 H ** | 6 E. coli (eae+) 11 E. coli (escV+) 6 E. coli (escV+, eae+) |
| Coyote (Canis latrans) | 2 E. coli O75H38 1 E. coli O17H18 4 E. coli O21H21 2 E. coli O110H28 13 E. coli O ** H11 | 17 E. coli (stx2+) 5 E. coli (stx1+, stx2+) |
| Hooded skunk (Mephitis macroura) | 7 E. coli O63H6 4 E. coli O96H49 3 E. coli O93H ** 2 E. coli nd | 12 E. coli (escV+) 3 E. coli (stx1+) 1 E. coli (escV+, eae+) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
López-Islas, J.J.; Méndez-Olvera, E.T.; Martínez-Gómez, D.; López-Pérez, A.M.; Orozco, L.; Suzan, G.; Eslava, C. Characterization of Salmonella spp. and E. coli Strains Isolated from Wild Carnivores in Janos Biosphere Reserve, Mexico. Animals 2022, 12, 1064. https://doi.org/10.3390/ani12091064
López-Islas JJ, Méndez-Olvera ET, Martínez-Gómez D, López-Pérez AM, Orozco L, Suzan G, Eslava C. Characterization of Salmonella spp. and E. coli Strains Isolated from Wild Carnivores in Janos Biosphere Reserve, Mexico. Animals. 2022; 12(9):1064. https://doi.org/10.3390/ani12091064
Chicago/Turabian StyleLópez-Islas, Jonathan J., Estela T. Méndez-Olvera, Daniel Martínez-Gómez, Andrés M. López-Pérez, Libertad Orozco, Gerardo Suzan, and Carlos Eslava. 2022. "Characterization of Salmonella spp. and E. coli Strains Isolated from Wild Carnivores in Janos Biosphere Reserve, Mexico" Animals 12, no. 9: 1064. https://doi.org/10.3390/ani12091064
APA StyleLópez-Islas, J. J., Méndez-Olvera, E. T., Martínez-Gómez, D., López-Pérez, A. M., Orozco, L., Suzan, G., & Eslava, C. (2022). Characterization of Salmonella spp. and E. coli Strains Isolated from Wild Carnivores in Janos Biosphere Reserve, Mexico. Animals, 12(9), 1064. https://doi.org/10.3390/ani12091064






