Candidate Genes and Their Expressions Involved in the Regulation of Milk and Meat Production and Quality in Goats (Capra hircus)
Abstract
Simple Summary
Abstract
1. Introduction
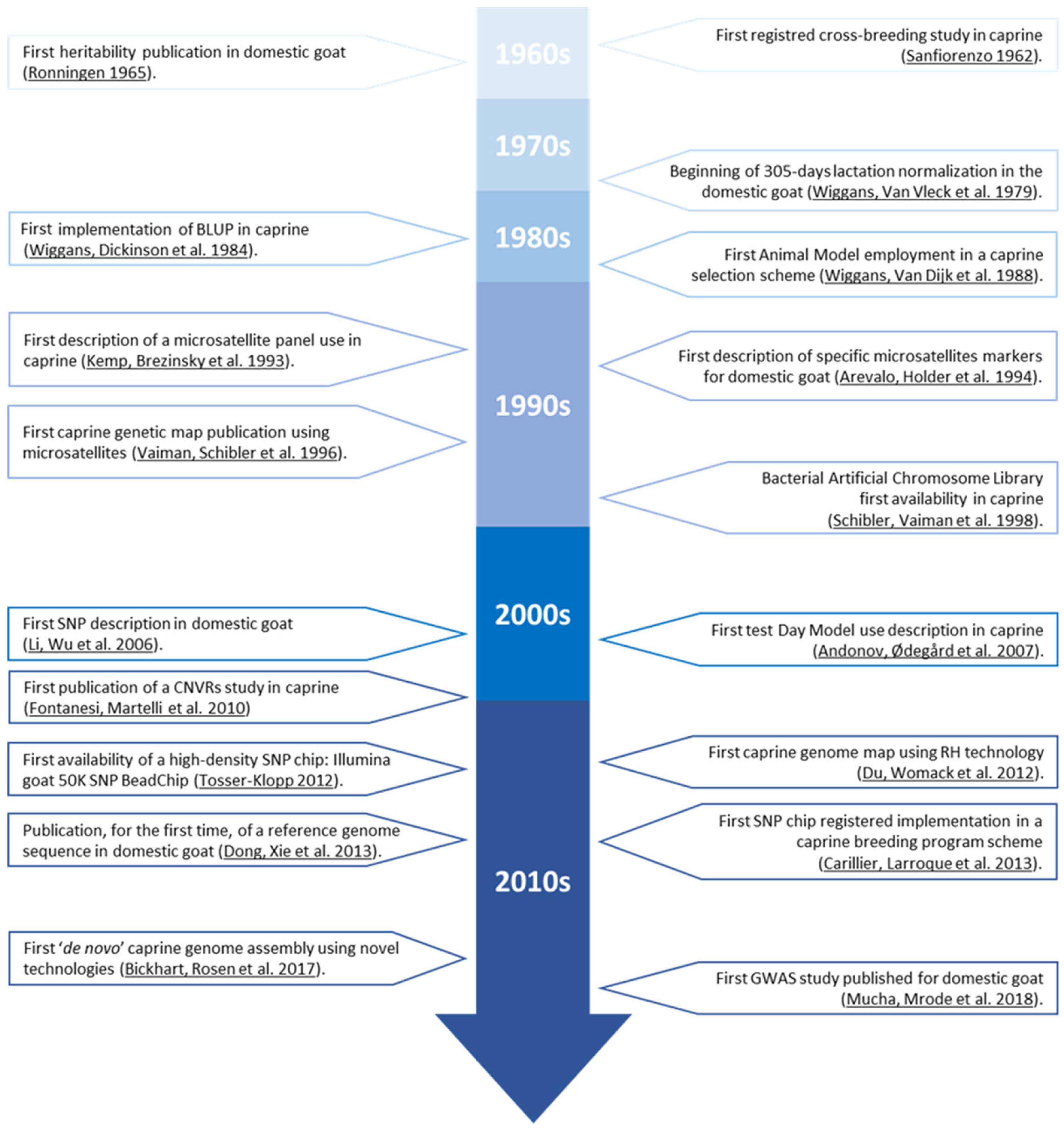
2. Data Collection
3. Candidate Genes Regulating the Expression of Economically Relevant Traits in Caprine
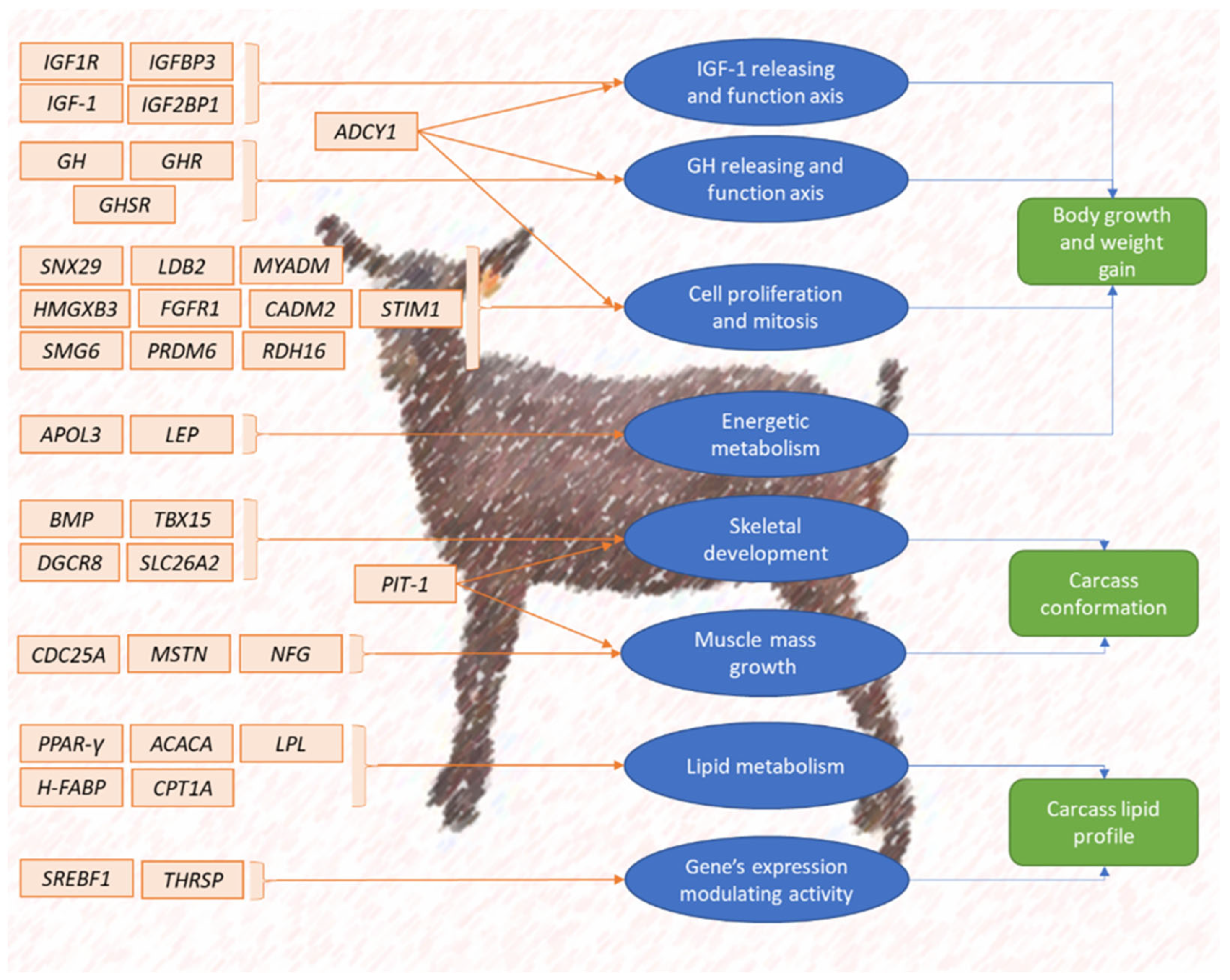
3.1. Candidate Genes Regulating the Expression of Goat Meat Breeding Criteria and Traits
3.1.1. Actual and Potential Candidate Genes Regulating the Expression of Body Growth and Weight Gain
3.1.2. Actual and Potential Candidate Genes Regulating the Expression of Body Size and Carcass Quality
3.1.3. Actual and Potential Candidate Genes Regulating the Expression of Carcass Lipidic Profile
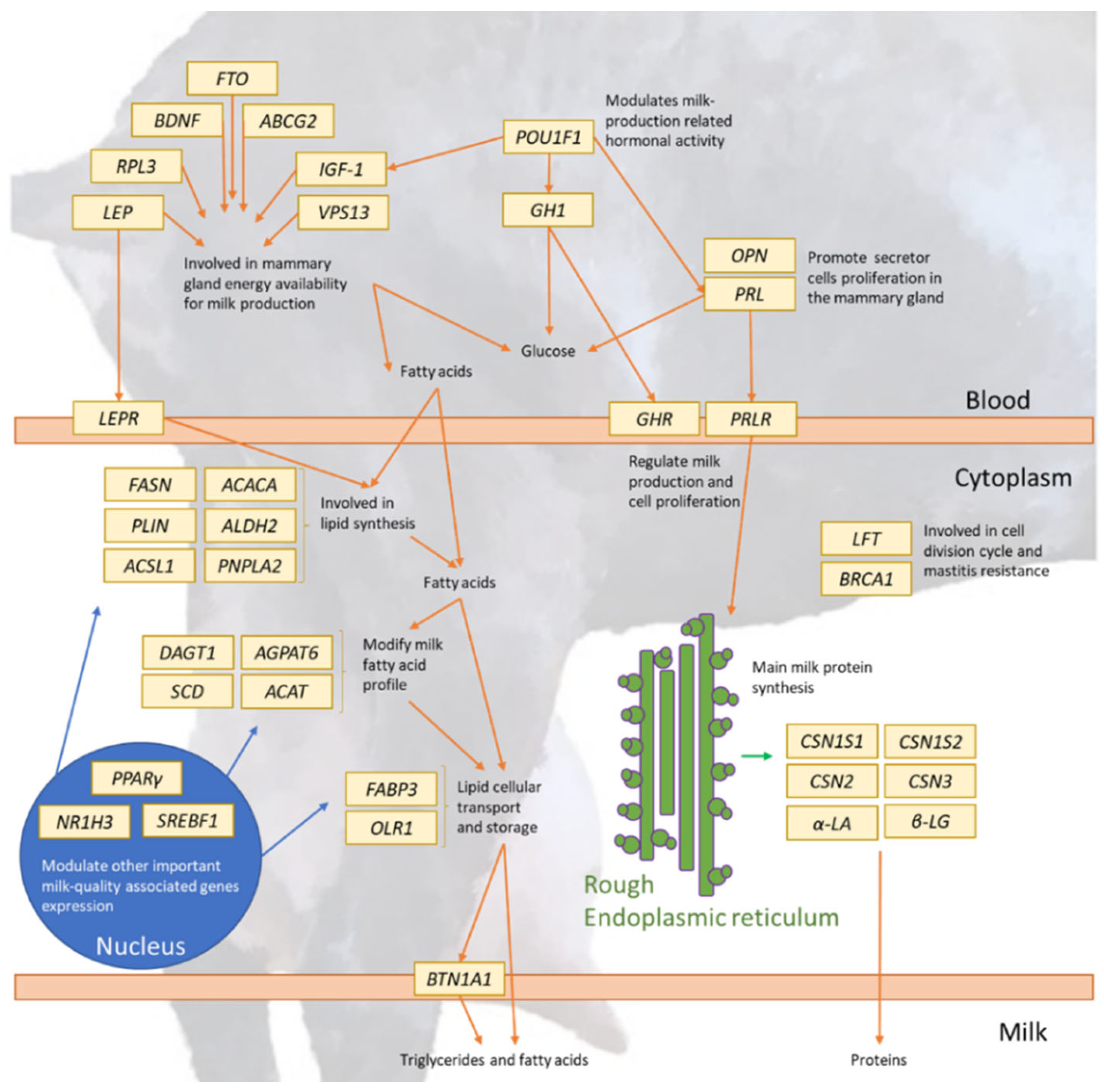
3.2. Candidate Genes Regulating the Expression of Goat Dairy Breeding Criteria and Traits
3.2.1. Actual and Potential Candidate Genes Regulating the Expression of Milk Production/Yield
3.2.2. Actual and Potential Candidate Genes Regulating the Expression of Milk Content/Composition
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Miller, B.A.; Lu, C.D. Current status of global dairy goat production: An overview. Asian Australas. J. Anim. Sci. 2019, 32, 1219. [Google Scholar] [CrossRef] [PubMed]
- Gama, L.; Bressan, M. Biotechnology applications for the sustainable management of goat genetic resources. Small Rumin. Res. 2011, 98, 133–146. [Google Scholar] [CrossRef]
- Amills, M.; Capote, J.; Tosser-Klopp, G. Goat domestication and breeding: A jigsaw of historical, biological and molecular data with missing pieces. Anim. Genet. 2017, 48, 631–644. [Google Scholar] [CrossRef] [PubMed]
- Fahmy, M.H.; Shrestha, J. Genetics for the improvement of goat meat production. In Proceedings of the 7th International Conference on Goats, Tours, France, 15–21 May 2000; pp. 187–190. [Google Scholar]
- Shrestha, J. Genetics and breeding of meat goats. In Goat Meat Production and Quality; Mahgoub, O., Kadim, I., Webb, E., Eds.; CAB International: London, UK, 2012; p. 52. [Google Scholar]
- Mazhangara, I.R.; Chivandi, E.; Mupangwa, J.F.; Muchenje, V. The potential of goat meat in the red meat industry. Sustainability 2019, 11, 3671. [Google Scholar] [CrossRef]
- Daly, K.G.; Delser, P.M.; Mullin, V.E.; Scheu, A.; Mattiangeli, V.; Teasdale, M.D.; Hare, A.J.; Burger, J.; Verdugo, M.P.; Collins, M.J. Ancient goat genomes reveal mosaic domestication in the Fertile Crescent. Science 2018, 361, 85–88. [Google Scholar] [CrossRef] [PubMed]
- Pidancier, N.; Jordan, S.; Luikart, G.; Taberlet, P. Evolutionary history of the genus Capra (Mammalia, Artiodactyla): Discordance between mitochondrial DNA and Y-chromosome phylogenies. Mol. Phylogenet. Evol. 2006, 40, 739–749. [Google Scholar] [CrossRef] [PubMed]
- Mirkena, T.; Duguma, G.; Haile, A.; Tibbo, M.; Okeyo, A.; Wurzinger, M.; Sölkner, J. Genetics of adaptation in domestic farm animals: A review. Livest. Sci. 2010, 132, 1–12. [Google Scholar] [CrossRef]
- Gootwine, E. Genetics and breeding of sheep and goats. In Animal Agriculture; Elsevier: College Station, TX, USA, 2020; pp. 183–198. [Google Scholar]
- Ferrando, A.; Manunza, A.; Jordana, J.; Capote, J.; Pons, A.; Pais, J.; Delgado, T.; Atoche, P.; Cabrera, B.; Martínez, A. A mitochondrial analysis reveals distinct founder effect signatures in Canarian and Balearic goats. Anim. Genet. 2015, 46, 452–456. [Google Scholar] [CrossRef]
- Yalcin, B. Sheep and Goats in Turkey; FAO: Rome, Italy, 1986. [Google Scholar]
- Lauvergne, J.; Renieri, C.; Machado, T.; Acharya, R. Goat genetic resources. In Proceedings of the 5th International Goat Conference, New Delhi, India, 2–8 March 1992. [Google Scholar]
- Jenot, F. Goat unions, FNEC and technical institutes, an intersecting destiny: Trajectories and dynamics since 1950 in France. Ethnozootechnie 2011, 91, 123–133. [Google Scholar]
- Casey, N.; Van Niekerk, W. The Boer goat. I. Origin, adaptability, performance testing, reproduction and milk production. Small Rumin. Res. 1988, 1, 291–302. [Google Scholar] [CrossRef]
- Sanfiorenzo, J.H. Evaluation of six breeding lines of milk goats. J. Agric. Univ. P. R. 1962, 46, 205–212. [Google Scholar] [CrossRef]
- Ronningen, K. Causes of variation in the flavour intensity of goat milk. Acta Agric. Scand. 1965, 15, 301–342. [Google Scholar] [CrossRef]
- Wiggans, G.; Van Vleck, L.D.; Dickinson, F. Projection factors for goat lactation records. J. Dairy Sci. 1979, 62, 797–801. [Google Scholar] [CrossRef]
- Gipson, T.A. Recent advances in breeding and genetics for dairy goats. Asian Australas. J. Anim. Sci. 2019, 32, 1275. [Google Scholar] [CrossRef] [PubMed]
- Wiggans, G.; Dickinson, F.; King, G.; Weller, J. Genetic evaluation of dairy goat bucks for daughter milk and fat. J. Dairy Sci. 1984, 67, 201–207. [Google Scholar] [CrossRef]
- Wiggans, G.; Van Dijk, J.; Misztal, I. Genetic evaluation of dairy goats for milk and fat yield with an animal model. J. Dairy Sci. 1988, 71, 1330–1337. [Google Scholar] [CrossRef]
- Kemp, S.J.; Brezinsky, L.; Teale, A. A panel of bovine, ovine and caprine polymorphic microsatellites. Anim. Genet. 1993, 24, 363–365. [Google Scholar] [CrossRef]
- Vaiman, D.; Schibler, L.; Bourgeois, F.; Oustry, A.; Amigues, Y.; Cribiu, E.P. A genetic linkage map of the male goat genome. Genetics 1996, 144, 279–305. [Google Scholar] [CrossRef]
- Arevalo, E.; Holder, D.; Derr, J.; Bhebhe, E.; Linn, R. Caprine microsatellite dinucleotide repeat polymorphisms at the Sr-Crsp-1, Sr-Crsp-2, Sr-Crsp-3, Sr-Crsp-4 and Sr-Crsp-5 loci. Anim. Genet. 1994, 25, 202. [Google Scholar] [CrossRef]
- Schibler, L.; Vaiman, D.; Oustry, A.; Guinec, N.; Dangy-Caye, A.L.; Billault, A.; Cribiu, E.P. Construction and extensive characterization of a goat bacterial artificial chromosome library with threefold genome coverage. Mamm. Genome 1998, 9, 119–124. [Google Scholar] [CrossRef]
- Mucha, S.; Mrode, R.; Coffey, M.; Kizilaslan, M.; Desire, S.; Conington, J. Genome-wide association study of conformation and milk yield in mixed-breed dairy goats. J. Dairy Sci. 2018, 101, 2213–2225. [Google Scholar] [CrossRef] [PubMed]
- Bickhart, D.M.; Rosen, B.D.; Koren, S.; Sayre, B.L.; Hastie, A.R.; Chan, S.; Lee, J.; Lam, E.T.; Liachko, I.; Sullivan, S.T. Single-molecule sequencing and chromatin conformation capture enable de novo reference assembly of the domestic goat genome. Nat. Genet. 2017, 49, 643–650. [Google Scholar] [CrossRef]
- Carillier, C.; Larroque, H.; Palhière, I.; Clément, V.; Rupp, R.; Robert-Granié, C. A first step toward genomic selection in the multi-breed French dairy goat population. J. Dairy Sci. 2013, 96, 7294–7305. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.; Xie, M.; Jiang, Y.; Xiao, N.; Du, X.; Zhang, W.; Tosser-Klopp, G.; Wang, J.; Yang, S.; Liang, J. Sequencing and automated whole-genome optical mapping of the genome of a domestic goat (Capra hircus). Nat. Biotechnol. 2013, 31, 135–141. [Google Scholar] [CrossRef] [PubMed]
- Tosser-Klopp, G. Goat genome assembly, Availability of an international 50K SNP chip and RH panel: An update of the International Goat Genome Consortium projects. In Proceedings of the International Plant and Animal Genome XX Conference, Town and Country Hotel, San Diego, CA, USA, 14–18 January 2012. [Google Scholar]
- Fontanesi, L.; Martelli, P.L.; Beretti, F.; Riggio, V.; Dall’Olio, S.; Colombo, M.; Casadio, R.; Russo, V.; Portolano, B. An initial comparative map of copy number variations in the goat (Capra hircus) genome. BMC Genom. 2010, 11, 639. [Google Scholar] [CrossRef]
- Li, X.-L.; Wu, Z.-L.; Liu, Z.-Z.; Gong, Y.-F.; Zhou, R.-Y.; Zheng, G.-R. SNP identification and analysis in part of intron 2 of goat MSTN gene and variation within and among species. J. Hered. 2006, 97, 285–289. [Google Scholar] [CrossRef]
- Andonov, S.; Ødegård, J.; Svendsen, M.; Ådnøy, T.; Vegara, M.; Klemetsdal, G. Comparison of random regression and repeatability models to predict breeding values from test-day records of Norwegian goats. J. Dairy Sci. 2013, 96, 1834–1843. [Google Scholar] [CrossRef]
- Gebreselassie, G.; Berihulay, H.; Jiang, L.; Ma, Y. Review on genomic regions and candidate genes associated with economically important production and reproduction traits in sheep (Ovies aries). Animals 2020, 10, 33. [Google Scholar] [CrossRef]
- Zhang, H.; Wang, Z.; Wang, S.; Li, H. Progress of genome wide association study in domestic animals. J. Anim. Sci. Biotechnol. 2012, 3, 26. [Google Scholar] [CrossRef] [PubMed]
- Lashmar, S.; Visser, C.; van Marle-Köster, E. Validation of the 50k Illumina goat SNP chip in the South African Angora goat. S. Afr. J. Anim. Sci. 2015, 45, 56–59. [Google Scholar] [CrossRef]
- Li, X.; Wu, Z.L.; Gong, Y.; Liu, Y.; Liu, Z.; Wang, X.; Xin, T.; Ji, Q. Single-nucleotide polymorphism identification in the caprine myostatin gene. J. Anim. Breed. 2006, 123, 141–144. [Google Scholar] [CrossRef] [PubMed]
- Andonov, S.; Ødegård, J.; Boman, I.; Svendsen, M.; Holme, I.; Ådnøy, T.; Vukovic, V.; Klemetsdal, G. Validation of test-day models for genetic evaluation of dairy goats in Norway. J. Dairy Sci. 2007, 90, 4863–4871. [Google Scholar] [CrossRef] [PubMed]
- Pizarro Inostroza, M.G.; Navas González, F.J.; Landi, V.; León Jurado, J.M.; Delgado Bermejo, J.V.; Fernández Álvarez, J.; Martínez Martínez, M.d.A. Software-Automatized Individual Lactation Model Fitting, Peak and Persistence and Bayesian Criteria Comparison for Milk Yield Genetic Studies in Murciano-Granadina Goats. Mathematics 2020, 8, 1505. [Google Scholar] [CrossRef]
- Tosser-Klopp, G.; Bardou, P.; Bouchez, O.; Cabau, C.; Crooijmans, R.; Dong, Y.; Donnadieu-Tonon, C.; Eggen, A.; Heuven, H.C.; Jamli, S. Design and characterization of a 52K SNP chip for goats. PLoS ONE 2014, 9, e86227. [Google Scholar] [CrossRef] [PubMed]
- Onzima, R.; Upadhyay, M.; Mukiibi, R.; Kanis, E.; Groenen, M.; Crooijmans, R. Genome-wide population structure and admixture analysis reveals weak differentiation among Ugandan goat breeds. Anim. Genet. 2018, 49, 59–70. [Google Scholar] [CrossRef]
- Schibler, L.; Vaiman, D.; Oustry, A.; Giraud-Delville, C.; Cribiu, E.P. Comparative gene mapping: A fine-scale survey of chromosome rearrangements between ruminants and humans. Genome Res. 1998, 8, 901–915. [Google Scholar] [CrossRef]
- Du, X.; Womack, J.; Owens, K.; Elliott, J.; Sayre, B.; Bottcher, P.; Milan, D.; Podesta, M.G.; Zhao, S.; Malek, M. A whole-genome radiation hybrid panel for goat. Small Rumin. Res. 2012, 105, 114–116. [Google Scholar] [CrossRef]
- Hirschhorn, J.N.; Daly, M.J. Genome-wide association studies for common diseases and complex traits. Nat. Rev. Genet. 2005, 6, 95–108. [Google Scholar] [CrossRef]
- González Ariza, A.; Arando Arbulu, A.; Navas González, F.J.; Nogales Baena, S.; Delgado Bermejo, J.V.; Camacho Vallejo, M.E. The Study of Growth and Performance in Local Chicken Breeds and Varieties: A Review of Methods and Scientific Transference. Animals 2021, 11, 2492. [Google Scholar] [CrossRef]
- McLean, A.K.; Navas Gonzalez, F.J. Can scientists influence donkey welfare? Historical perspective and a contemporary view. J. Equine Vet. Sci. 2018, 65, 25–32. [Google Scholar] [CrossRef]
- Iglesias Pastrana, C.; Navas González, F.J.; Ciani, E.; Barba Capote, C.J.; Delgado Bermejo, J.V. Effect of research impact on emerging camel husbandry, welfare and social-related awareness. Animals 2020, 10, 780. [Google Scholar] [CrossRef] [PubMed]
- Zonaed Siddiki, A.; Miah, G.; Islam, M.; Kumkum, M.; Rumi, M.H.; Baten, A.; Hossain, M.A. Goat Genomic Resources: The Search for Genes Associated with Its Economic Traits. Int. J. Genom. 2020, 2020, 5940205. [Google Scholar] [CrossRef] [PubMed]
- Webb, E.; Casey, N.; Simela, L. Growth, development and growth manipulation in goats. In Goat Meat Production and Quality; Mahgoub, O., Kadim, I.T., Cottington Webb, E., Ovid Technologies, Inc., Eds.; CAB International: Cambridge, MA, USA, 2012; pp. 196–199. [Google Scholar]
- Shrestha, J.; Fahmy, M. Breeding goats for meat production: 3. Selection and breeding strategies. Small Rumin. Res. 2007, 67, 113–125. [Google Scholar] [CrossRef]
- Shrestha, J.; Fahmy, M. Breeding goats for meat production: 2. Crossbreeding and formation of composite population. Small Rumin. Res. 2007, 67, 93–112. [Google Scholar] [CrossRef]
- Fernández Álvarez, J.; León Jurado, J.; Navas González, F.J.; Iglesias Pastrana, C.; Delgado Bermejo, J.V. CAPRIGRAN Linear Appraisal Evidences Dairy Selection Signs in Murciano-Granadina Goats and Bucks: Presentation of the New Linear Appraisal Scale. Arch. Zootecn. 2021, 70, 240–245. [Google Scholar] [CrossRef]
- Webb, E.; Casey, N.; Simela, L. Goat meat quality. Small Rumin. Res. 2005, 60, 153–166. [Google Scholar] [CrossRef]
- Webb, E. Goat meat production, composition, and quality. Anim. Front. 2014, 4, 33–37. [Google Scholar] [CrossRef]
- Raes, K.; Balcaen, A.; Dirinck, P.; De Winne, A.; Claeys, E.; Demeyer, D.; De Smet, S. Meat quality, fatty acid composition and flavour analysis in Belgian retail beef. Meat Sci. 2003, 65, 1237–1246. [Google Scholar] [CrossRef]
- Herrera, P.Z.; Bermejo, J.V.D.; Henríquez, A.A.; Vallejo, M.E.C.; Costa, R.G. Effects of extensive system versus semi-intensive and intensive systems on growth and carcass quality of dairy kids. Rev. Bras. Zootec. 2011, 40, 2613–2620. [Google Scholar] [CrossRef][Green Version]
- Jembere, T.; Dessie, T.; Rischkowsky, B.; Kebede, K.; Okeyo, A.; Haile, A. Meta-analysis of average estimates of genetic parameters for growth, reproduction and milk production traits in goats. Small Rumin. Res. 2017, 153, 71–80. [Google Scholar] [CrossRef]
- Huson, H.; Sonstegard, T.; Silverstein, J.; Woodward Greene, J.; Masiga, C.; Muchadeyi, F.; Rees, J.; Sayre, B.; Elbeltagy, A.; Rothschild, M.F. Genetic and phenotypic characterization of African goat populations to prioritize conservation and production efforts for small-holder farmers in Sub-Saharan africa. In Proceedings of the 10th World Congress on Genetics Applied to Livestock Production, Vancouver, BC, Canada, 17–22 August 2014; American Society of Animal Science: Champaign, IL, USA, 2014. [Google Scholar]
- Zhang, N.; Teng, Z.; Qi, Q.; Hu, G.; Lian, H.; Gao, T. Carcass traits, meat quality characteristics, and lipid metabolism-related gene expression pattern of Yaoshan white goats raised in traditional extensive production system: Effects of slaughter age and meat cuts. Small Rumin. Res. 2020, 182, 29–36. [Google Scholar] [CrossRef]
- Liu, M.; Woodward-Greene, J.; Kang, X.; Pan, M.G.; Rosen, B.; Van Tassell, C.P.; Chen, H.; Liu, G.E. Genome-wide CNV analysis revealed variants associated with growth traits in African indigenous goats. Genomics 2020, 112, 1477–1480. [Google Scholar] [CrossRef] [PubMed]
- Yarwood, S.J.; Kilgour, E.; Anderson, N.G. Cyclic AMP potentiates growth hormone-dependent differentiation of 3T3-F442A preadipocytes: Possible involvement of the transcription factor CREB. Mol. Cell. Endocrinol. 1998, 138, 41–50. [Google Scholar] [CrossRef]
- Harwood, J.P.; Grewe, C.; Aguilera, G. Actions of growth hormone-releasing factor and somatostatin on adenylate cyclase and growth hormone release in rat anterior pituitary. Mol. Cell. Endocrinol. 1984, 37, 277–284. [Google Scholar] [CrossRef]
- Bi, Y.; Chen, Y.; Xin, D.; Liu, T.; He, L.; Kang, Y.; Pan, C.; Shen, W.; Lan, X.; Liu, M. Effect of indel variants within the sorting nexin 29 (SNX29) gene on growth traits of goats. Anim. Biotechnol. 2020, 19, 1–6. [Google Scholar] [CrossRef]
- Hägg, S.; Skogsberg, J.; Lundström, J.; Noori, P.; Nilsson, R.; Zhong, H.; Maleki, S.; Shang, M.M.; Brinne, B.; Bradshaw, M. Multi-organ expression profiling uncovers a gene module in coronary artery disease involving transendothelial migration of leukocytes and LIM domain binding 2: The Stockholm Atherosclerosis Gene Expression (STAGE) study. PLoS Genet. 2009, 5, e1000754. [Google Scholar] [CrossRef]
- Liu, R.; Sun, Y.; Zhao, G.; Wang, F.; Wu, D.; Zheng, M.; Chen, J.; Zhang, L.; Hu, Y.; Wen, J. Genome-wide association study identifies loci and candidate genes for body composition and meat quality traits in Beijing-You chickens. PLoS ONE 2013, 8, e61172. [Google Scholar] [CrossRef]
- Gonzalez, M.V.; Mousel, M.R.; Herndon, D.R.; Jiang, Y.; Dalrymple, B.P.; Reynolds, J.O.; Johnson, W.C.; Herrmann-Hoesing, L.M.; White, S.N. A divergent Artiodactyl MYADM-like repeat is associated with erythrocyte traits and weight of lamb weaned in domestic sheep. PLoS ONE 2013, 8, e74700. [Google Scholar] [CrossRef]
- Hoopes, B.C.; Rimbault, M.; Liebers, D.; Ostrander, E.A.; Sutter, N.B. The insulin-like growth factor 1 receptor (IGF1R) contributes to reduced size in dogs. Mamm. Genome 2012, 23, 780–790. [Google Scholar] [CrossRef]
- Bickhart, D.M.; Hou, Y.; Schroeder, S.G.; Alkan, C.; Cardone, M.F.; Matukumalli, L.K.; Song, J.; Schnabel, R.D.; Ventura, M.; Taylor, J.F. Copy number variation of individual cattle genomes using next-generation sequencing. Genome Res. 2012, 22, 778–790. [Google Scholar] [CrossRef]
- Wu, J.; Peng, X.; Li, F.; Qiao, M.; Wu, H.; Mei, S. Association of porcine STIM1 gene with litter size traits. Anim. Sci. Pap. Rep. 2016, 34, 165–173. [Google Scholar]
- Zhang, B.; Chang, L.; Lan, X.; Asif, N.; Guan, F.; Fu, D.; Li, B.; Yan, C.; Zhang, H.; Zhang, X. Genome-wide definition of selective sweeps reveals molecular evidence of trait-driven domestication among elite goat (Capra species) breeds for the production of dairy, cashmere, and meat. GigaScience 2018, 7, giy105. [Google Scholar] [CrossRef] [PubMed]
- Granados-Principal, S.; Liu, Y.; Rodriguez-Aguayo, C.; Sood, A.K.; Chang, J.C. HMG Box Domain Containing 3 (HMGXB3), a novel TGFβ-induced cancer gene that influences cancer stem cells and metastasis in triple negative breast cancer. Cancer Res. 2015, 75, 4086. [Google Scholar]
- Li, M.Y.; Min, L.J.; Sun, G.Q.; Pan, Q.J.; Shen, W.; Wang, G.L. Polymorphism analysis of the goat growth hormone gene in the 5’regulatory sequence. Hereditas 2004, 26, 831–835. [Google Scholar] [PubMed]
- Jin, Q.; Fang, X.T.; Yang, L.; Zhang, C.L.; Sun, J.J.; Chen, D.X.; Shi, X.; Du, Y.; Lei, C.; Chen, H. Novel SNPs of the caprine growth hormone secretagogue receptor (GHSR) gene and their association with growth traits in goats. Biochem. Genet. 2010, 48, 847–856. [Google Scholar] [CrossRef] [PubMed]
- Lucy, M.; Johnson, G.; Shibuya, H.; Boyd, C.; Herring, W. Rapid communication: Polymorphic (GT) n microsatellite in the bovine somatotropin gene promoter. J. Anim. Sci. 1998, 76, 2209. [Google Scholar] [CrossRef]
- Mullen, M.P.; Berry, D.P.; Howard, D.J.; Diskin, M.G.; Lynch, C.O.; Giblin, L.; Kenny, D.A.; Magee, D.A.; Meade, K.G.; Waters, S.M. Single nucleotide polymorphisms in the insulin-like growth factor 1 (IGF-1) gene are associated with performance in Holstein-Friesian dairy cattle. Front. Genet. 2011, 2, 3. [Google Scholar] [CrossRef]
- Van Laere, A.-S.; Nguyen, M.; Braunschweig, M.; Nezer, C.; Collette, C.; Moreau, L.; Archibald, A.L.; Haley, C.S.; Buys, N.; Tally, M. A regulatory mutation in IGF2 causes a major QTL effect on muscle growth in the pig. Nature 2003, 425, 832–836. [Google Scholar] [CrossRef]
- Bale, L.K.; Conover, C.A. Regulation of insulin-like growth factor binding protein-3 messenger ribonucleic acid expression by insulin-like growth factor I. Endocrinology 1992, 131, 608–614. [Google Scholar]
- Ji, W.; Yu, Y.; Li, Z.; Wang, G.; Li, F.; Xia, W.; Lu, S. FGFR1 promotes the stem cell-like phenotype of FGFR1-amplified non-small cell lung cancer cells through the Hedgehog pathway. Oncotarget 2016, 7, 15118. [Google Scholar] [CrossRef]
- Fang, X.; Xu, H.; Zhang, C.; Chen, H.; Hu, X.; Gao, X.; Gu, C.; Yue, W. Polymorphism in BMP4 gene and its association with growth traits in goats. Mol. Biol. Rep. 2009, 36, 1339–1344. [Google Scholar] [CrossRef] [PubMed]
- Chu, M.; Jiao, C.; He, Y.; Wang, J.; Liu, Z.; Chen, G. Association between PCR-SSCP of bone morphogenetic protein 15 gene and prolificacy in Jining Grey goats. Anim. Biotechnol. 2007, 18, 263–274. [Google Scholar] [CrossRef] [PubMed]
- García-de Teresa, B.; Hernández-Gómez, M.; Frías, S. DNA damage as a driver for growth delay: Chromosome instability syndromes with intrauterine growth retardation. BioMed Res. Int. 2017, 2017, 8193892. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Wang, X.; Zhang, Z.; An, Q.; Wen, Y.; Wang, D.; Liu, X.; Li, Z.; Lyu, S.; Li, L. Copy number variation of CADM2 gene revealed its association with growth traits across Chinese Capra hircus (goat) populations. Gene 2020, 741, 144519. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Tao, H.; Li, P.; Li, L.; Zhong, T.; Wang, L.; Ma, J.; Chen, X.; Song, T.; Zhang, H. Whole-genome sequencing reveals selection signatures associated with important traits in six goat breeds. Sci. Rep. 2018, 8, 10405. [Google Scholar] [CrossRef]
- Dong, Y.; Zhang, X.; Xie, M.; Arefnezhad, B.; Wang, Z.; Wang, W.; Feng, S.; Huang, G.; Guan, R.; Shen, W. Reference genome of wild goat (Capra aegagrus) and sequencing of goat breeds provide insight into genic basis of goat domestication. BMC Genom. 2015, 16, 431. [Google Scholar] [CrossRef]
- Supakorn, C. The important candidate genes in goats—A review. Walailak J. Sci. Technol. 2009, 6, 17–36. [Google Scholar]
- Naicy, T.; Venkatachalapathy, R.; Aravindakshan, T.; Kurian, E. Association of a Cac8I polymorphism in the IGF1 gene with growth traits in Indian goats. J. Genet. Eng. Biotechnol. 2017, 15, 7–11. [Google Scholar] [CrossRef]
- Wang, Z.; Zhang, X.; Jiang, E.; Yan, H.; Zhu, H.; Chen, H.; Liu, J.; Qu, L.; Pan, C.; Lan, X. InDels within caprine IGF 2 BP 1 intron 2 and the 3′-untranslated regions are associated with goat growth traits. Anim. Genet. 2020, 51, 117–121. [Google Scholar] [CrossRef]
- Wang, Z.; Wang, C.; Guo, Y.; She, S.; Wang, B.; Jiang, Y.; Bai, Y.; Song, X.; Li, L.; Shi, L. Screening of deletion variants within the goat PRDM6 gene and its effects on growth traits. Animals 2020, 10, 208. [Google Scholar] [CrossRef]
- Naicy, T.; Venkatachalapathy, T.; Aravindakshan, T.; Bosewell, A.; Silpa, M. Association of a SacII polymorphism in the Nerve Growth Factor (NGF) gene exon 3 with growth traits in Indian goats. Small Rumin. Res. 2018, 158, 19–21. [Google Scholar] [CrossRef]
- Lan, X.; Pan, C.; Chen, H.; Zhang, C.; Li, J.; Zhao, M.; Lei, C.; Zhang, A.; Zhang, L. An AluI PCR-RFLP detecting a silent allele at the goat POU1F1 locus and its association with production traits. Small Rumin. Res. 2007, 73, 8–12. [Google Scholar] [CrossRef]
- Renaville, R.; Gengler, N.; Vrech, E.; Prandi, A.; Massart, S.; Corradini, C.; Bertozzi, C.; Mortiaux, F.; Burny, A.; Portetelle, D. Pit-1 gene polymorphism, milk yield, and conformation traits for Italian Holstein-Friesian bulls. J. Dairy Sci. 1997, 80, 3431–3438. [Google Scholar] [CrossRef]
- Houseknecht, K.L.; Baile, C.A.; Matteri, R.L.; Spurlock, M.E. The biology of leptin: A review. J. Anim. Sci. 1998, 76, 1405–1420. [Google Scholar] [CrossRef] [PubMed]
- Barzekar, R.; Salehi, A.; Mahjoubi, F. Polymorphisms of the ovine leptin gene and its association with growth and carcass traits in three Iranian sheep breeds. Iran. J. Biotechnol. 2009, 7, 241–246. [Google Scholar]
- Singh, M.K.; Petry, M.; Haenig, B.; Lescher, B.; Leitges, M.; Kispert, A. The T-box transcription factor Tbx15 is required for skeletal development. Mech. Dev. 2005, 122, 131–144. [Google Scholar] [CrossRef]
- Sugatani, T.; Hildreth III, B.E.; Toribio, R.E.; Malluche, H.H.; Hruska, K.A. Expression of DGCR8-dependent microRNAs is indispensable for osteoclastic development and bone-resorbing activity. J. Cell. Biochem. 2014, 115, 1043–1047. [Google Scholar] [CrossRef]
- Sarkar, S.; Dey, B.K.; Dutta, A. MiR-322/424 and-503 are induced during muscle differentiation and promote cell cycle quiescence and differentiation by down-regulation of Cdc25A. Mol. Biol. Cell. 2010, 21, 2138–2149. [Google Scholar] [CrossRef]
- Barreda-Bonis, A.C.; Barraza-García, J.; Parrón, M.; Pastor, I.; Heath, K.E.; González-Casado, I. Multiple SLC26A2 mutations occurring in a three-generational family. Eur. J. Med. Genet. 2018, 61, 24–28. [Google Scholar] [CrossRef]
- Wang, X.; Liu, J.; Zhou, G.; Guo, J.; Yan, H.; Niu, Y.; Li, Y.; Yuan, C.; Geng, R.; Lan, X. Whole-genome sequencing of eight goat populations for the detection of selection signatures underlying production and adaptive traits. Sci. Rep. 2016, 6, 38932. [Google Scholar] [CrossRef]
- Liu, S.H.; Chiu, C.Y.; Wang, L.P.; Chiang, M.T. Omega-3 fatty acids-enriched fish oil activates AMPK/PGC-1α signaling and prevents obesity-related skeletal muscle wasting. Mar. Drugs 2019, 17, 380. [Google Scholar] [CrossRef] [PubMed]
- Oh, D.; La, B.; Lee, Y.; Byun, Y.; Lee, J.; Yeo, G.; Yeo, J. Identification of novel single nucleotide polymorphisms (SNPs) of the lipoprotein lipase (LPL) gene associated with fatty acid composition in Korean cattle. Mol. Biol. Rep. 2013, 40, 3155–3163. [Google Scholar] [CrossRef] [PubMed]
- Shin, S.C.; Heo, J.P.; Chung, E.R. Effect of Single Nucleotide Polymorphisms of Acetyl-CoA Carboxylase α (ACACA) gene on carcass traits in Hanwoo (Korean cattle). Asian Australas. J. Anim. Sci. 2011, 24, 744–751. [Google Scholar] [CrossRef]
- Eberlé, D.; Hegarty, B.; Bossard, P.; Ferré, P.; Foufelle, F. SREBP transcription factors: Master regulators of lipid homeostasis. Biochimie 2004, 86, 839–848. [Google Scholar] [CrossRef]
- Yang, C.; Liu, J.; Wu, X.; Bao, P.; Long, R.; Guo, X.; Ding, X.; Yan, P. The response of gene expression associated with lipid metabolism, fat deposition and fatty acid profile in the longissimus dorsi muscle of Gannan yaks to different energy levels of diets. PLoS ONE 2017, 12, e0187604. [Google Scholar] [CrossRef] [PubMed]
- Shimano, H. Sterol regulatory element-binding proteins (SREBPs): Transcriptional regulators of lipid synthetic genes. Prog. Lipid Res. 2001, 40, 439–452. [Google Scholar] [CrossRef]
- Oh, D.Y.; Lee, Y.S.; La, B.M.; Lee, J.Y.; Park, Y.S.; Lee, J.H.; Ha, J.J.; Yi, J.K.; Kim, B.K.; Yeo, J.S. Identification of exonic nucleotide variants of the thyroid hormone responsive protein gene associated with carcass traits and Fatty Acid composition in korean cattle. Asian-Australas. J. Anim. Sci. 2014, 27, 1373. [Google Scholar] [CrossRef]
- van der Leij, F.R.; Huijkman, N.C.; Boomsma, C.; Kuipers, J.R.; Bartelds, B. Genomics of the human carnitine acyltransferase genes. Mol. Genet. Metab. 2000, 71, 139–153. [Google Scholar] [CrossRef]
- Sodhi, S.S.; Park, W.C.; Ghosh, M.; Kim, J.N.; Sharma, N.; Shin, K.Y.; Cho, I.C.; Ryu, Y.C.; Oh, S.J.; Kim, S.H. Comparative transcriptomic analysis to identify differentially expressed genes in fat tissue of adult Berkshire and Jeju Native Pig using RNA-seq. Mol. Biol. Rep. 2014, 41, 6305–6315. [Google Scholar] [CrossRef]
- Wei-Jun, P.; Liang, B.; Gong-She, Y. Relationship among H-FABP gene polymorphism, intramuscular fat content, and adipocyte lipid droplet content in main pig breeds with different genotypes in western China. Acta Genet. Sin. 2006, 33, 515–524. [Google Scholar]
- Benjelloun, B.; Alberto, F.J.; Streeter, I.; Boyer, F.; Coissac, E.; Stucki, S.; BenBati, M.; Ibnelbachyr, M.; Chentouf, M.; Bechchari, A. Characterizing neutral genomic diversity and selection signatures in indigenous populations of Moroccan goats (Capra hircus) using WGS data. Front. Genet. 2015, 6, 107. [Google Scholar] [CrossRef] [PubMed]
- FAO. FAOSTAT Statistical Database. 2019. Available online: http://www.fao.org/faostat/en/ (accessed on 17 July 2021).
- García, V.; Rovira, S.; Boutoial, K.; López, M. Improvements in goat milk quality: A review. Small Rumin. Res. 2014, 121, 51–57. [Google Scholar] [CrossRef]
- Pazzola, M.; Stocco, G.; Dettori, M.L.; Bittante, G.; Vacca, G.M. Effect of goat milk composition on cheesemaking traits and daily cheese production. J. Dairy Sci. 2019, 102, 3947–3955. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Yu, Q.; Zhang, Q.; Yang, Q. Construction of mammary gland specific expression plasmid pIN and its expression in vitro and in vivo. Afr. J. Biotechnol. 2012, 11, 6946–6955. [Google Scholar]
- Lemley, C.; Butler, S.; Butler, W.; Wilson, M. Insulin alters hepatic progesterone catabolic enzymes cytochrome P450 2C and 3A in dairy cows. J. Dairy Sci. 2008, 91, 641–645. [Google Scholar] [CrossRef] [PubMed]
- Capitan, A.; Michot, P.; Baur, A.; Saintilan, R.; Hoze, C.; Valour, D.; Guillaume, F.; Boichon, D.; Barbat, A.; Boichard, D. Genetic tools to improve reproduction traits in dairy cattle. Reprod. Fert. Dev. 2015, 27, 14–21. [Google Scholar] [CrossRef] [PubMed]
- Liefers, S.; Te Pas, M.; Veerkamp, R.; Van Der Lende, T. Associations between leptin gene polymorphisms and production, live weight, energy balance, feed intake, and fertility in Holstein heifers. J. Dairy Sci. 2002, 85, 1633–1638. [Google Scholar] [CrossRef]
- Banos, G.; Woolliams, J.; Woodward, B.; Forbes, A.; Coffey, M. Impact of single nucleotide polymorphisms in leptin, leptin receptor, growth hormone receptor, and diacylglycerol acyltransferase (DGAT1) gene loci on milk production, feed, and body energy traits of UK dairy cows. J. Dairy Sci. 2008, 91, 3190–3200. [Google Scholar] [CrossRef] [PubMed]
- Ron, M.; Cohen-Zinder, M.; Peter, C.; Weller, J.; Erhardt, G. A polymorphism in ABCG2 in Bos indicus and Bos taurus cattle breeds. J. Dairy Sci. 2006, 89, 4921–4923. [Google Scholar] [CrossRef]
- Rahmatalla, S.A.; Müller, U.; Strucken, E.M.; Reissmann, M.; Brockmann, G.A. The F279Y polymorphism of the GHR gene and its relation to milk production and somatic cell score in German Holstein dairy cattle. J. Appl. Genet. 2011, 52, 459–465. [Google Scholar] [CrossRef]
- Dybus, A. Associations of growth hormone (GH) and prolactin (PRL) genes polymorphisms with milk production traits in Polish Black-and-White cattle. Anim. Sci. Pap. Rep. 2002, 20, 203–212. [Google Scholar]
- Lü, A.; Hu, X.; Chen, H.; Dong, Y.; Zhang, Y.; Wang, X. Novel SNPs of the bovine PRLR gene associated with milk production traits. Biochem. Genet. 2011, 49, 177–189. [Google Scholar] [CrossRef] [PubMed]
- Wesolowski, S.R.; Allan, M.F.; Nielsen, M.K.; Pomp, D. Evaluation of hypothalamic gene expression in mice divergently selected for heat loss. Physiol. Genom. 2003, 13, 129–137. [Google Scholar] [CrossRef] [PubMed]
- Leonard, S.; Khatib, H.; Schutzkus, V.; Chang, Y.; Maltecca, C. Effects of the osteopontin gene variants on milk production traits in dairy cattle. J. Dairy Sci. 2005, 88, 4083–4086. [Google Scholar] [CrossRef]
- Mullen, M.P.; Lynch, C.; Waters, S.; Howard, D.J.; O’boyle, P.; Kenny, D.; Buckley, F.; Horan, B.; Diskin, M. Single nucleotide polymorphisms in the growth hormone and insulin-like growth factor-1 genes are associated with milk production, body condition score and fertility traits in dairy cows. Genet. Mol. Res. 2011, 26, 1819–1830. [Google Scholar] [CrossRef]
- Ward, P.; Paz, E.; Conneely, O. Lactoferrin. Cell. Mol. Life Sci. 2005, 62, 2540–2548. [Google Scholar] [CrossRef]
- Yuan, Z.; Chu, G.; Dan, Y.; Li, J.; Zhang, L.; Gao, X.; Gao, H.; Li, J.; Xu, S.; Liu, Z. BRCA1: A new candidate gene for bovine mastitis and its association analysis between single nucleotide polymorphisms and milk somatic cell score. Mol. Biol. Rep. 2012, 39, 6625–6631. [Google Scholar] [CrossRef]
- Moioli, B.; D’Andrea, M.; Pilla, F. Candidate genes affecting sheep and goat milk quality. Small Rumin. Res. 2007, 68, 179–192. [Google Scholar] [CrossRef]
- Dagnachew, B.S.; Ådnøy, T. Additive and dominance effects of casein haplotypes on milk composition and quality in Norwegian dairy goats. Small Rumin. Res. 2014, 122, 59–69. [Google Scholar] [CrossRef]
- Dagnachew, B.S.; Thaller, G.; Lien, S.; Ådnøy, T. Casein SNP in Norwegian goats: Additive and dominance effects on milk composition and quality. Genet. Sel. Evol. 2011, 43, 31. [Google Scholar] [CrossRef]
- Devold, T.G.; Nordbø, R.; Langsrud, T.; Svenning, C.; Brovold, M.J.; Sørensen, E.S.; Christensen, B.; Ådnøy, T.; Vegarud, G.E. Extreme frequencies of the αs1-casein “null” variant in milk from Norwegian dairy goats—Implications for milk composition, micellar size and renneting properties. Dairy Sci. Technol. 2011, 91, 39–51. [Google Scholar] [CrossRef]
- Hayes, B.; Hagesæther, N.; Adnøy, T.; Pellerud, G.; Berg, P.R.; Lien, S. Effects on production traits of haplotypes among casein genes in Norwegian goats and evidence for a site of preferential recombination. Genetics 2006, 174, 455–464. [Google Scholar] [CrossRef] [PubMed]
- Pizarro Inostroza, M.G.; Navas González, F.J.; Landi, V.; León Jurado, J.M.; Delgado Bermejo, J.V.; Fernández Álvarez, J.; Martínez Martínez, M.d.A. Bayesian Analysis of the Association between Casein Complex Haplotype Variants and Milk Yield, Composition, and Curve Shape Parameters in Murciano-Granadina Goats. Animals 2020, 10, 1845. [Google Scholar] [CrossRef] [PubMed]
- Nilsen, H.; Olsen, H.G.; Hayes, B.; Sehested, E.; Svendsen, M.; Nome, T.; Meuwissen, T.; Lien, S. Casein haplotypes and their association with milk production traits in Norwegian Red cattle. Genet. Sel. Evol. 2009, 41, 24. [Google Scholar] [CrossRef]
- Pizarro Inostroza, M.G.; Landi, V.; Navas González, F.J.; León Jurado, J.M.; Delgado Bermejo, J.V.; Fernández Álvarez, J.; Martínez Martínez, M.d.A. Integrating casein complex SNPs additive, dominance and epistatic effects on genetic parameters and breeding values estimation for murciano-granadina goat milk yield and components. Genes 2020, 11, 309. [Google Scholar] [CrossRef]
- Pizarro Inostroza, M.G.; Landi, V.; Navas González, F.J.; León Jurado, J.M.; Martínez Martínez, A.; Fernández Álvarez, J.; Delgado Bermejo, J.V. Does the acknowledgement of αS1-casein genotype affect the estimation of genetic parameters and prediction of breeding values for milk yield and composition quality-related traits in Murciano-Granadina? Animals 2019, 9, 679. [Google Scholar] [CrossRef]
- Pizarro Inostroza, M.G.; Landi, V.; Navas Gonzalez, F.J.; León Jurado, J.M.; Martínez Martínez, M.d.A.; Fernández Álvarez, J.; Delgado Bermejo, J.V. Non-parametric association analysis of additive and dominance effects of casein complex SNPs on milk content and quality in Murciano-Granadina goats. J. Anim. Breed. Genet. 2020, 137, 407–422. [Google Scholar] [CrossRef]
- Pizarro, M.; Landi, V.; Navas, F.; León, J.; Martínez, A.; Fernández, J.; Delgado, J. Nonparametric analysis of casein complex genes’ epistasis and their effects on phenotypic expression of milk yield and composition in Murciano-Granadina goats. J. Dairy Sci. 2020, 103, 8274–8291. [Google Scholar] [CrossRef]
- Pizarro, M.; Landi, V.; Navas, F.; León, J.; Martínez, A.; Fernández, J.; Delgado, J. Non-parametric analysis of the effects of nongenetic factors on milk yield, fat, protein, lactose, dry matter content and somatic cell count in Murciano-Granadina goats. Ital. J. Anim. Sci. 2020, 19, 960–973. [Google Scholar] [CrossRef]
- Vacca, G.M.; Dettori, M.L.; Piras, G.; Manca, F.; Paschino, P.; Pazzola, M. Goat casein genotypes are associated with milk production traits in the Sarda breed. Anim. Genet. 2014, 45, 723–731. [Google Scholar] [CrossRef]
- Rijnkels, M.; Elnitski, L.; Miller, W.; Rosen, J.M. Multispecies comparative analysis of a mammalian-specific genomic domain encoding secretory proteins. Genomics 2003, 82, 417–432. [Google Scholar] [CrossRef]
- Bleck, G.T.; White, B.R.; Miller, D.J.; Wheeler, M.B. Production of bovine α-lactalbumin in the milk of transgenic pigs. J. Anim. Sci. 1998, 76, 3072–3078. [Google Scholar] [CrossRef] [PubMed]
- Kahilo, K.; El-Shazly, S.; El-Khadrawy, A.; Fattouh, I. Genetic polymorphism in β-lactoglobulin gene of some goat breeds in Egypt and its influence on milk yield. Life Sci. J. 2014, 11, 232–238. [Google Scholar]
- Strucken, E.M.; Rahmatalla, S.; Koning, D.-J.D.; Brockmann, G.A. Haplotype analysis and linkage disequilibrium for DGAT1. Arch. Anim. Breed. 2010, 53, 247–255. [Google Scholar] [CrossRef]
- He, C.; Wang, C.; Chang, Z.; Guo, B.; Li, R.; Yue, X.; Lan, X.; Chen, H.; Lei, C. AGPAT6 polymorphism and its association with milk traits of dairy goats. Genet. Mol. Res. 2011, 10, 2747–2756. [Google Scholar] [CrossRef]
- Ogg, S.L.; Weldon, A.K.; Dobbie, L.; Smith, A.J.; Mather, I.H. Expression of butyrophilin (Btn1a1) in lactating mammary gland is essential for the regulated secretion of milk–lipid droplets. Proc. Natl. Acad. Sci. USA 2004, 101, 10084–10089. [Google Scholar] [CrossRef]
- Morris, C.A.; Cullen, N.G.; Glass, B.C.; Hyndman, D.L.; Manley, T.R.; Hickey, S.M.; McEwan, J.C.; Pitchford, W.S.; Bottema, C.D.; Lee, M.A. Fatty acid synthase effects on bovine adipose fat and milk fat. Mamm. Genome 2007, 18, 64–74. [Google Scholar] [CrossRef]
- Federica, S.; Francesco, N.; Giovanna, D.M.; Carmela, S.M.; Gennaro, C.; Carmela, T.; Bianca, M. Identification of novel single nucleotide polymorphisms in promoter III of the acetyl-CoA carboxylase-α gene in goats affecting milk production traits. J. Hered. 2009, 100, 386–389. [Google Scholar] [CrossRef]
- Tudisco, R.; Calabrò, S.; Cutrignelli, M.; Moniello, G.; Grossi, M.; Gonzalez, O.; Piccolo, V.; Infascelli, F. Influence of organic systems on Stearoyl-CoA desaturase gene expression in goat milk. Small Rumin. Res. 2012, 106, S37–S42. [Google Scholar] [CrossRef]
- Shi, H.; Luo, J.; Zhu, J.; Li, J.; Sun, Y.; Lin, X.; Zhang, L.; Yao, D.; Shi, H. PPARγ regulates genes involved in triacylglycerol synthesis and secretion in mammary gland epithelial cells of dairy goats. PPAR Res. 2013, 2013, 310948. [Google Scholar] [CrossRef]
- Chong, B.M.; Reigan, P.; Mayle-Combs, K.D.; Orlicky, D.J.; McManaman, J.L. Determinants of adipophilin function in milk lipid formation and secretion. Trends Endocrinol. Metab. 2011, 22, 211–217. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wolins, N.E.; Rubin, B.; Brasaemle, D.L. TIP47 associates with lipid droplets. J. Biol. Chem. 2001, 276, 5101–5108. [Google Scholar] [CrossRef]
- Xu, H.; Luo, J.; Zhao, W.; Yang, Y.; Tian, H.; Shi, H.; Bionaz, M. Overexpression of SREBP1 (sterol regulatory element binding protein 1) promotes de novo fatty acid synthesis and triacylglycerol accumulation in goat mammary epithelial cells. J. Dairy Sci. 2016, 99, 783–795. [Google Scholar] [CrossRef] [PubMed]
- Shen, B.; Pan, Q.; Yang, Y.; Gao, Y.; Liu, X.; Li, W.; Han, Y.; Yuan, X.; Qu, Y.; Zhao, Z. miR-224 affects mammary epithelial cell apoptosis and triglyceride production by downregulating ACADM and ALDH2 genes. DNA Cell Biol. 2017, 36, 26–33. [Google Scholar] [CrossRef]
- Dubey, P.K.; Goyal, S.; Mishra, S.K.; Arora, R.; Mukesh, M.; Niranjan, S.K.; Kathiravan, P.; Kataria, R.S. Identification of polymorphism in fatty acid binding protein 3 (FABP3) gene and its association with milk fat traits in riverine buffalo (Bubalus bubalis). Trop. Anim. Health. Prod. 2016, 48, 849–853. [Google Scholar] [CrossRef]
- Yao, D.; Luo, J.; He, Q.; Xu, H.; Li, J.; Shi, H.; Wang, H.; Chen, Z.; Loor, J. Liver X receptor α promotes the synthesis of monounsaturated fatty acids in goat mammary epithelial cells via the control of stearoyl-coenzyme A desaturase 1 in an SREBP-1-dependent manner. J. Dairy Sci. 2016, 99, 6391–6402. [Google Scholar] [CrossRef]
- Khatib, H.; Leonard, S.; Schutzkus, V.; Luo, W.; Chang, Y. Association of the OLR1 gene with milk composition in Holstein dairy cattle. J. Dairy Sci. 2006, 89, 1753–1760. [Google Scholar] [CrossRef]
- Zielke, L.G.; Bortfeldt, R.H.; Tetens, J.; Brockmann, G.A. BDNF contributes to the genetic variance of milk fat yield in German Holstein cattle. Front. Genet. 2011, 2, 16. [Google Scholar] [CrossRef]
- Zielke, L.G.; Bortfeldt, R.H.; Reissmann, M.; Tetens, J.; Thaller, G.; Brockmann, G.A. Impact of variation at the FTO locus on milk fat yield in Holstein dairy cattle. PLoS ONE 2013, 8, e63406. [Google Scholar]
- Cochran, S.D.; Cole, J.B.; Null, D.J.; Hansen, P.J. Discovery of single nucleotide polymorphisms in candidate genes associated with fertility and production traits in Holstein cattle. BMC Genet. 2013, 14, 49. [Google Scholar] [CrossRef]
- Miltiadou, D.; Hager-Theodorides, A.; Symeou, S.; Constantinou, C.; Psifidi, A.; Banos, G.; Tzamaloukas, O. Variants in the 3’UTR of the ovine Acetyl-Coenzyme A Acyltransferase 2 gene are associated with dairy traits and exhibit differential allelic expression. J. Dairy Sci. 2017, 100, 6285–6297. [Google Scholar] [CrossRef] [PubMed]
- Yao, D.; Luo, J.; He, Q.; Shi, H.; Li, J.; Wang, H.; Xu, H.; Chen, Z.; Yi, Y.; Loor, J.J. SCD1 alters long-chain fatty acid (LCFA) composition and its expression is directly regulated by SREBP-1 and PPARγ1 in dairy goat mammary cells. J. Cell. Physiol. 2017, 232, 635–649. [Google Scholar] [CrossRef] [PubMed]
- Strucken, E.M.; Laurenson, Y.C.; Brockmann, G.A. Go with the flow—Biology and genetics of the lactation cycle. Front. Genet. 2015, 6, 118. [Google Scholar] [CrossRef] [PubMed]
- Hou, J.; An, X.; Song, Y.; Wang, J.; Ma, T.; Han, P.; Fang, F.; Cao, B. Combined effects of four SNPs within goat PRLR gene on milk production traits. Gene 2013, 529, 276–281. [Google Scholar] [CrossRef] [PubMed]
- Cohen-Zinder, M.; Seroussi, E.; Larkin, D.M.; Loor, J.J.; Everts-Van Der Wind, A.; Lee, J.-H.; Drackley, J.K.; Band, M.R.; Hernandez, A.; Shani, M. Identification of a missense mutation in the bovine ABCG2 gene with a major effect on the QTL on chromosome 6 affecting milk yield and composition in Holstein cattle. Genome Res. 2005, 15, 936–944. [Google Scholar] [CrossRef] [PubMed]
- Le Provost, F.; Leroux, C.; Martin, P.; Gaye, P.; Djiane, J. Prolactin gene expression in ovine and caprine mammary gland. Neuroendocrinology 1994, 60, 305–313. [Google Scholar] [CrossRef]
- Yakan, A.; Özkan, H.; ŞAKAR, A.E.; Ünal, N.; Özbeyaz, C. Gene expression levels in some candidate genes for mastitis resistance, milk yield, and milk quality of goats reared under different feeding systems. Turk. J. Vet. Anim. Sci. 2018, 42, 18–28. [Google Scholar] [CrossRef]
- Pizarro, M.G.; Landi, V.; Navas González, F.J.; León, J.M.; Delgado, J.V. Non-parametric analysis of the effects of αS1-casein genotype and parturition non-genetic factors on milk yield and composition in Murciano-Granadina goats. Ital. J. Anim. Sci. 2019, 18, 1021–1034. [Google Scholar] [CrossRef]
- Lan, X.; Pan, C.; Chen, H.; Zhang, C.; Zhang, A.; Zhang, L.; Li, J.; Lei, C. An MspI PCR-RFLP detecting a single nucleotide polymorphism at alpha-lactalbumin gene in goat. Czech J. Anim. Sci. 2007, 52, 138. [Google Scholar] [CrossRef]
- Winter, A.; Krämer, W.; Werner, F.A.; Kollers, S.; Kata, S.; Durstewitz, G.; Buitkamp, J.; Womack, J.E.; Thaller, G.; Fries, R. Association of a lysine-232/alanine polymorphism in a bovine gene encoding acyl-CoA: Diacylglycerol acyltransferase (DGAT1) with variation at a quantitative trait locus for milk fat content. Proc. Natl. Acad. Sci. USA 2002, 99, 9300–9305. [Google Scholar] [CrossRef]
- Zhang, C.-L.; Gao, X.-Y.; Shao, R.-Y.; Wang, Y.-H.; Fang, X.-T.; Chen, H. Stearoyl-CoA desaturase (SCD) gene polymorphism in goat breeds. Biochem. Genet. 2010, 48, 822–828. [Google Scholar] [CrossRef] [PubMed]
- Dixit, S.; Sivalingam, J.; Tyagi, A.; Saroha, V.; Sharma, A.; Nagda, R. Association of novel SNPs in the candidate genes affecting caprine milk fatty acids related to human health. Meta Gene 2015, 4, 45–56. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Zhu, J.; Luo, J.; Cao, W.; Shi, H.; Yao, D.; Li, J.; Sun, Y.; Xu, H.; Yu, K. Genes regulating lipid and protein metabolism are highly expressed in mammary gland of lactating dairy goats. Funct. Integr. Genom. 2015, 15, 309–321. [Google Scholar] [CrossRef] [PubMed]

| Breeding Criteria | Traits | Reference |
|---|---|---|
| Body weight | Birth weight, body weight at 7, 14, 21 and 28 days, monthly weighing until 18 months of age | [56,57] |
| Growth | Average daily gain (ADG) before weaning, from 3 to 6 months of age, from 6 to 12 months of age | [57] |
| Conformation/corporal structure | Body length, height at the withers, chest girth, shoulder width and pin-bone width | [58] |
| Carcass quality | Hot (immediately after slaughter) and cold (before chilling) carcass weight; weights of the head, skin, heart, and thoracic and abdominal viscera; carcass dressing percentage; leg length and carcass width; and carcass fat coverage and lean meat and bone yield percentages | [56,59] |
| Meat quality | Muscle pH measurements, colour, water retention capacity, nutritional composition (protein, fat, and collagen percentages), and fatty acid profile in samples of longissimus thoracis et lumborum, triceps brachii and semimembranosus | [56] |
| Acronym | Gene Name | Chromosome | Exon Count | Transcripts | Physiological Function | Goat Breeds in Which the Gene Has Been Described | Reference |
|---|---|---|---|---|---|---|---|
| ADCY1 | Adenylate Cyclase 1 | 4 | 20 | 2 | Involved in energetic metabolism, cellular mitosis and GH releasing. | Cameroon, West African Dwarf, Small East African and Landim goat | [60] |
| SNX29 | Sorting Nexin 29 | 25 | 21 | 2 | Regulates myoblast differentiation and proliferation. | ||
| LDB2 | LIM Domain Binding Factor 2 | 6 | 9 | 7 | Modulates transendothelial leucocyte migration. | Nanjiang yellow goat | [83] |
| MYADM | Myeloid-associated Differentiation Marker | 18 | 1 | 1 | Involved in cellular membrane formation in a wide variety of cellular lines. | Bamu wild goat, Khonj wild goat, Australian feral Rangeland goats, Boer goats and Australian cashmere goat | [84] |
| IGF1R | Insulin-like Growth Factor 1 Receptor | 21 | 21 | 1 | Regulates IGF-1 activity. | ||
| APOL3 | Apolipoprotein L3 | 5 | 4 | 6 | Involved in lipid blood transport and metabolism. | Leizhou goat | [70] |
| STIM1 | Stromal Interaction Molecule 1 | 15 | 14 | 4 | Associated with body weight gain. | ||
| HMGXB3 | HMG-Box Containing 3 | 7 | 21 | 3 | Demonstrated relationship with cellular proliferation in neoplasia. | ||
| GH | Growth Hormone | 19 | 5 | 1 | Related to corporal development. | Thai Native, Anglo-Nubian, Boer and Saanen goat | [85] |
| GHR | Growth Hormone Receptor | 20 | 13 | 6 | |||
| IGFBP3 | Insulin-like Growth Factor Binding Protein 3 | 4 | 5 | 1 | |||
| BMP4 | Bone Morphogenic Protein 4 | 10 | 6 | 5 | Boer goat, Chinese Xuhuai white goat and Chinese Haimen goat | [85] | |
| BMP15 | Bone Morphogenic Protein 15 | 10 | 2 | 1 | Jining Grey goat | [85] | |
| IGF-1 | Insulin-like Growth Factor | 5 | 7 | 9 | Involved in corporal metabolism, growth and development. | Malabari and Black Attappady goat | [86] |
| IGF2BP1 | Insulin-like Growth Factor Binding Protein 1 | 19 | 15 | 2 | Involved in endocrine routes associated with body growth and development. | Shaanbei White Cashmere goat | [87] |
| FGFR1 | Fibroblastic Growth Factor Receptor 1 | 27 | 20 | 13 | |||
| SMG6 | Nonsense-mediated mRNA Decay Factor | 19 | 20 | 2 | Genome stabilization functions | ||
| CADM2 | Cell Adhesion Molecule 2 | 1 | 11 | 3 | Involved in cellular migration and proliferation. | White and Black Guizhou, Nubiann, Boer and Huai goat. | [82] |
| GHSR | Growth Hormone Secretagogue Receptor | 1 | 3 | 2 | Regulates GH realise | Boer, Xuhuai and Haimen goat | [73] |
| Acronym | Gene Name | Chromosome | Exon Count | Transcripts | Physiological Function | Goat Breeds in Which the Gene Has Been Described | Reference |
|---|---|---|---|---|---|---|---|
| TBX15 | T-box Transcription Factor 15 | 3 | 9 | 2 | Related to body size and chondrocytes and mesenchymal cell precursors regulator. | Gizhou Small goat | [98] |
| DGCR8 | Drosha and DiGeorge Syndrome Critical Region microprocessor complex subunit gene 8 | 17 | 16 | 1 | Associated with body size and involved in the osteoclastic development and remodelling bone activity. | ||
| CDC25A | Cell Division Cycle 25 Homolog A | 22 | 15 | 1 | Responsible for corporal development and involved in myoblast differentiation and G1 quiescence. | ||
| LEP | Leptin | 4 | 3 | 1 | Related to corporal and muscle mass development. | ||
| MSTN | Myostatin | 2 | 3 | 1 | |||
| PRDM6 | PR/SET Domain 6 | 7 | 8 | 1 | Regulates skeletal development and body mass index. | Shaanbei White Cashmere goat | [88] |
| NGF | Nerve Growth Factor | 3 | 3 | 2 | Involved in muscle and nervous tissue development. | Black Attappady and Malabari goat | [89] |
| SLC26A2 | Solute Carrier Family 26 Member 2 | 7 | 9 | 8 | Involved in bone tissue development. | Leizhou goat | [70] |
| POU1F1 | Pituitary-specific Positive Transcription Factor 1 | 1 | 6 | 2 | Plays an important role in corporal metabolism, growth and corporal development. | Thai Native, Ango-nubian, Boer and Saanen goat | [85] |
| Acronym | Gene Name | Chromosome | Exon Count | Transcripts | Physiological Function | Goat Breeds in Which the Gene Has Been Described | Reference |
|---|---|---|---|---|---|---|---|
| PPAR-γ | Peroxisome Proliferator-activated Receptors Gamma | 22 | 6 | 1 | Regulates adipose differentiation and lipid metabolism. | White Yaoshan goat | [59] |
| LPL | Lipoprotein Lipase | 8 | 10 | 2 | Involved in triglycerides degradation, obtaining glycerol and fatty acids. | ||
| H-FABP (FABP3) | Heart Fatty Acid-Binding Protein (fatty acid binding protein 3) | 2 | 5 | 2 | Related to lipid metabolism. | ||
| SREBF1 | Sterol Regulatory Element-binding Protein 1 | 19 | 8 | 1 | Modulates expression of other lipid-metabolism-related genes. | ||
| ACACA | Acetyl-Coenzyme A Carboxylase | 19 | 60 | 7 | Involved in liver fatty acid synthesis. | ||
| THRSP (SPOT14) | Thyroid Hormone Responsive | 29 | 2 | 1 | Regulates expression of other lipid-metabolism-related genes. | ||
| CPT1A | Carnitine Palmitoyl-transferase 1A | 29 | 19 | 1 | Responsible for acyl-carnitine obtention, product of fatty acid degradation. | Moroccan goat breeds | [109] |
| RDH16 | Retinol deshidrogenase-16 | 5 | 4 | 1 | Involved in energetic metabolism. | Gizhou goat | [98] |
| Acronym | Gene Name | Chromosome | Exon Count | Transcripts | Physiological Function | Goat Breeds in Which the Gene Has Been Described | Reference |
|---|---|---|---|---|---|---|---|
| LEP | Leptin | 4 | 3 | 1 | Regulates glycaemia, milk production and milk fat percentage. | Dairy cattle | [117] |
| LEPR | Leptin receptor | 3 | 22 | 3 | |||
| BDNF | Brain-derived Neurotrophy Factor | 15 | 6 | 5 | Plays different roles in daily food intake and, in consequence, in nutrient and energy availability in the mammary gland. | Dairy cattle | [162] |
| FTO | Fat Mass and Obesity-associated Protein | 18 | 9 | 1 | |||
| IGF-1 | Insulin-like Growth Factor 1 | 5 | 7 | 9 | |||
| ABCG2 | ATP Binding Cassete Subfamily G Member 2 | 6 | 22 | 8 | Related to milk production and milk fat percentage. | Dairy cattle | [163] |
| GHR | Growth Hormone Receptor | 20 | 13 | 6 | Regulates cell growth, proliferation and apoptosis. | Dairy cattle | [117] |
| PRLR | Prolactin Receptor | 20 | 11 | 7 | [164] | ||
| PRL | Prolactin | 23 | 5 | 1 | Involved in mammary gland tissue preparation before lactation and regulates milk production. | Alpine goat | [165] |
| RPL3 | Ribosomal Protein L3 | 5 | 10 | 1 | Modulates energetic balance during the lactation yield peak. | Saanen goat | [70] |
| VPS13 | Vacuolar Protein Sorting 13 | 8 | 73 | 3 | Gene family associated with milk production. | ||
| VPS13B | Vacuolar Protein Sorting 13 Homolog B | 14 | 65 | 10 | Related to leg’s morphological traits associated with fertility and milk production. | Dairy cattle | [115] |
| VPS13C | Vacuolar Protein Sorting 13 Homolog C | 10 | 85 | 1 | Regulates glycaemia increasing milk production. | Dairy cattle | [114] |
| SPP1 (OPN) | Osteopontin | 6 | 7 | 2 | Involved in milk yield production and milk fat percentage. | Dairy cattle | [123] |
| GH1 | Growth Hormone 1 | 19 | 5 | 1 | Seems to stimulate udder development and is associated with milk dairy traits. | Dairy cattle | [124] |
| LTF | Lactoferrin | 22 | 17 | 1 | Associated with mastitis resistance somatic cell count. | Damascus goat | [166] |
| BRCA1 | Breast Cancer Type 1 | 19 | 23 | 5 | |||
| POU1F1 (PIT1) | Pituitary-specific Positive Transcription Factor 1 (POU class 1 homeobox 1) | 1 | 6 | 2 | Modulates many milk-related hormones’ action. | ||
| CSN1S1 | αs1-casein | 6 | 19 | 8 | Associated with total milk protein composition and milk yield production. | Murciano-Granadina and Norwegian goats | [128,129,130,131,132,133,134,135,136,137,138,167] |
| CSN1S2 | αs2-casein | 6 | 19 | 7 | Encodes for most of the important milk proteins. | Sarda goat | [128,129,130,131,132,133,134,135,136,137,138,167] |
| CSN2 | β-Casein | 6 | 9 | 2 | |||
| CSN3 | Ƙ-Casein | 6 | 5 | 1 | Norwegian, Saanen, Canaria, Malagueña, Murciano-Granadina and Payoya goat | ||
| αLA LALBA (ALA) | α-lactalbumin | 5 | 4 | 1 | Encodes for most of the important milk serum proteins. | White Inner Mongolia Cashmere, Xinong, Guanzhong, Laoshan, Leizhou, White and Black Guizhou and Banjiao, Matou goat | [168] |
| β-LG LGB (PAEP) | β-lactoglobulin/progestagen-associated endometrial protein | 11 | 8 | 4 | Zarayby, Damascus, Albino and Balady Hybrid goat | [142] | |
| DGAT1 | Diacylglycerol Acyltransferase | 14 | 18 | 2 | Involved in triglycerides synthesis. | Xinong, Saanen and Guanzhong goat | [169] |
| AGPAT6 (AGPAT6) | 1-acylglicerol-3-phosohate-O-acyltransferase | 27 | 14 | 2 | [144] | ||
| BTN1A1 | Butyrophilin Subfamily 1 Member A1 | 23 | 9 | 5 | Essential in milk lipid micelles secretion in the mammary gland. | Bamu wild goat, Khonj wild goat, Australian feral rangeland goat, Boer and Australian Cashmere goat | [84] |
| FASN | Fatty Acid Synthase | 19 | 42 | 1 | Plays an important role in fatty acids synthesis. | [84] | |
| ACACA | Acetyl-CoA Carboxylase | 19 | 60 | 7 | Involved in liver fatty acid synthesis. | Saanen and ‘Grey local’ goat | [147] |
| SCD | Stearoyl-CoA Desaturase | 26 | 6 | 1 | Catalyze unsaturated to mono-satured fatty acids transformation reaction. | Boer, Xuhuai White and Haimen goat | [170] |
| PPARγ (PPARG) | Peroxisome Proliferator-activated Receptor Gamma | 22 | 6 | 1 | Regulates lipogenesis and adipogenesis. | Damascus goat | [166] |
| PLIN3 | Perilipin 3 | 7 | 8 | 2 | Involved in tissue fatty acid synthesis. | Not specified/Dairy goats | [149] |
| PLIN2 | Perilipin 2 | 8 | 9 | 4 | |||
| FABP3 | Fatty Acid Binding Protein 3 | 2 | 5 | 2 | Involved in cytoplasmatic fatty acid storage and transport. | ||
| PNPLA2 | Patatin-like Phospholipase Domain Containing 2 | 29 | 11 | 3 | Encodes for one of the proteins involved in fat micelles formation. | ||
| SREBF1 | Sterol Regulatory Element-binding Protein 1 | 19 | 8 | 1 | Transcription factors involved in lipid homeostasis. | ||
| NR1H3 | Liver X Receptor-α (nuclear receptor subfamily 1 group H member 3) | 15 | 10 | 2 | |||
| OLR1 | Oxidized Low Density Lipoprotein Receptor | 5 | 6 | 2 | Involved in lipid transport and oxided-LDL form degradation. | Sirohi goat | [171] |
| ALDH2 | Aldehyde Dehydrogenase 2 | 17 | 13 | 1 | Associated with triglycerides synthesis in mammary glandular tissue. | Xinong Saanen goat | [172] |
| ACAT1 | Acetyl-CoA Acetyltransferase 1 | 15 | 12 | 1 | Involved in cholesterol metabolism. | ||
| ACAT2 | Acetyl-CoA Acetyltransferase 2 | 9 | 9 | 1 | |||
| ACSL1 | Long-chain Acyl-CoA Synthetase Isoform 1 | 27 | 24 | 5 | Participates in triglycerides, phospholipids and cholesterol synthesis. | ||
| BDNF | Brain-derived Neurotrophy Factor | 15 | 6 | 5 | Plays different roles in daily food intake and, in consequence, in nutrient and energy availability in the mammary gland. | Dairy cattle | [162] |
| FTO | Fat Mass and Obesity-associated Protein | 18 | 9 | 1 | |||
| ABCG2 | ATP Binding Cassete Subfamily G Member 2 | 6 | 22 | 8 | Related to milk production and milk fat percentage. | Dairy cattle | [163] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Salgado Pardo, J.I.; Delgado Bermejo, J.V.; González Ariza, A.; León Jurado, J.M.; Marín Navas, C.; Iglesias Pastrana, C.; Martínez Martínez, M.d.A.; Navas González, F.J. Candidate Genes and Their Expressions Involved in the Regulation of Milk and Meat Production and Quality in Goats (Capra hircus). Animals 2022, 12, 988. https://doi.org/10.3390/ani12080988
Salgado Pardo JI, Delgado Bermejo JV, González Ariza A, León Jurado JM, Marín Navas C, Iglesias Pastrana C, Martínez Martínez MdA, Navas González FJ. Candidate Genes and Their Expressions Involved in the Regulation of Milk and Meat Production and Quality in Goats (Capra hircus). Animals. 2022; 12(8):988. https://doi.org/10.3390/ani12080988
Chicago/Turabian StyleSalgado Pardo, Jose Ignacio, Juan Vicente Delgado Bermejo, Antonio González Ariza, José Manuel León Jurado, Carmen Marín Navas, Carlos Iglesias Pastrana, María del Amparo Martínez Martínez, and Francisco Javier Navas González. 2022. "Candidate Genes and Their Expressions Involved in the Regulation of Milk and Meat Production and Quality in Goats (Capra hircus)" Animals 12, no. 8: 988. https://doi.org/10.3390/ani12080988
APA StyleSalgado Pardo, J. I., Delgado Bermejo, J. V., González Ariza, A., León Jurado, J. M., Marín Navas, C., Iglesias Pastrana, C., Martínez Martínez, M. d. A., & Navas González, F. J. (2022). Candidate Genes and Their Expressions Involved in the Regulation of Milk and Meat Production and Quality in Goats (Capra hircus). Animals, 12(8), 988. https://doi.org/10.3390/ani12080988







