CircEZH2 Regulates Milk Fat Metabolism through miR-378b Sponge Activity
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. RNA Extraction and Real-Time Quantitative PCR
2.2. Identification of circRNA
2.3. Plasmid Construction and Oligonucleotide Synthesis
2.4. Cell Culture and Transfection
2.5. Western Blot Analysis
2.6. Cell Proliferation Analysis
2.7. Flow Cytometry Analysis
2.8. Fluorescent Fatty Acid Uptake Assay
2.9. Double Luciferase Activity Analysis
2.10. Prediction of Translation Function
2.11. Statistical Analysis
3. Results
3.1. Homology of Mouse CircEZH2 with Bovine Sequences
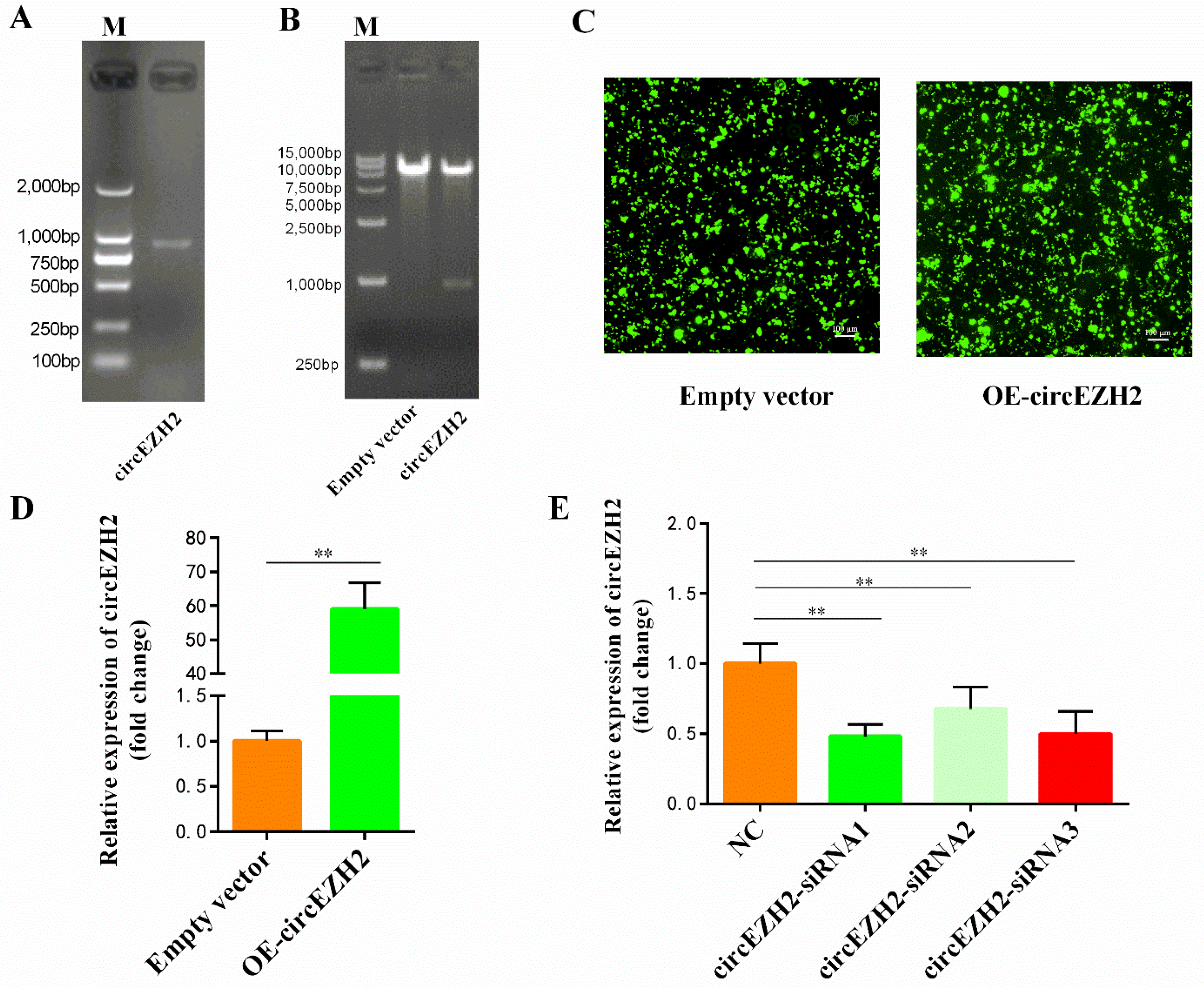
3.2. Construction of the CircEZH2 Overexpression Vector and Interference Sequences
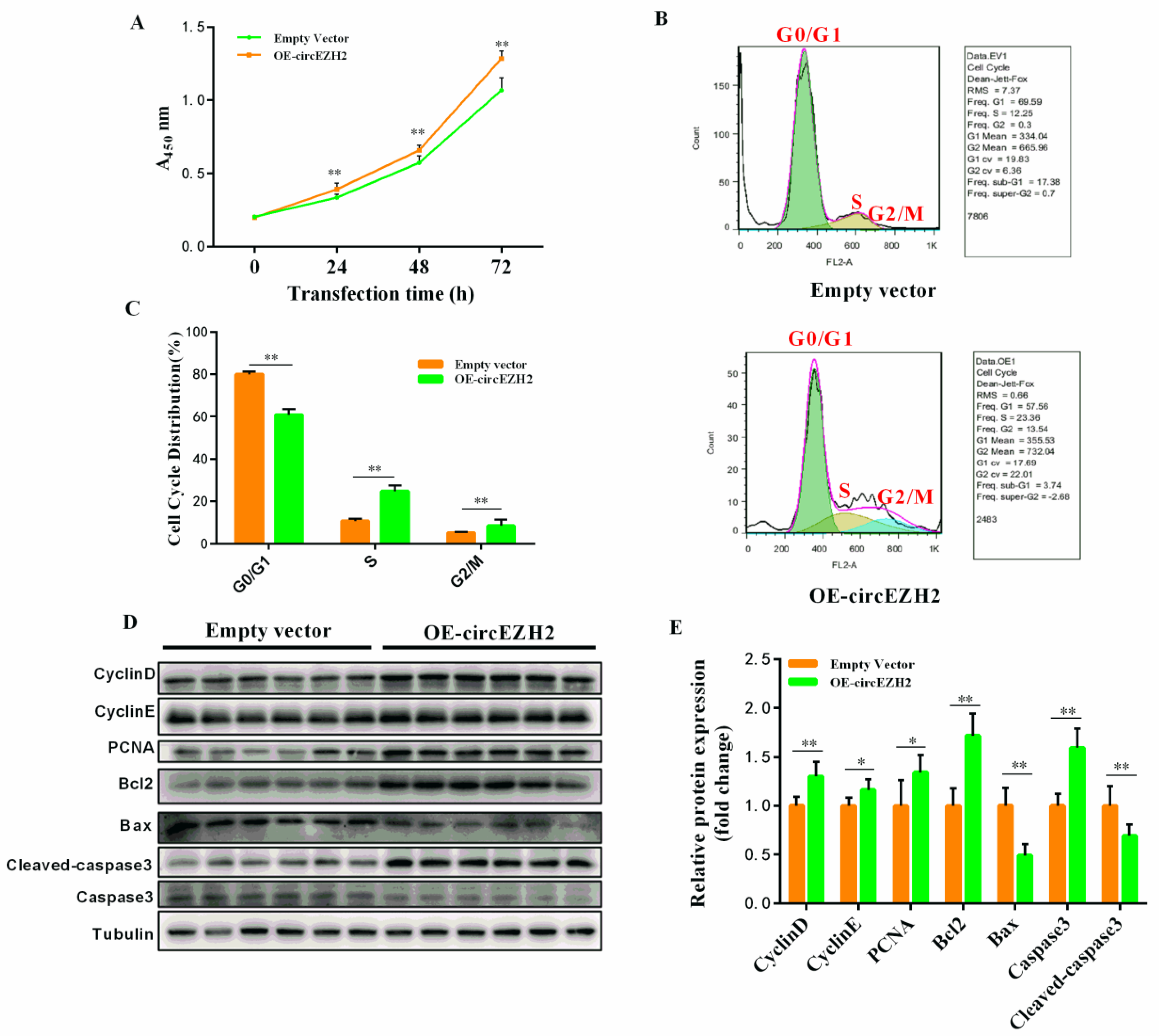
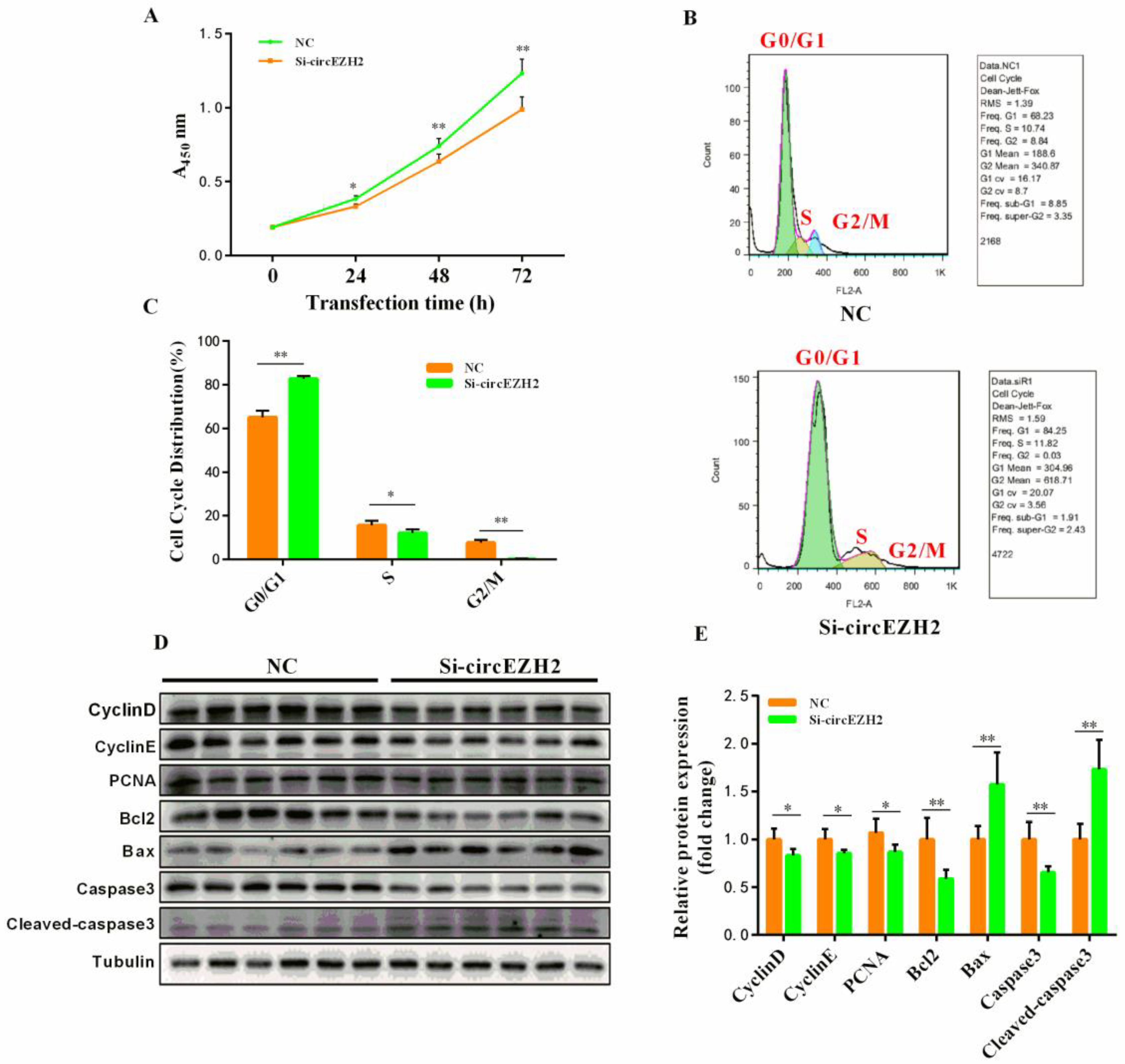
3.3. CircEZH2 Affects HC11 Cell Proliferation and Apoptosis
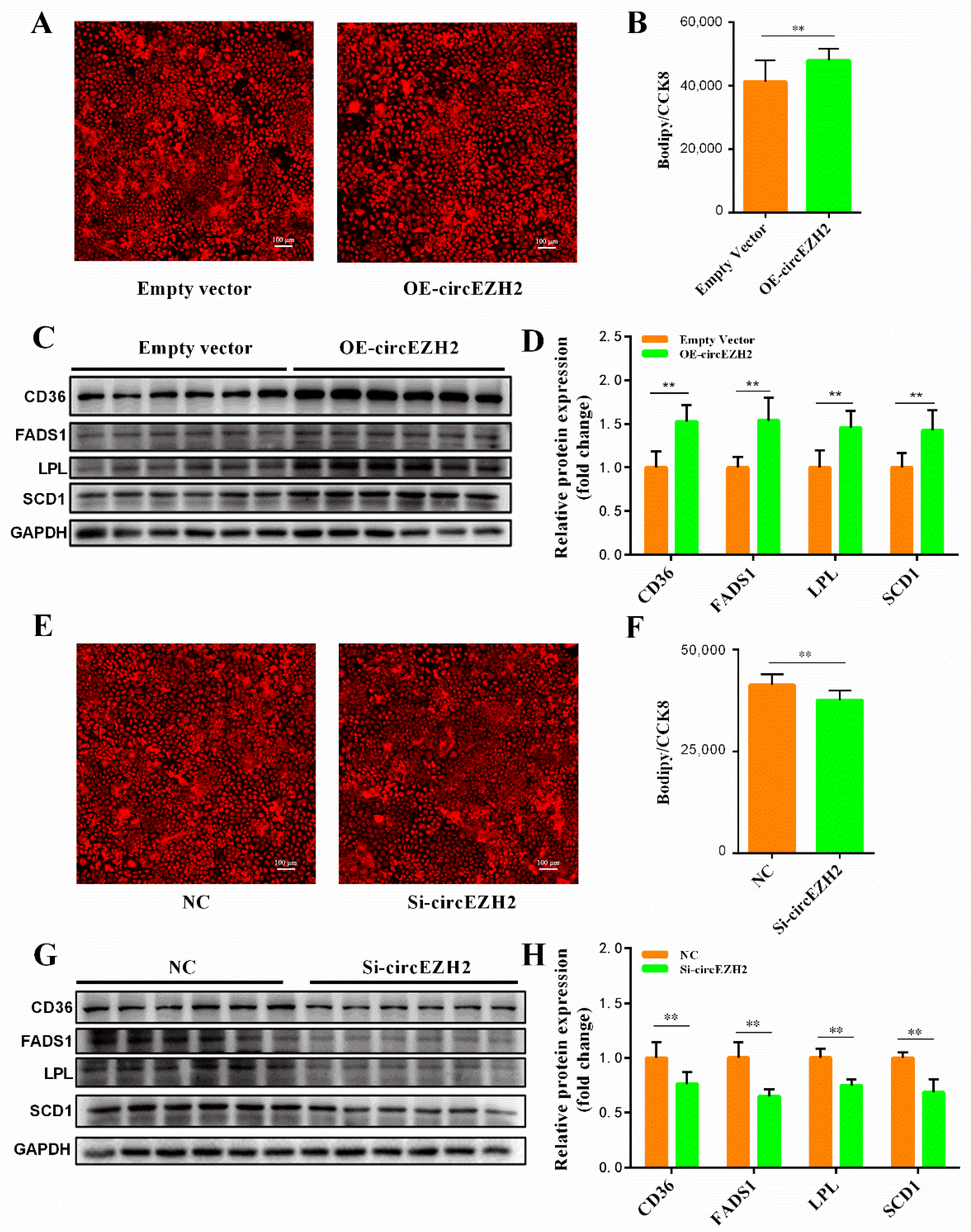
3.4. CircEZH2 Affects Cell Lipid Metabolism
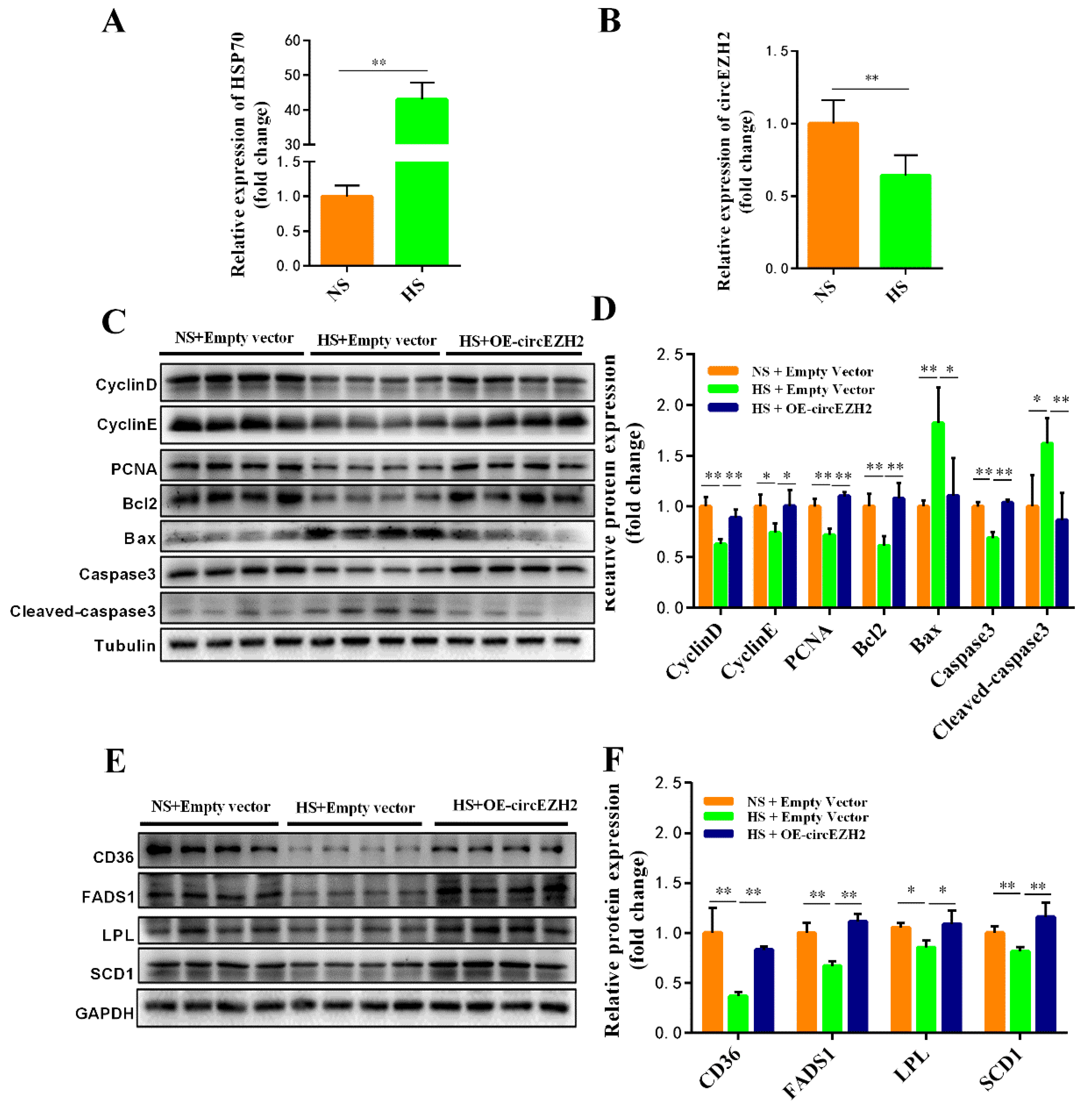
3.5. CircEZH2 Relieves the Negative Effects of Heat Stress
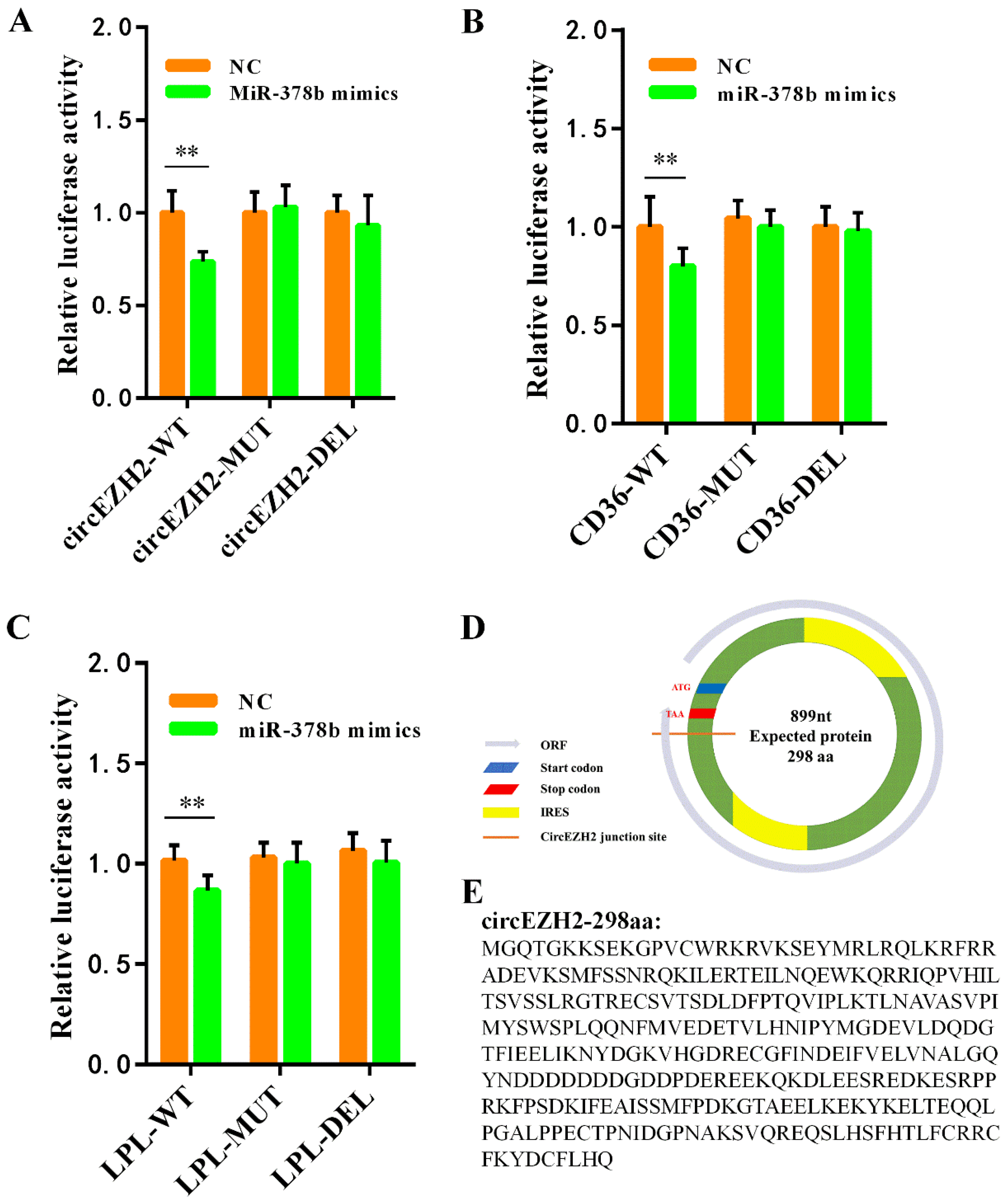
3.6. Analysis of the CircEZH2 Competitive Regulatory Network
3.7. Verification of Target Relationship and Prediction of Translation Ability
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| circRNAs | circular RNAs |
| ceRNA | competing endogenous RNAs |
| EZH2 | Enhancer of zeste 2 polycomb repressive complex 2 subunit |
| LPL | Lipoprotein lipase |
| PCNA | Proliferating cell nuclear antigen |
| siRNA | Small interfering RNA |
| FADS1 | Fatty acid desaturase 1 |
| SCD1 | Stearoyl-Coenzyme A desaturase 1 |
| ncRNA | non-coding RNA |
| RT-qPCR | Real-time quantitative PCR |
| UTR | Untranslated regions |
| EGF | Epidermal growth factor |
| SDS | Sodium dodecyl sulfate |
| PVDF | Polyvinylidene fluoride |
| CCK-8 | Cell Counting Kit-8 |
| EDTA | Ethylenediamine tetraacetic acid |
| FBS | Fetal bovine serum |
| PBS | Phosphate balanced normal saline |
| DMEM | Dulbecco’s modified eagle medium |
| IRES | Internal ribosome entry site |
| ORF | Open reading frame |
| gDNA | genomic DNA |
| GFP | Green fluorescent protein |
| HSP70 | Heat shock protein 70 |
References
- Lauridsen, C. Effects of Dietary Fatty Acids on Gut Health and Function of Pigs Pre- and Post-Weaning. J. Anim. Sci. 2020, 98, 1–12. [Google Scholar] [CrossRef] [PubMed]
- German, J.B.; Dillard, C.J. Composition, Structure and Absorption of Milk Lipids: A Source of Energy, Fat-Soluble Nutrients and Bioactive Molecules. Crit. Rev. Food Sci. Nutr. 2006, 46, 57–92. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Chu, S.; Wang, X.; Fan, Y.; Zhan, T.; Arbab, A.A.I.; Li, M.; Zhang, H.; Mao, Y.; Loor, J.J.; et al. MicroRNA-106b regulates milk fat metabolism via ATP binding cassette subfamily A member 1 (ABCA1) in bovine mammary epithelial cells. J. Agric. Food Chem. 2019, 67, 3981–3990. [Google Scholar] [CrossRef]
- Xu, B.; Gerin, I.; Miao, H.; Vu-Phan, D.; Johnson, C.N.; Xu, R.; Chen, X.W.; Cawthorn, W.P.; MacDougald, O.A.; Koenig, R.J. Multiple Roles for the Non-Coding RNA SRA in Regulation of Adipogenesis and Insulin Sensitivity. PLoS ONE 2010, 5, e14199. [Google Scholar] [CrossRef] [PubMed]
- Ibeagha-Awemu, E.M.; Li, R.; Ammah, A.A.; Dudemaine, P.L.; Bissonnette, N.; Benchaar, C.; Zhao, X. Transcriptome adaptation of the bovine mammary gland to diets rich in unsaturated fatty acids shows greater impact of linseed oil over safflower oil on gene expression and metabolic pathways. BMC Genom. 2016, 17, 104. [Google Scholar] [CrossRef] [PubMed]
- Jeck, W.R.; Sorrentino, J.A.; Wang, K.; Slevin, M.K.; Burd, C.E.; Liu, J.; Marzluff, W.F.; Sharpless, N.E. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 2013, 19, 141–157. [Google Scholar] [CrossRef]
- Zeng, B.; Chen, T.; Luo, J.; Xie, M.; Wei, L.; Xi, Q.; Sun, J.; Zhang, Y. Exploration of Long Non-coding RNAs and Circular RNAs in Porcine Milk Exosomes. Front. Genet. 2020, 11, 652. [Google Scholar] [CrossRef]
- Yang, R.; Xing, L.; Zheng, X.; Sun, Y.; Wang, X.; Chen, J. The circRNA circAGFG1 acts as a sponge of miR-195-5p to promote triple-negative breast cancer progression through regulating CCNE1 expression. Mol. Cancer 2019, 18, 4. [Google Scholar] [CrossRef]
- Zhang, C.; Wu, H.; Wang, Y.; Zhu, S.; Liu, J.; Fang, X.; Chen, H. Circular RNA of cattle casein genes are highly expressed in bovine mammary gland. J. Dairy Sci. 2016, 99, 4750–4760. [Google Scholar] [CrossRef]
- Hao, Z.; Zhou, H.; Hickford, J.G.H.; Gong, H.; Wang, J.; Hu, J.; Liu, X.; Li, S.; Zhao, M.; Luo, Y. Identification and characterization of circular RNA in lactating mammary glands from two breeds of sheep with different milk production profiles using RNA-Seq. Genomics 2020, 112, 2186–2193. [Google Scholar] [CrossRef]
- Zhu, C.; Jiang, Y.; Zhu, J.; He, Y.; Yin, H.; Duan, Q.; Zhang, L.; Cao, B.; An, X. CircRNA8220 Sponges MiR-8516 to Regulate Cell Viability and Milk Synthesis via Ras/MEK/ERK and PI3K/AKT/mTOR Pathways in Goat Mammary Epithelial Cells. Animals 2020, 10, 1347. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Zhang, H.; Hu, B.; Xie, Y.; Wang, D.; Zhang, J.; Chen, T.; Luo, J.; Wang, S.; Jiang, Q.; et al. Emerging Roles of Heat-Induced circRNAs Related to Lactogenesis in Lactating Sows. Front. Genet. 2020, 10, 1347. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Chen, Z.; Zhuang, X.; Luo, J.; Chen, T.; Xi, Q.; Zhang, Y.; Sun, J. Identification of circRNA-Associated-ceRNA Networks Involved in Milk Fat Metabolism under Heat Stress. Int. J. Mol. Sci. 2020, 21, 4162. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Wu, J.; Xu, T.; Yang, Q.; He, J.; Song, X. IRESfinder: Identifying RNA internal ribosome entry site in eukaryotic cell using framed k-mer features. J. Genet. Genom. 2018, 45, 403–406. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Han, P.; Zhou, T.; Guo, X.; Song, X.; Li, Y. circRNADb: A comprehensive database for human circular RNAs with protein-coding annotations. Sci. Rep. 2016, 6, 34985. [Google Scholar] [CrossRef] [PubMed]
- Kong, S.; Tao, M.; Shen, X.; Ju, S. Translatable circRNAs and lncRNAs: Driving mechanisms and functions of their translation products. Cancer Lett. 2020, 483, 59–65. [Google Scholar] [CrossRef]
- Zganiacz, D.; Milanowski, R. Characteristics of circular ribonucleic acid molecules (circRNA). Postepy Biochem. 2017, 63, 221–232. [Google Scholar]
- Kolahi, K.S.; Amy, V.; Thornburg, K.L. Real-time microscopic assessment of fatty acid uptake kinetics in the human term placenta. Placenta 2018, 72–73, 1–9. [Google Scholar] [CrossRef]
- Rambold, A.S.; Cohen, S.; Lippincott-Schwartz, J. Fatty acid trafficking in starved cells: Regulation by lipid droplet lipolysis, autophagy, and mitochondrial fusion dynamics. Dev. Cell 2015, 32, 678–692. [Google Scholar] [CrossRef]
- Xu, D.; Li, Y.; Wu, L.; Li, Y.; Zhao, D.; Yu, J.; Huang, T.; Ferguson, C.; Parton, R.G.; Yang, H.; et al. Rab18 promotes lipid droplet (LD) growth by tethering the ER to LDs through SNARE and NRZ interactions. J. Cell Biol. 2018, 217, 975–995. [Google Scholar] [CrossRef]
- Isosaki, M.; Nakashima, T. Psychological stress induces heat shock protein 70 expression in rat aorta. Jpn. J. Pharmacol. 1998, 76, 305. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Huang, C.; Shan, G. What happens at or after transcription: Insights into circRNA biogenesis and function. Transcription 2015, 6, 61–64. [Google Scholar] [CrossRef] [PubMed]
- Barrett, S.P.; Salzman, J. Circular RNAs: Analysis, expression and potential functions. Development 2016, 143, 1838. [Google Scholar] [CrossRef] [PubMed]
- Geng, X.; Jia, Y.; Zhang, Y.; Shi, L.; Li, Q.; Zang, A.; Wang, H. Circular RNA: Biogenesis, degradation, functions and potential roles in mediating resistance to anticarcinogens. Epigenomics 2020, 12, 267–283. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Zhao, K.; Xu, X.; Yang, Y.; Yan, S.; Wei, P.; Liu, H.; Xu, J.; Xiao, F.; Zhou, H.; et al. A peptide encoded by circular form of LINC-PINT suppresses oncogenic transcriptional elongation in glioblastoma. Nat. Commun. 2018, 9, 4475. [Google Scholar] [CrossRef]
- Godet, A.C.; David, F.; Hantelys, F.; Tatin, F.; Lacazette, E.; Garmy-Susini, B.; Prats, A.C. IRES Trans-Acting Factors, Key Actors of the Stress Response. Int. J. Mol. Sci. 2019, 20, 924. [Google Scholar] [CrossRef]
- Xiang, L.; Li, Y.; Chen, L.L. The Biogenesis, Functions, and Challenges of Circular RNAs. Mol. Cell 2018, 71, 428–442. [Google Scholar]
- West, J.W. Effects of heat-stress on production in dairy cattle. J. Dairy Sci. 2003, 86, 2131–2144. [Google Scholar] [CrossRef]
- Jin, L.; Yan, S.; Shi, B.; Bao, H.; Gong, J.; Guo, X.; Li, J. Effects of vitamin A on the milk performance, antioxidant functions and immune functions of dairy cows. Anim. Feed Sci. Technol. 2014, 192, 15–23. [Google Scholar] [CrossRef]
- Lacasse, P.; Lollivier, V.; Dessauge, F.; Bruckmaier, R.M.; Ollier, S.; Boutinaud, M. New developments on the galactopoietic role of prolactin in dairy ruminants. Domest. Anim. Endocrin. 2012, 43, 154–160. [Google Scholar] [CrossRef]
- Cowley, F.C.; Barber, D.G.; Houlihan, A.V.; Poppi, D.P. Immediate and residual effects of heat stress and restricted intake on milk protein and casein composition and energy metabolism. J. Dairy Sci. 2015, 98, 2356–2368. [Google Scholar] [CrossRef] [PubMed]
- Pan, T.; Sun, X.; Liu, Y.; Li, H.; Deng, G.; Lin, H.; Wang, S. Heat stress alters genome-wide profiles of circular RNAs in Arabidopsis. Plant. Mol. Biol. 2018, 96, 217–229. [Google Scholar] [CrossRef] [PubMed]
- Dou, C.; Cao, Z.; Yang, B.; Ding, N.; Hou, T.; Luo, F.; Kang, F.; Li, J.; Yang, X.; Jiang, H.; et al. Changing expression profiles of lncRNAs, mRNAs, circRNAs and miRNAs during osteoclastogenesis. Sci. Rep. 2016, 6, 21499. [Google Scholar] [CrossRef] [PubMed]
- Jin, W.; Ibeagha-Awemu, E.M.; Liang, G.; Beaudoin, F.; Zhao, X.; Guan, L.L. Transcriptome microRNA profiling of bovine mammary epithelial cells challenged with Escherichia coli or Staphylococcus aureus bacteria reveals pathogen directed microRNA expression profiles. BMC Genom. 2014, 15, 181. [Google Scholar] [CrossRef]
- Lin, X.; Luo, J.; Zhang, L.; Wang, W.; Gou, D. MiR-103 controls milk fat accumulation in goat (Capra hircus) mammary gland during lactation. PLoS ONE 2013, 8, e79258. [Google Scholar]
- Chen, Z.; Zhou, J.; Wang, M.; Liu, J.; Zhang, L.; Loor, J.J.; Liang, Y.; Wu, H.; Yang, Z. Circ09863 Regulates Unsaturated Fatty Acid Metabolism by Adsorbing miR-27a-3p in Bovine Mammary Epithelial Cells. J. Agric. Food Chem. 2020, 68, 8589–8601. [Google Scholar] [CrossRef]
- Petridis, I.G.; Fthenakis, G.C. Mammary involution and relevant udder health management in sheep. Small Rumin. Res. 2019, 181, 66–75. [Google Scholar] [CrossRef]
- Collier, R.J.; Collier, J.L.; Rhoads, R.P.; Baumgard, L.H. Invited review: Genes involved in the bovine heat stress response. J. Dairy Sci. 2008, 91, 445–454. [Google Scholar] [CrossRef]
- Li, L.; Sun, Y.; Wu, J.; Li, X.; Luo, M.; Wang, G. The global effect of heat on gene expression in cultured bovine mammary epithelial cells. Cell Stress Chaperon. 2015, 20, 381–389. [Google Scholar] [CrossRef]
- Pantschenko, A.G.; Woodcock-Mitchell, J.; Bushmich, S.L.; Yang, T.J. Establishment and characterization of a caprine mammary epithelial cell line (CMEC). In Vitro Cell Dev. Biol. Anim. 2000, 36, 26–37. [Google Scholar] [CrossRef]
- Kadegowda, A.K.; Bionaz, M.; Piperova, L.S.; Erdman, R.A.; Loor, J.J. Peroxisome proliferator-activated receptor-gamma activation and long-chain fatty acids alter lipogenic gene networks in bovine mammary epithelial cells to various extents. J. Dairy Sci. 2009, 92, 4276–4289. [Google Scholar] [CrossRef] [PubMed]
- Faulconnier, Y.; Bonnet, M.; Bocquier, F.; Leroux, C.; Chilliard, Y. Effects of photoperiod and feeding level on adipose tissue and muscle lipoprotein lipase activity and mRNA level in dry non-pregnant sheep. Brit. J. Nutr. 2001, 85, 299–306. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Xu, S.; Jay, A.; Brunaldi, K.; Huang, N.; Hamilton, J.A. CD36 enhances fatty acid uptake by increasing the rate of intracellular esterification but not transport across the plasma membrane. Biochemistry 2013, 52, 7254–7261. [Google Scholar] [CrossRef] [PubMed]
- Macciotta, N.P.; Mele, M.; Conte, G.; Serra, A.; Cassandro, M.; Zotto, R.D.; Borlino, A.C.; Pagnacco, G.; Secchiari, P. Association Between a Polymorphism at the Stearoyl CoA Desaturase Locus and Milk Production Traits in Italian Holsteins. J. Dairy Sci. 2008, 91, 3184–3189. [Google Scholar] [CrossRef]
- Glaser, C.; Heinrich, J.; Koletzko, B. Role of FADS1 and FADS2 polymorphisms in polyunsaturated fatty acid metabolism. Metabolism 2010, 59, 993–999. [Google Scholar] [CrossRef]
- Kulcheski, F.R.; Christoff, A.P.; Margis, R. Circular RNAs are miRNA sponges and can be used as a new class of biomarker. J. Biotechnol. 2016, 238, 42–51. [Google Scholar] [CrossRef]
- Lu, Y.L.; Jing, W.; Feng, L.S.; Zhang, L.; Xu, J.F.; You, T.J.; Zhao, J. Effects of hypoxic exercise training on microRNA expression and lipid metabolism in obese rat livers. J. Zhejiang Univ. Sci. B 2014, 15, 820–829. [Google Scholar] [CrossRef]
- Gerin, I.; Bommer, G.T.; McCoin, C.S.; Sousa, K.M.; Krishnan, V.; MacDougald, O.A. Roles for miRNA-378/378* in adipocyte gene expression and lipogenesis. Am. J. Physiol. Endocrinol. Metab. 2010, 299, E198–E206. [Google Scholar] [CrossRef]
- Kulyté, A.; Lorente-Cebrián, S.; Gao, H.; Mejhert, N.; Agustsson, T.; Arner, P.; Rydén, M.; Dahlman, I. MicroRNA profiling links miR-378 to enhanced adipocyte lipolysis in human cancer cachexia. Am. J. Physiol. Endocrinol. Metab. 2014, 306, E267–E274. [Google Scholar] [CrossRef]
- Shi, X.; Sun, M.; Liu, H.; Yao, Y.; Song, Y. Long non-coding RNAs: A new frontier in the study of human diseases. Cancer Lett. 2013, 339, 159–166. [Google Scholar] [CrossRef]
- Yang, Y.; Fan, X.; Mao, M.; Song, X.; Wu, P.; Zhang, Y.; Jin, Y.; Yang, Y.; Chen, L.L.; Wang, Y.; et al. Extensive translation of circular RNAs driven by N6-methyladenosine. Cell Res. 2017, 27, 626–641. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, D.; Zhao, Z.; Shi, Y.; Luo, J.; Chen, T.; Xi, Q.; Zhang, Y.; Sun, J. CircEZH2 Regulates Milk Fat Metabolism through miR-378b Sponge Activity. Animals 2022, 12, 718. https://doi.org/10.3390/ani12060718
Wang D, Zhao Z, Shi Y, Luo J, Chen T, Xi Q, Zhang Y, Sun J. CircEZH2 Regulates Milk Fat Metabolism through miR-378b Sponge Activity. Animals. 2022; 12(6):718. https://doi.org/10.3390/ani12060718
Chicago/Turabian StyleWang, Dongyang, Zhengjiang Zhao, Yiru Shi, Junyi Luo, Ting Chen, Qianyun Xi, Yongliang Zhang, and Jiajie Sun. 2022. "CircEZH2 Regulates Milk Fat Metabolism through miR-378b Sponge Activity" Animals 12, no. 6: 718. https://doi.org/10.3390/ani12060718
APA StyleWang, D., Zhao, Z., Shi, Y., Luo, J., Chen, T., Xi, Q., Zhang, Y., & Sun, J. (2022). CircEZH2 Regulates Milk Fat Metabolism through miR-378b Sponge Activity. Animals, 12(6), 718. https://doi.org/10.3390/ani12060718





