Mitochondrial DNA Variation Contributes to the Aptitude for Dressage and Show Jumping Ability in the Holstein Horse Breed
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling, Sequencing, and Enlargement of the Data Set
2.2. Phenotypic Data
2.3. Data Analysis
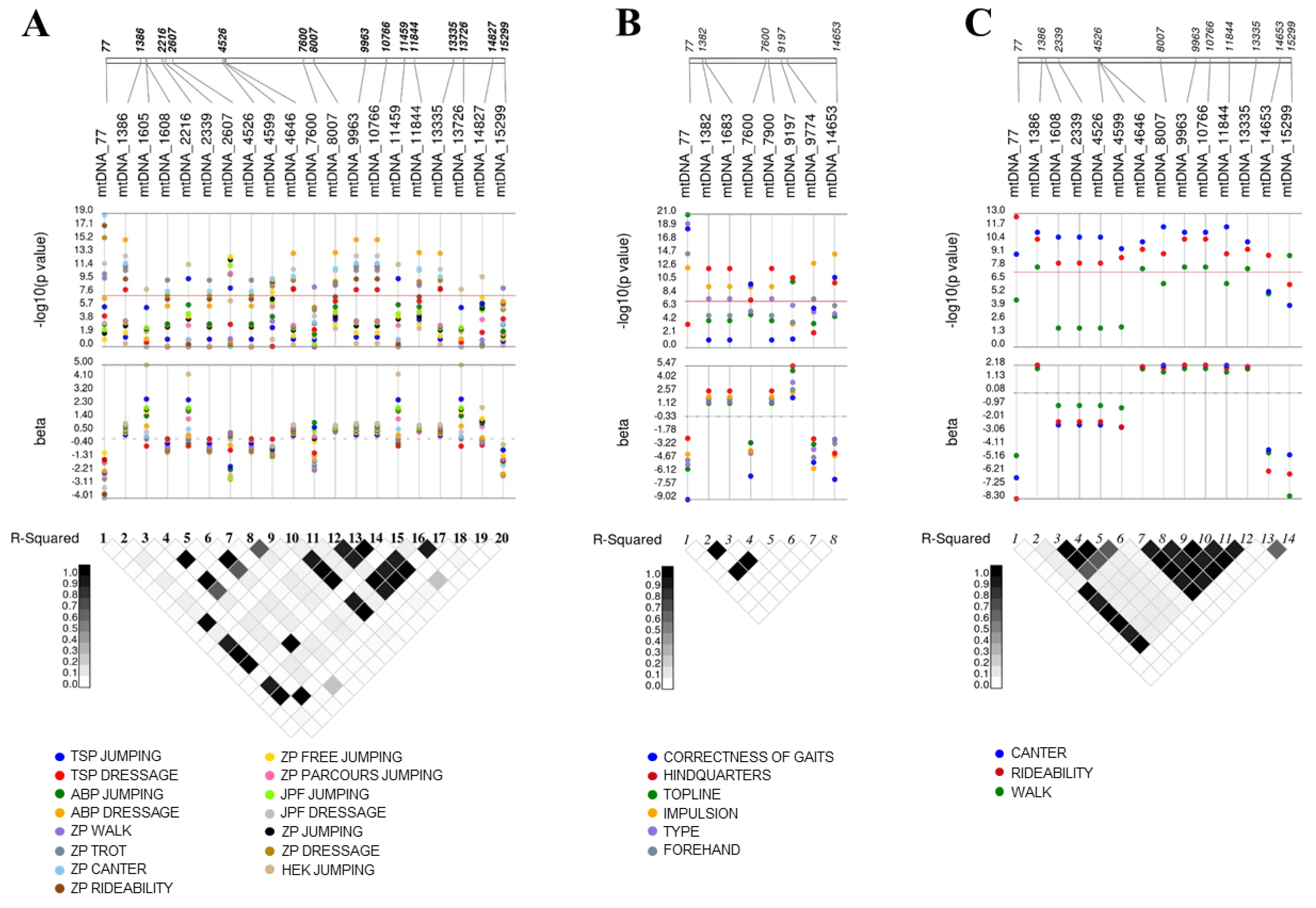
3. Results
4. Discussion
4.1. Mitochondrial Variants
4.2. Mitochondrial Haplogroups and Performance
4.3. Limitations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| Trait | mtSNP a | n | Alleles b | MAF | Ø EBV Major Allele (SD) | Ø EBV Minor Allele (SD) | Occurrence of Minor Allele in Haplogroups (%) c | p-Value | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| B | D | G | I | K | L | N | P | ||||||||
| TSP jumping | mtDNA_2216 | 12,656 | C/T | 0.023 | 105.74 (13.92) | 111.03 (14.23) | 100 | 1.983 × 10−10 | |||||||
| mtDNA_2607 | 12,656 | C/T | 0.039 | 106.01 (13.92) | 102.28 (14.24) | 18.75 | 4.293 × 10−09 | ||||||||
| TSP dressage | mtDNA_9963 | 11,045 | G/A | 0.224 | 86.29 (9.11) | 87.48 (9.35) | 100 | 7.325 × 10−09 | |||||||
| ABP jumping | mtDNA_2607 | 12,971 | C/T | 0.039 | 99.25 (13.80) | 95.12 (14.76) | 18.75 | 4.380 × 10−11 | |||||||
| ABP dressage | mtDNA_4599 | 12,231 | C/G | 0.124 | 85.08 (11.96) | 83.03 (12.12) | 76.92 | 2.638 × 10−10 | |||||||
| mtDNA_8007 | 12,231 | A/G | 0.269 | 84.34 (11.98) | 86.17 (11.96) | 100 | 100 | 3.821 × 10−14 | |||||||
| mtDNA_9963 | 12,231 | G/A | 0.246 | 84.34 (11.98) | 86.37 (11.92) | 100 | 5.709 × 10−16 | ||||||||
| ZP walk | mtDNA_77 | 12,482 | C/T | 0.017 | 86.63 (10.40) | 82.00 (9.77) | 6.25 | 1.620 × 10−10 | |||||||
| mtDNA_7600 | 12,482 | G/C | 0.018 | 86.62 (10.39) | 82.52 (10.48) | 4.17 | 3.067 × 10−09 | ||||||||
| mtDNA_8007 | 12,482 | A/G | 0.265 | 86.21 (10.30) | 87.50 (10.64) | 100 | 100 | 9.512 × 10−10 | |||||||
| mtDNA_9963 | 12,482 | G/A | 0.239 | 86.19 (10.30) | 87.67 (10.67) | 100 | 1.260 × 10−11 | ||||||||
| mtDNA_15299 | 12,482 | C/T | 0.013 | 86.61 (10.41) | 81.84 (9.25) | 11.11 | 4.391 × 10−09 | ||||||||
| ZP trot | mtDNA_77 | 12,482 | C/T | 0.017 | 82.58 (11.99) | 77.16 (12.25) | 6.25 | 4.254 × 10−11 | |||||||
| mtDNA_1608 | 12,482 | A/T | 0.173 | 82.79 (12.04) | 81.01 (11.78) | 100 | 3.203 × 10−10 | ||||||||
| mtDNA_4599 | 12,482 | C/G | 0.125 | 82.73 (11.98) | 80.76 (12.08) | 76.92 | 1.158 × 10−09 | ||||||||
| mtDNA_8007 | 12,482 | A/G | 0.268 | 82.10 (11.88) | 83.56 (12.32) | 100 | 100 | 1.755 × 10−09 | |||||||
| mtDNA_9963 | 12,482 | G/A | 0.242 | 82.07 (11.87) | 83.82 (12.37) | 100 | 2.748 × 10−12 | ||||||||
| ZP canter | mtDNA_77 | 12,724 | C/T | 0.017 | 88.59 (12.99) | 80.57 (13.89) | 6.25 | 2.200 × 10−16 | |||||||
| mtDNA_1608 | 12,724 | A/T | 0.169 | 88.75 (13.09) | 87.00 (12.73) | 100 | 1.273 × 10−08 | ||||||||
| mtDNA_4599 | 12,724 | C/G | 0.123 | 88.73 (13.05) | 86.49 (12.83) | 76.92 | 1.994 × 10−10 | ||||||||
| mtDNA_8007 | 12,724 | A/G | 0.263 | 87.98 (12.98) | 89.77 (13.12) | 100 | 100 | 7.456 × 10−12 | |||||||
| mtDNA_9963 | 12,724 | G/A | 0.238 | 87.99 (12.99) | 89.92 (13.11) | 100 | 1.260 × 10−12 | ||||||||
| ZP rideability | mtDNA_77 | 12,701 | C/T | 0.017 | 85.45 (12.61) | 78.01 (12.60) | 6.25 | 2.200 × 10−16 | |||||||
| mtDNA_4599 | 12,701 | C/G | 0.123 | 85.58 (12.63) | 83.47 (12.60) | 76.92 | 5.726 × 10−10 | ||||||||
| mtDNA_4646 | 12,701 | C/T | 0.241 | 84.96 (12.56) | 86.48 (12.87) | 100 | 6.596 × 10−09 | ||||||||
| mtDNA_9963 | 12,701 | G/A | 0.238 | 84.92 (12.57) | 86.59 (17.82) | 100 | 2.243 × 10−10 | ||||||||
| ZP free jumping | mtDNA_2607 | 12,587 | C/T | 0.039 | 104.78 (15.87) | 99.39 (18.22) | 18.75 | 1.588 × 10−13 | |||||||
| mtDNA_4599 | 12,587 | C/G | 0.123 | 104.86 (15.98) | 102.42 (16.03) | 76.92 | 1.565 × 10−08 | ||||||||
| ZP parcours jumping | mtDNA_2607 | 12,599 | C/T | 0.039 | 102.34 (13.53) | 98.21 (14.79) | 18.75 | 3.446 × 10−11 | |||||||
| JPf jumping | mtDNA_2607 | 12,918 | C/T | 0.039 | 101.65 (16.48) | 96.39 (18.02) | 18.75 | 2.553 × 10−12 | |||||||
| JPf dressage | mtDNA_77 | 12,823 | C/T | 0.017 | 82.38 (13.71) | 75.76 (14.08) | 6.25 | 1.398 × 10−12 | |||||||
| mtDNA_4599 | 12,823 | C/G | 0.123 | 82.54 (13.73) | 80.27 (13.68) | 76.92 | 7.841 × 10−10 | ||||||||
| mtDNA_8007 | 12,823 | A/G | 0.264 | 81.76 (13.65) | 83.63 (13.91) | 100 | 100 | 1.348 × 10−11 | |||||||
| mtDNA_9963 | 12,823 | G/A | 0.238 | 81.76 (13.65) | 83.87 (13.90) | 100 | 1.136 × 10−13 | ||||||||
| ZP jumping | mtDNA_2607 | 12,696 | C/T | 0.039 | 103.67 (16.28) | 98.26 (18.27) | 18.75 | 3.863 × 10−13 | |||||||
| ZP dressage | mtDNA_77 | 12,801 | C/T | 0.017 | 82.91 (13.55) | 75.37 (13.86) | 6.25 | 2.900 × 10−16 | |||||||
| mtDNA_4599 | 12,801 | C/G | 0.123 | 83.05 (13.59) | 80.87 (13.41) | 76.92 | 2.268 × 10−09 | ||||||||
| mtDNA_8007 | 12,801 | A/G | 0.264 | 82.34 (13.46) | 84.03 (13.87) | 100 | 100 | 5.207 × 10−10 | |||||||
| mtDNA_9963 | 12,801 | G/A | 0.238 | 82.32 (13.46) | 84.27 (13.89) | 100 | 4.479 × 10−12 | ||||||||
| HEK jumping | mtDNA_1605 | 10,711 | G/A | 0.011 | 109.26 (18.93) | 119.26 (18.74) | 20.00 | 6.616 × 10−09 | |||||||
| mtDNA_2216 | 10,711 | C/T | 0.022 | 109.28 (18.88) | 117.92 (20.22) | 100 | 1.877 × 10−12 | ||||||||
| mtDNA_14827 | 10,711 | G/A | 0.085 | 109.01 (18.92) | 113.25 (18.94) | 100 | 11.11 | 20.00 | 37.50 | 7.69 | 1.061 × 10−10 | ||||
| Trait | mtSNP a | n | Alleles b | MAF | Ø EBV Major Allele (SD) | Ø EBV Minor Allele (SD) | Occurrence of Minor Allele in Haplogroups (%) c | p-Value | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| B | D | G | I | K | L | N | P | ||||||||
| Studbook inspection | |||||||||||||||
| HOL type | mtDNA_77 | 16,447 | C/T | 0.019 | 95.54 (19.88) | 85.09 (17.44) | 6.25 | 2.200 × 10−16 | |||||||
| mtDNA_1382 | 16,447 | C/T | 0.056 | 95.13 (19.89) | 98.90 (19.48) | 100 | 1.977 × 10−08 | ||||||||
| HOL topline | mtDNA_77 | 16,447 | C/T | 0.019 | 94.79 (20.96) | 83.36 (18.23) | 6.25 | 2.200 × 10−16 | |||||||
| mtDNA_9197 | 16,447 | T/C | 0.012 | 94.46 (20.97) | 104.37 (19.26) | 11.11 | 4.124 × 10−11 | ||||||||
| HOL forehand | mtDNA_77 | 16,447 | C/T | 0.019 | 92.66 (20.86) | 83.18 (18.26) | 6.25 | 1.688 × 10−15 | |||||||
| mtDNA_7600 | 16,447 | G/C | 0.017 | 92.62 (20.16) | 84.60 (22.16) | 4.17 | 1.774 × 10−10 | ||||||||
| mtDNA_9774 | 16,447 | G/A | 0.011 | 92.58 (20.76) | 83.67 (26.86) | 4.17 | 2.089 × 10−08 | ||||||||
| HOL hindquarters | mtDNA_1382 | 16,447 | C/T | 0.056 | 90.51 (22.42) | 96.02 (22.31) | 100 | 3.562 × 10−13 | |||||||
| mtDNA_7600 | 16,447 | G/C | 0.017 | 90.95 (22.44) | 83.46 (21.91) | 4.17 | 3.122 × 10−08 | ||||||||
| mtDNA_9197 | 16,447 | T/C | 0.012 | 90.69 (22.45) | 101.64 (20.11) | 11.11 | 1.004 × 10−11 | ||||||||
| mtDNA_14653 | 16,447 | A/G | 0.021 | 90.99 (22.46) | 83.01 (20.67) | 33.33 | 6.133 × 10−11 | ||||||||
| HOL correctness of gaits | mtDNA_77 | 16,447 | C/T | 0.019 | 95.13 (19.66) | 85.01 (18.09) | 6.25 | 2.200 × 10−16 | |||||||
| mtDNA_7600 | 16,447 | G/C | 0.017 | 95.07 (19.66) | 87.40 (19.30) | 4.17 | 9.645 × 10−11 | ||||||||
| mtDNA_14653 | 16,447 | A/G | 0.021 | 95.09 (19.69) | 87.79 (17.72) | 33.33 | 8.903 × 10−12 | ||||||||
| HOL impulsion | mtDNA_77 | 16,447 | C/T | 0.019 | 95.34 (19.76) | 87.12 (19.06) | 6.25 | 2.666 × 10−13 | |||||||
| mtDNA_1382 | 16,447 | C/T | 0.056 | 94.95 (19.74) | 99.15 (18.52) | 100 | 2.482 × 10−10 | ||||||||
| mtDNA_7600 | 16,447 | G/C | 0.017 | 95.31 (19.68) | 87.83 (18.92) | 4.17 | 2.829 × 10−10 | ||||||||
| mtDNA_9774 | 16,447 | G/A | 0.011 | 95.31 (19.62) | 84.03 (23.45) | 4.17 | 5.447 × 10−14 | ||||||||
| mtDNA_14653 | 16,447 | A/G | 0.021 | 95.37 (19.69) | 86.86 (17.61) | 33.33 | 1.897 × 10−15 | ||||||||
| Mare performance test | |||||||||||||||
| HOL canter | mtDNA_77 | 6334 | C/T | 0.016 | 94.41 (20.08) | 81.09 (20.55) | 6.25 | 9.199 × 10−10 | |||||||
| mtDNA_1608 | 6334 | A/T | 0.160 | 94.99 (22.23) | 89.97 (21.25) | 100 | 2.009 × 10−11 | ||||||||
| mtDNA_4599 | 6334 | C/G | 0.151 | 91.24 (22.12) | 85.94 (21.73) | 76.92 | 2.666 × 10−10 | ||||||||
| mtDNA_4646 | 6334 | C/T | 0.258 | 93.12 (22.05) | 97.24 (22.17) | 100 | 5.784 × 10−11 | ||||||||
| mtDNA_8007 | 6334 | A/G | 0.277 | 92.99 (22.06) | 97.31 (22.09) | 100 | 100 | 2.000 × 10−12 | |||||||
| HOL walk | mtDNA_4646 | 6334 | C/T | 0.258 | 97.84 (22.54) | 101.61 (24.12) | 100 | 2.351 × 10−08 | |||||||
| mtDNA_15299 | 6334 | C/T | 0.012 | 99.01 (22.99) | 82.84 (17.19) | 11.11 | 1.249 × 10−09 | ||||||||
| HOL rideability | mtDNA_77 | 6334 | C/T | 0.016 | 94.85 (22.27) | 78.25 (21.13) | 6.25 | 2.088 × 10−13 | |||||||
| mtDNA_1608 | 6334 | A/T | 0.160 | 95.31 (22.34) | 90.79 (22.19) | 100 | 6.809 × 10−09 | ||||||||
| mtDNA_4599 | 6334 | C/G | 0.151 | 95.21 (22.23) | 89.88 (22.85) | 76.92 | 4.052 × 10−12 | ||||||||
| mtDNA_4646 | 6334 | C/T | 0.258 | 93.52 (22.20) | 97.64 (22.64) | 100 | 3.055 × 10−10 | ||||||||
| mtDNA_8007 | 6334 | A/G | 0.277 | 93.50 (22.18) | 97.42 (22.67) | 100 | 100 | 8.262 × 10−10 | |||||||
| mtDNA_14653 | 6334 | A/G | 0.021 | 94.83 (22.38) | 82.56 (19.07) | 33.33 | 1.183 × 10−09 | ||||||||
References
- World Breeding Federation for Sport Horses: Breeder and Studbook Rankings. Available online: http://www.wbfsh.org/GB/Rankings/Breeder%20and%20Studbook%20rankings.aspx (accessed on 1 August 2021).
- Fédération Équestre Nationale (FN). Annual Report 2020; FN-Verlag: Warendorf, Germany, 2020. [Google Scholar]
- Lin, X.; Zheng, H.X.; Davie, A.; Zhou, S.; Wen, L.; Meng, J.; Zhang, Y.; Aladaer, Q.; Liu, B.; Liu, W.J.; et al. Association of low race performance with mtDNA haplogroup L3b of Australian thoroughbred horses. Mitochondrial DNA Part A 2018, 29, 323–330. [Google Scholar] [CrossRef] [PubMed]
- Art, T.; Amory, H.; Desmecht, D.; Lekeux, P. Effect of show jumping on heart rate, blood lactate and other plasma biochemical values. Equine Vet. J. 1990, 22, 78–82. [Google Scholar] [CrossRef] [PubMed]
- Engel, L.; Becker, D.; Nissen, T.; Russ, I.; Thaller, G.; Krattenmacher, N. Exploring the Origin and Relatedness of Maternal Lineages Through Analysis of Mitochondrial DNA in the Holstein Horse. Front. Genet. 2021, 12, 1242. [Google Scholar] [CrossRef] [PubMed]
- Bowling, A.T.; Del Valle, A.; Bowling, M. A pedigree-based study of mitochondrial D-loop DNA sequence variation among Arabian horses. Anim. Genet. 2000, 31, 1–7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kavar, T.; Habe, F.; Brem, G.; Dovč, P. Mitochondrial D-loop sequence variation among the 16 maternal lines of the Lipizzan horse breed. Anim. Genet. 1999, 30, 423–430. [Google Scholar] [CrossRef] [PubMed]
- Welker, V.; Stock, K.F.; Schöpke, K.; Swalve, H.H. Genetic parameters of new comprehensive performance traits for dressage and show jumping competitions performance of German riding horses. Livest. Sci. 2018, 212, 93–98. [Google Scholar] [CrossRef]
- Jaitner, J. Genetic Evaluation for Horses. 2019. Available online: https://www.vit.de/fileadmin/DE/Zuchtwertschaetzung/FN_ZWS_2019_description.pdf (accessed on 22 June 2021).
- Miller, S.A.; Dykes, D.D.; Polesky, H.F.R.N. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988, 16, 1215. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rozen, S.; Skaletsky, H. Primer3 on the WWW for general users and for biologist programmers. In Bioinformatics Methods and Protocols: Methods in Molecular Biology; Krawetz, S., Misener, S., Eds.; Humana Press: Totowa, NJ, USA, 2000; Volume 132, pp. 365–386. [Google Scholar]
- Achilli, A.; Olivieri, A.; Soares, P.; Lancioni, H.; Kashani, B.H.; Perego, U.A.; Nergadze, S.G.; Carossa, V.; Santagostino, M.; Capomaccio, S.; et al. Mitochondrial genomes from modern horses reveal the major haplogroups that underwent domestication. Proc. Natl. Acad. Sci. USA 2012, 109, 2449–2454. [Google Scholar] [CrossRef] [Green Version]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.; Bender, D.; Maller, J.; Sklar, P.; De Bakker, P.I.; Daly, M.J.; et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef] [Green Version]
- Barrett, J.C.; Fry, B.; Maller, J.; Daly, M.J. Haploview: Analysis and visualization of LD and haplotype maps. Bioinformatics 2005, 21, 263–265. [Google Scholar] [CrossRef] [Green Version]
- Pendergrass, S.A.; Dudek, S.M.; Crawford, D.C.; Ritchie, M.D. Synthesis-View: Visualization and interpretation of SNP association results for multi-cohort, multi-phenotype data and meta-analysis. BioData Min. 2010, 3, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2020. [Google Scholar]
- Ekine, C.C.; Rowe, S.J.; Bishop, S.C.; de Koning, D.J. Why breeding values estimated using familial data should not be used for genome-wide association studies. G3 Genes Genomes Genet. 2014, 4, 341–347. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boore, J.L. Animal mitochondrial genomes. Nucleic Acids Res. 1999, 27, 1767–1780. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lührs-Behnke, H.; Röhe, R.; Kalm, E. Genetic analyses of riding test and their connections with traits of stallion performance and breeding mare tests. Züchtungskunde 2006, 78, 119–128. [Google Scholar]
- Bray, M.S.; Hagberg, J.M.; Perusse, L.; Rankinen, T.; Roth, S.M.; Wolfarth, B.; Bouchard, C. The human gene map for performance and health-related fitness phenotypes: The 2006-2007 update. Med. Sci. Sports Exerc. 2009, 41, 34–72. [Google Scholar] [CrossRef] [PubMed]
- Niemi, A.K.; Majamaa, K. Mitochondrial DNA and ACTN3 genotypes in Finnish elite endurance and sprint athletes. Eur. J. Hum. Genet. 2005, 13, 965–969. [Google Scholar] [CrossRef] [PubMed]
- Mikami, E.; Fuku, N.; Takahashi, H.; Ohiwa, N.; Scott, R.A.; Pitsiladis, Y.P.; Higuchi, M.; Kawahara, T.; Tanaka, M. Mitochondrial haplogroups associated with elite Japanese athlete status. Br. J. Sports Med. 2011, 45, 1179–1183. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nogales-Gadea, G.; Pinós, T.; Ruiz, J.R.; Marzo, P.F.; Fiuza-Luces, C.; López-Gallardo, E.; Ruiz-Pesini, E.; Martín, M.A.; Arenas, J.; Morán, M.; et al. Are mitochondrial haplogroups associated with elite athletic status? A study on a Spanish cohort. Mitochondrion 2011, 11, 905–908. [Google Scholar] [CrossRef] [PubMed]
- Harrison, S.P.; Turrion-Gomez, J.L. Mitochondrial DNA: An important female contribution to thoroughbred racehorse performance. Mitochondrion 2006, 6, 53–66. [Google Scholar] [CrossRef]
- Próchniak, T.; Rozempolska-Rucińska, I.; Zięba, G. Maternal effect on sports performance traits in horses. Czech J. Anim. Sci. 2019, 64, 361–365. [Google Scholar] [CrossRef]
- Krattenmacher, N.; Tetens, J.; Hedt, S.; Stamer, E.; Thaller, G. The role of maternal lineages in horse breeding: Effects on conformation and performance traits. In Proceedings of the 10th World Congress on Genetics Applied to Livestock Production, Vancouver, BC, Canada, 17–22 August 2014; Available online: http://wcgalp.org/system/files/proceedings/2014/role-maternal-lineages-horse-breeding-effects-conformation-and-performance-traits.pdf (accessed on 8 June 2021).
- Børte, S.; Zwart, J.A.; Skogholt, A.H.; Gabrielsen, M.E.; Thomas, L.F.; Fritsche, L.G.; Surakka, I.; Nielsen, J.B.; Zhou, W.; Wolford, B.N.; et al. Mitochondrial genome-wide association study of migraine–the HUNT Study. Cephalalgia 2020, 40, 625–634. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Schröder, R.; Ni, S.; Madea, B.; Stoneking, M. Extensive tissue-related and allele-related mtDNA heteroplasmy suggests positive selection for somatic mutations. Proc. Natl. Acad. Sci. USA 2015, 112, 2491–2496. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| Haplogroup a | Number of Lineages | Number of Sequenced Mares | Number of Total Mares with b | ||
|---|---|---|---|---|---|
| FN Breeding Values c | SBI Breeding Values | MPT Breeding Values | |||
| B | 24 | 72 | 3091 | 3952 | 1618 |
| D | 4 | 13 | 290 | 352 | 120 |
| G | 9 | 33 | 2099 | 2590 | 1057 |
| I | 5 | 14 | 817 | 1053 | 420 |
| K | 1 | 3 | 112 | 150 | 42 |
| L | 16 | 78 | 3646 | 4650 | 1716 |
| N | 13 | 11 | 2209 | 2770 | 1016 |
| P | 3 | 47 | 707 | 930 | 345 |
| Total | 75 | 271 | 12,971 | 16,447 | 6334 |
| Breeding Value a | Reliability | ||||
|---|---|---|---|---|---|
| Trait b | n | Mean (SD c) | Range | Mean (SD c) | Range |
| FN genetic evaluation | |||||
| TSP jumping | 12,656 | 105.86 (13.95) | 48−148 | 41.82 (5.50) | 30−75 |
| TSP dressage | 11,045 | 86.58 (9.18) | 56−136 | 35.96 (3.92) | 30−66 |
| ABP jumping | 12,971 | 99.09 (13.87) | 31−137 | 48.75 (7.64) | 30−78 |
| ABP dressage | 12,231 | 84.83 (12.00) | 34−147 | 43.41 (7.19) | 30−68 |
| ZP walk | 12,482 | 86.55 (10.41) | 39−136 | 44.51 (8.59) | 30−71 |
| ZP trot | 12,482 | 82.48 (12.01) | 31−141 | 48.12 (10.82) | 30−76 |
| ZP canter | 12,724 | 88.45 (13.04) | 29−143 | 48.04 (10.33) | 30−75 |
| ZP rideability | 12,701 | 85.32 (12.65) | 34−143 | 47.15 (9.83) | 30−74 |
| ZP free jumping | 12,587 | 104.56 (16.01) | 45−149 | 43.60 (7.01) | 30−71 |
| ZP parcours jumping | 12,599 | 102.17 (13.61) | 44−142 | 40.59 (4.59) | 30−70 |
| JPf jumping | 12,918 | 101.45 (16.58) | 31−148 | 48.09 (6.83) | 30−79 |
| JPf dressage | 12,918 | 82.26 (13.74) | 24−149 | 49.81 (11.06) | 30−77 |
| ZP jumping | 12,823 | 103.46 (16.39) | 43−149 | 44.20 (5.95) | 30−72 |
| ZP dressage | 12,696 | 82.78 (13.59) | 24−149 | 51.13 (12.38) | 30−80 |
| HEK jumping | 10,711 | 109.37 (18.95) | 50−178 | 48.15 (7.96) | 30−81 |
| HEK dressage | 5302 | 89.95 (12.35) | 60−161 | 35.03 (4.77) | 30−63 |
| Studbook inspection | |||||
| HOL type | 16,447 | 95.34 (19.88) | 10−169 | 58.27 (3.83) | 30−83 |
| HOL topline | 16,447 | 94.58 (20.97) | 5−168 | 58.27 (3.83) | 30−83 |
| HOL forehand | 16,447 | 92.48 (20.85) | 4−165 | 58.27 (3.83) | 30−83 |
| HOL hindquarters | 16,447 | 90.82 (22.45) | 4−173 | 58.27 (3.83) | 30−83 |
| HOL correctness of gaits | 16,447 | 94.94 (19.68) | 11−187 | 58.27 (3.83) | 30−83 |
| HOL impulsion | 16,447 | 95.19 (19.69) | 13−189 | 58.27 (3.83) | 30−83 |
| Mare performance test | |||||
| HOL walk | 6334 | 98.81 (23.54) | 3−214 | 52.13 (7.83) | 30−69 |
| HOL trot | 6334 | 96.83 (22.19) | 12−187 | 52.13 (7.83) | 30−69 |
| HOL canter | 6334 | 94.19 (21.91) | 4−189 | 52.13 (7.83) | 30−69 |
| HOL rideability | 6334 | 94.59 (22.81) | 11−200 | 52.13 (7.83) | 30−69 |
| HOL free jumping | 6334 | 85.10 (22.14) | 3−149 | 52.13 (7.83) | 30−69 |
| Genetic Data | |
|---|---|
| Total number of substitutions | 467 |
| Non-synonymous mtSNPs | 101 |
| Non-synonymous mtSNPs with MAF > 0.01 | 51 |
| Results | |
| Total number of associated mtSNPs | 24 |
| Number of mtSNPs associated with FN-EBVs a | 20 |
| Number of mtSNPs associated with HOL-EBVs b | 20 |
| Number of mtSNP pairs in total LD (r2 = 1) | 4 |
| Number of mtSNPs triplets in total LD (r2 = 1) | 2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Engel, L.; Becker, D.; Nissen, T.; Russ, I.; Thaller, G.; Krattenmacher, N. Mitochondrial DNA Variation Contributes to the Aptitude for Dressage and Show Jumping Ability in the Holstein Horse Breed. Animals 2022, 12, 704. https://doi.org/10.3390/ani12060704
Engel L, Becker D, Nissen T, Russ I, Thaller G, Krattenmacher N. Mitochondrial DNA Variation Contributes to the Aptitude for Dressage and Show Jumping Ability in the Holstein Horse Breed. Animals. 2022; 12(6):704. https://doi.org/10.3390/ani12060704
Chicago/Turabian StyleEngel, Laura, Doreen Becker, Thomas Nissen, Ingolf Russ, Georg Thaller, and Nina Krattenmacher. 2022. "Mitochondrial DNA Variation Contributes to the Aptitude for Dressage and Show Jumping Ability in the Holstein Horse Breed" Animals 12, no. 6: 704. https://doi.org/10.3390/ani12060704
APA StyleEngel, L., Becker, D., Nissen, T., Russ, I., Thaller, G., & Krattenmacher, N. (2022). Mitochondrial DNA Variation Contributes to the Aptitude for Dressage and Show Jumping Ability in the Holstein Horse Breed. Animals, 12(6), 704. https://doi.org/10.3390/ani12060704







