Reaching the Monophyly: Re-Evaluation of the Enigmatic Species Tenuibiotus hyperonyx (Maucci, 1983) and the Genus Tenuibiotus (Eutardigrada)
Abstract
:Simple Summary
Abstract
1. Introduction
2. Material and Methods
2.1. Samples and Specimens
2.2. DNA Sequencing
2.3. Phylogenetic Analyses
2.4. Microscopy and Imaging
2.5. Morphometry and Morphological Nomenclature
2.6. Comparative Material
2.7. Availability of Data and Materials
3. Results
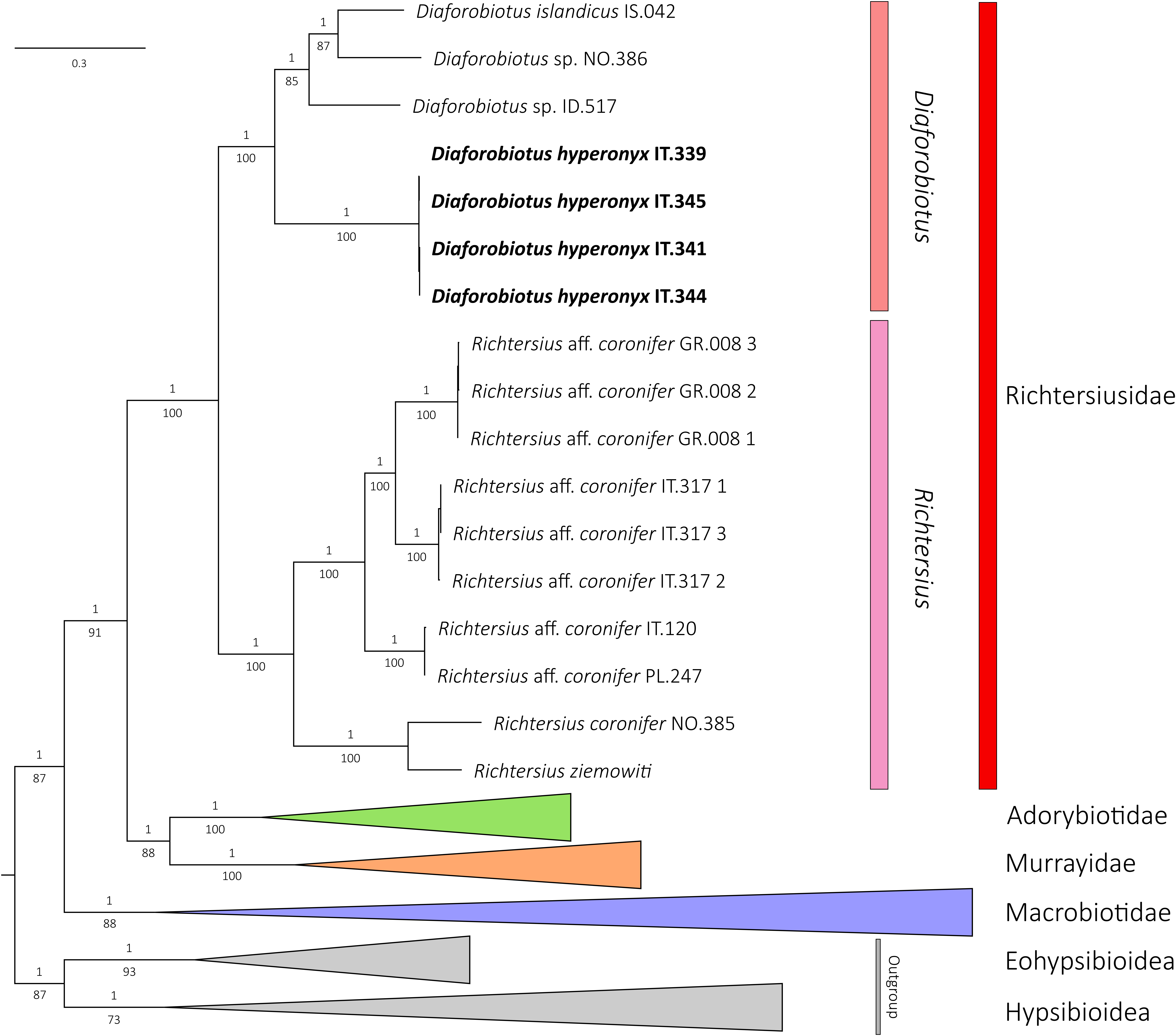
3.1. Phylogenetic Position of T. hyperonyx
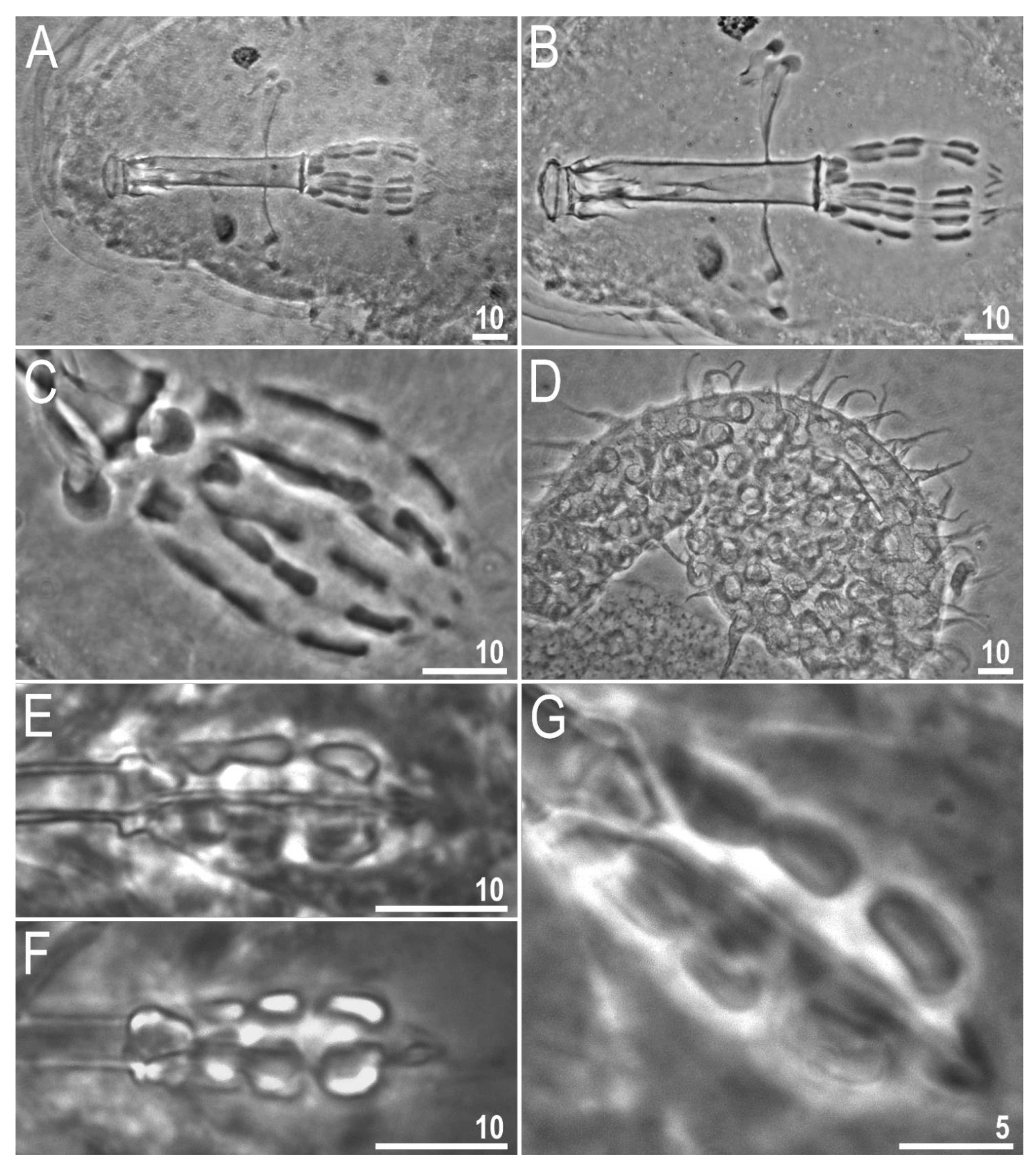
3.2. Amended Description of D. hyperonyx comb. nov.
3.2.1. Systematic and Taxonomic Account
3.2.2. Material Examined
3.2.3. Slide and SEM Stubs Depositories
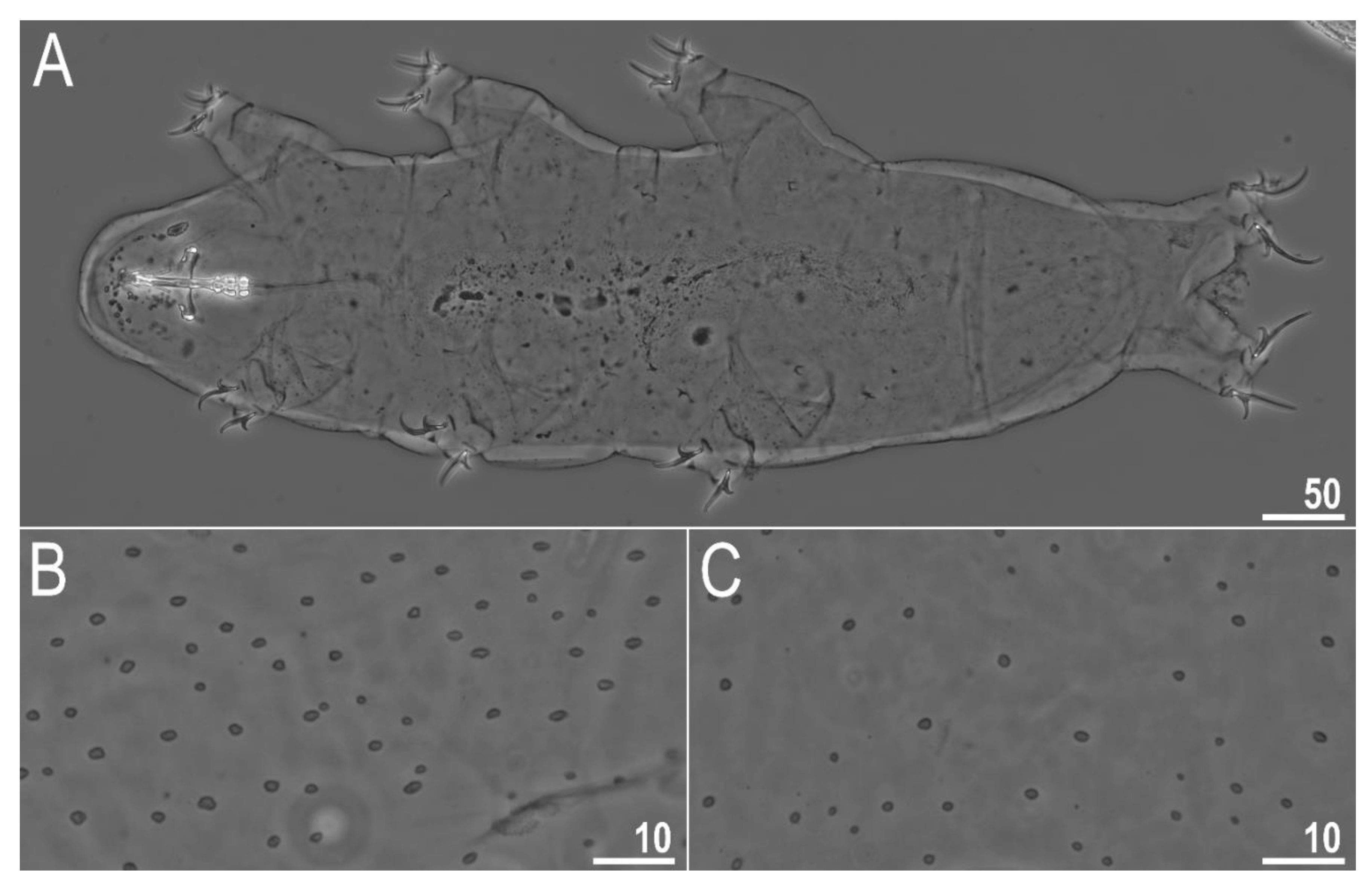
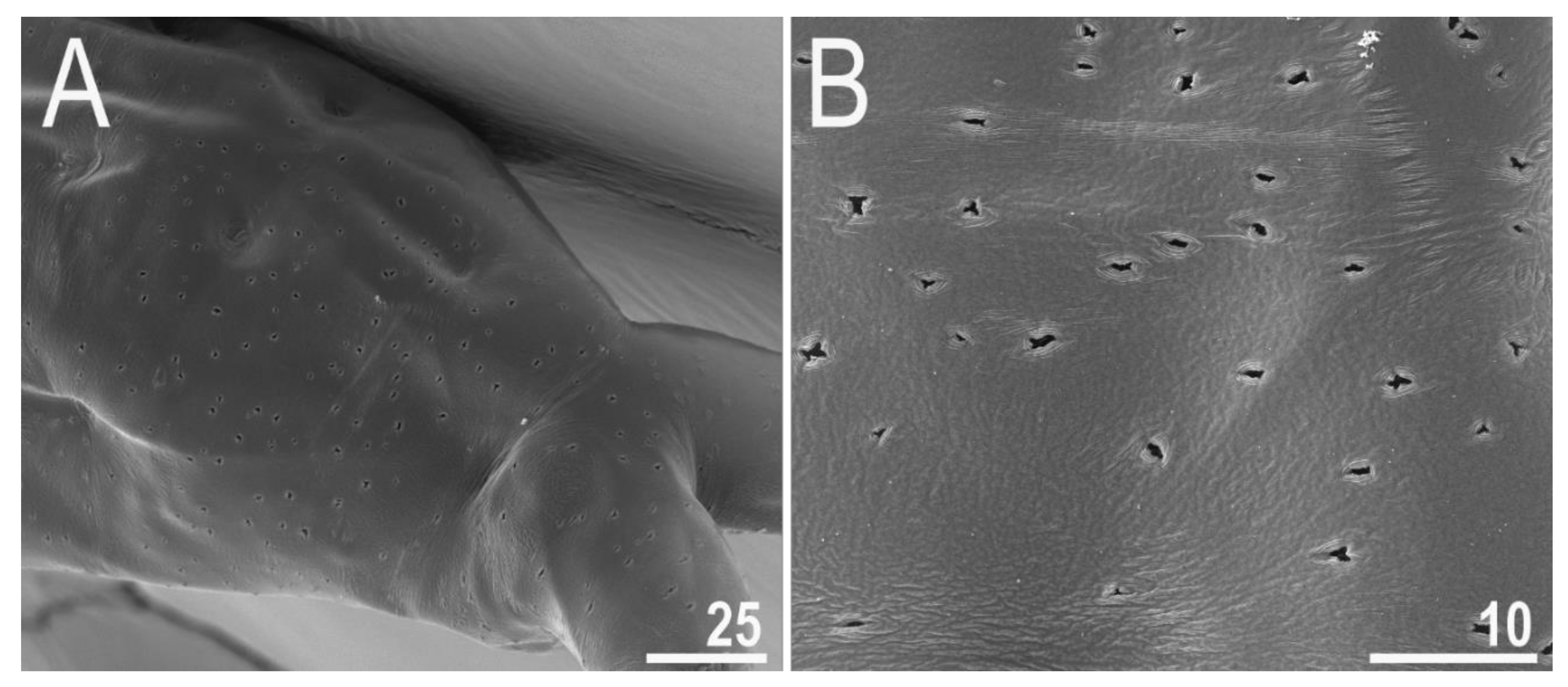
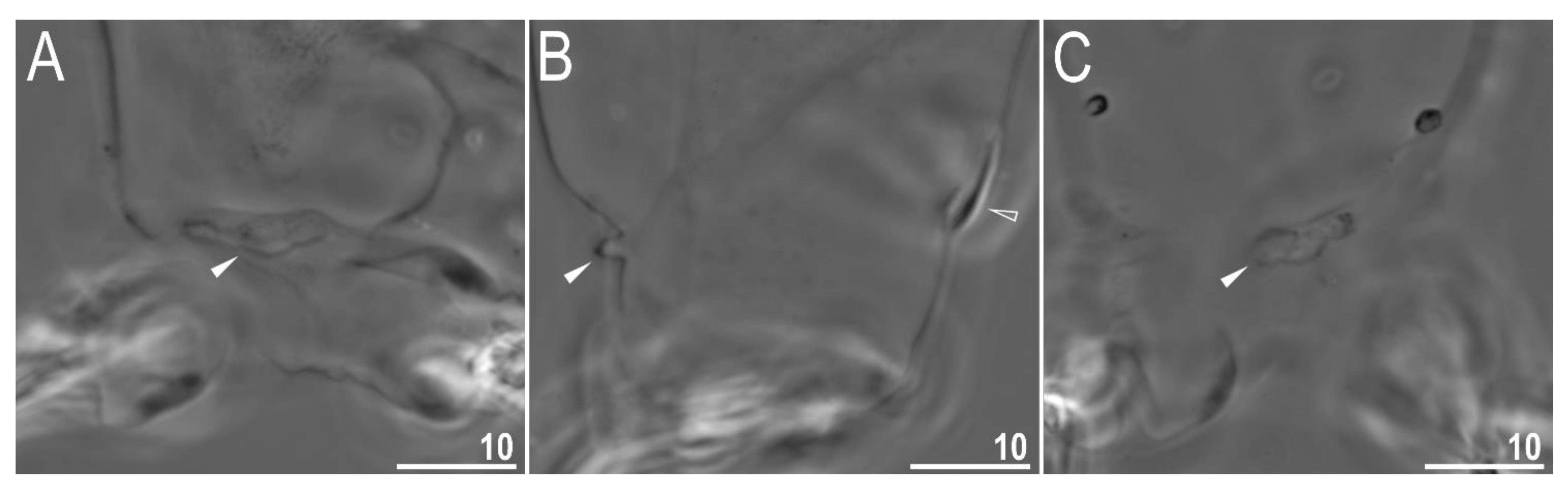
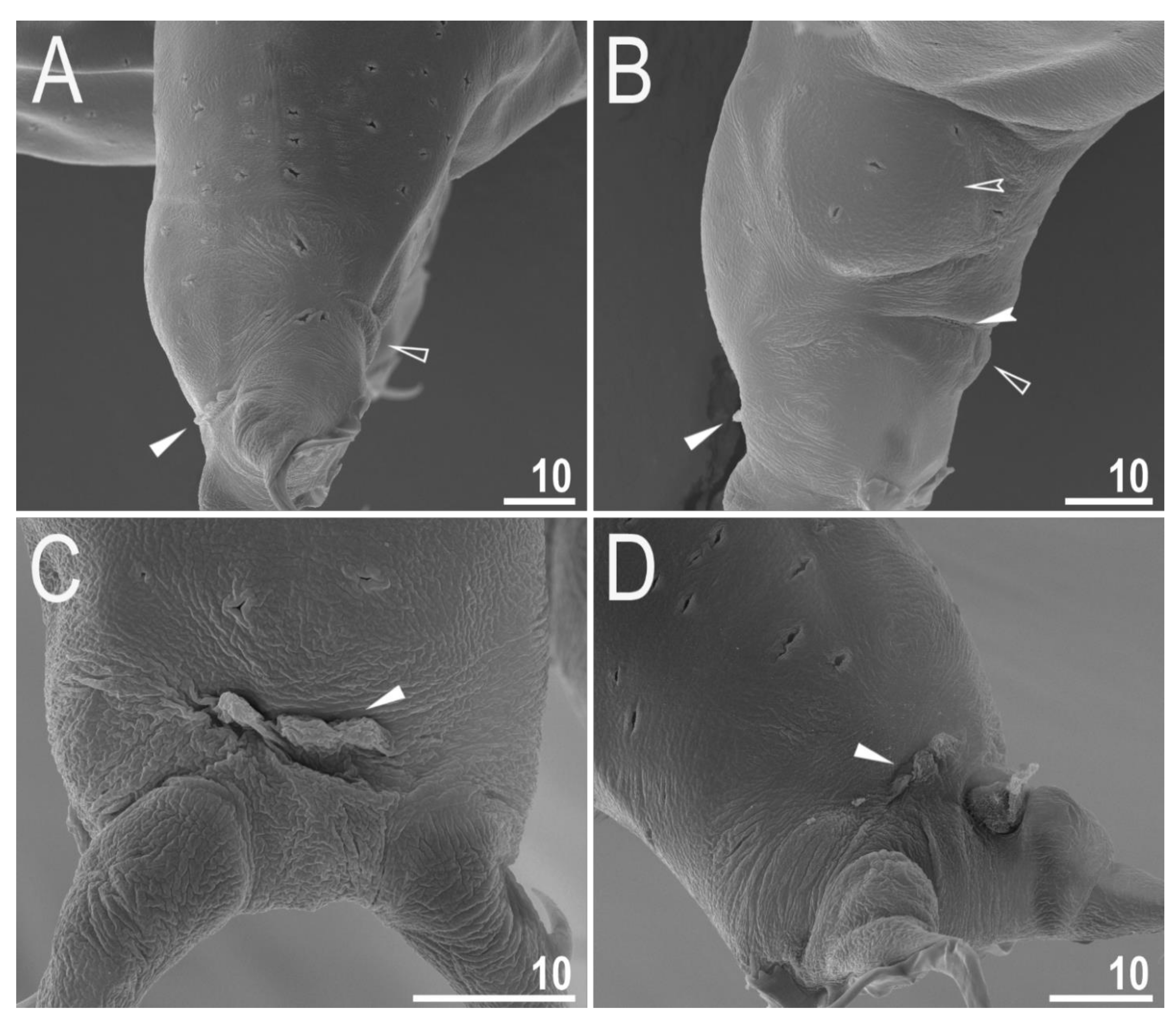
3.2.4. Animals
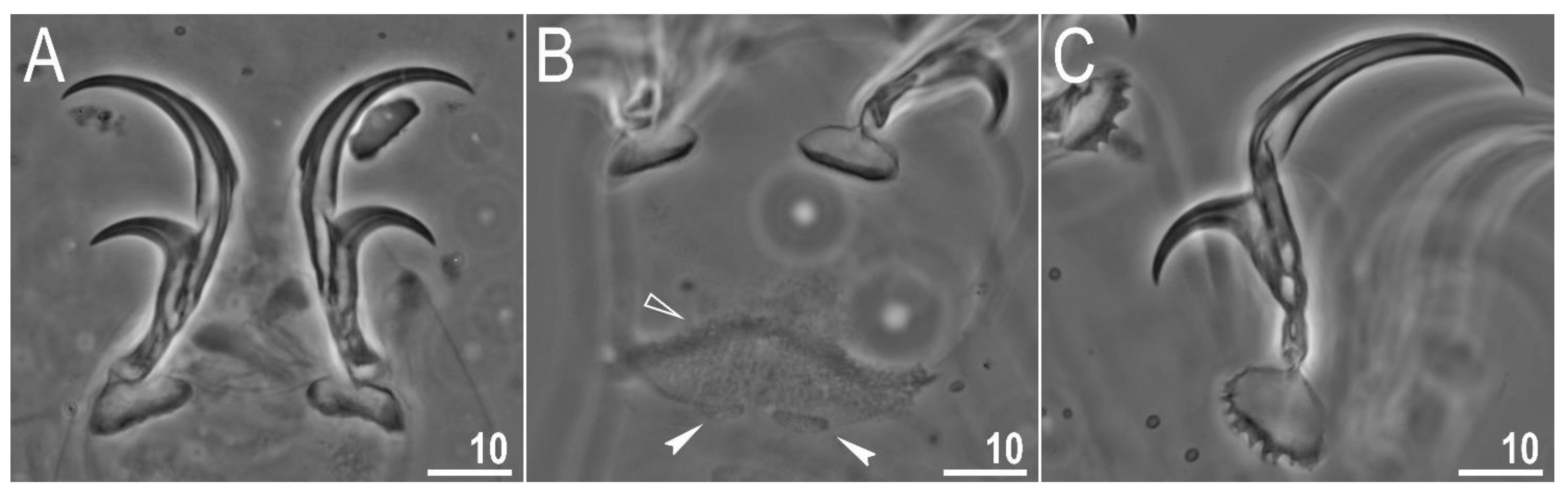
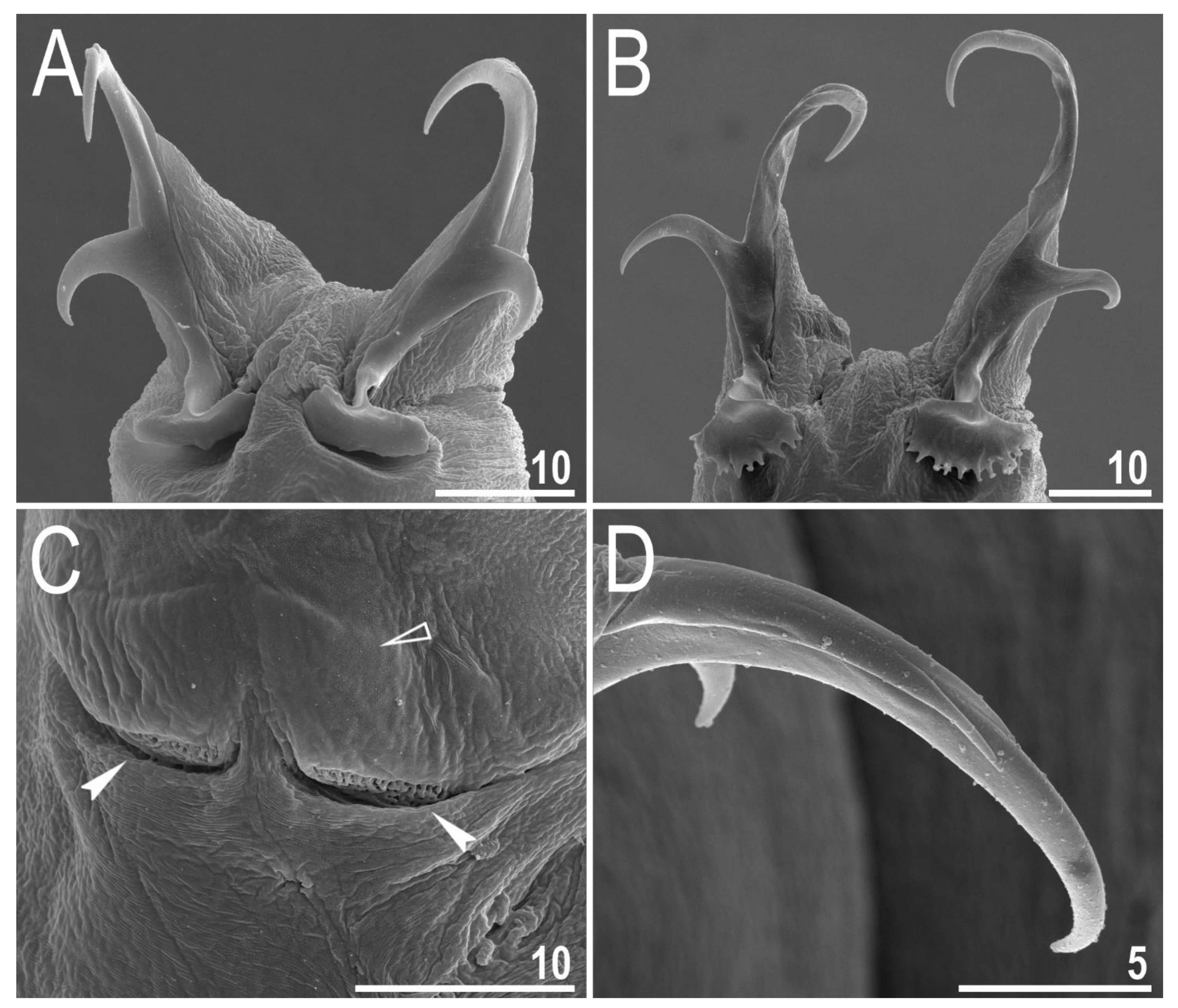

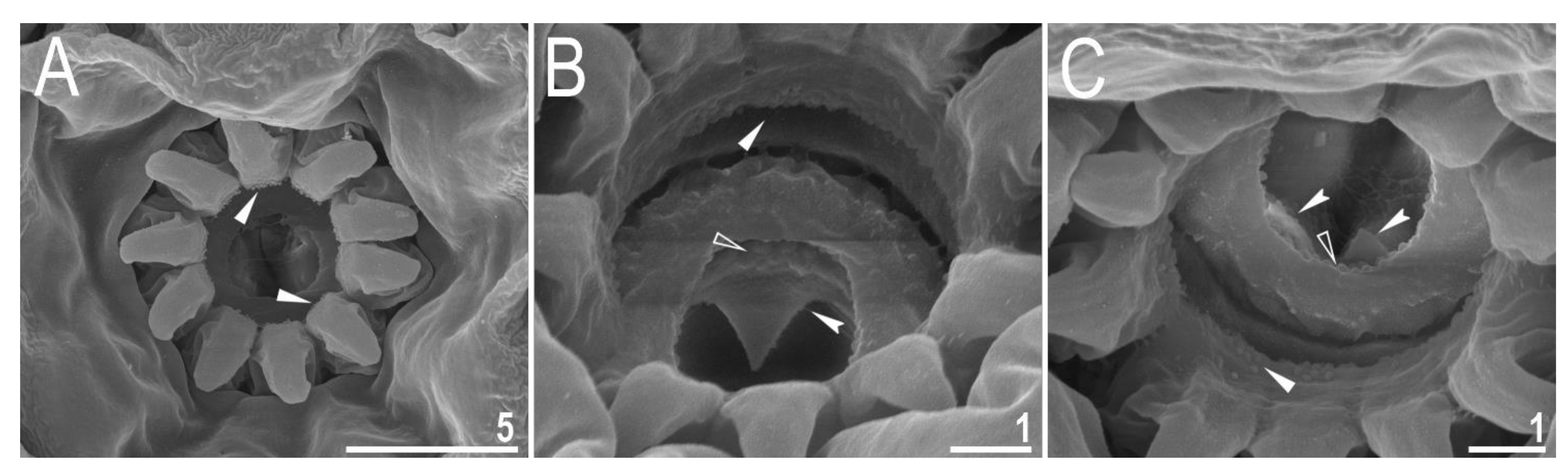
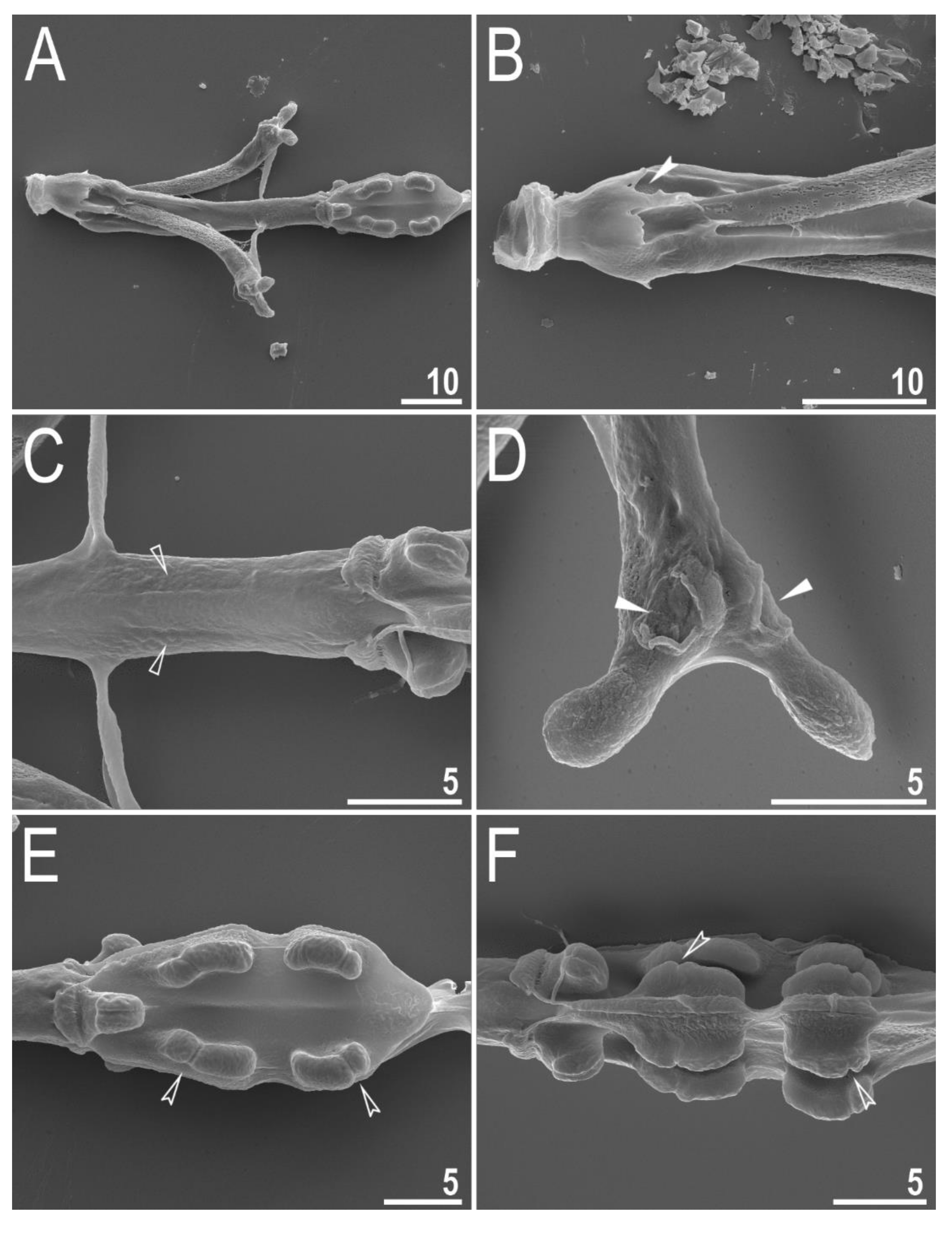
| Character | N | Range | Mean | SD | |||
|---|---|---|---|---|---|---|---|
| µm | pt | µm | pt | µm | pt | ||
| Body length | 18 | 449–961 | 1095–1888 | 663 | 1394 | 137 | 186 |
| Buccal tube | |||||||
| Buccal tube length | 18 | 34.8–52.6 | 47.2 | 5.1 | |||
| Stylet support insertion point | 18 | 25.8–39.3 | 72.0–74.7 | 34.6 | 73.3 | 3.8 | 0.9 |
| Buccal tube external width | 18 | 3.7–6.1 | 10.2–11.8 | 5.2 | 10.9 | 0.7 | 0.5 |
| Buccal tube internal width | 18 | 2.0–3.3 | 4.6–6.5 | 2.7 | 5.7 | 0.4 | 0.6 |
| Ventral lamina length | 17 | 19.0–28.7 | 49.1–56.6 | 25.7 | 53.6 | 2.7 | 2.5 |
| Placoid lengths | |||||||
| Macroplacoid 1 | 18 | 5.4–9.9 | 12.7–19.4 | 7.5 | 15.8 | 1.1 | 1.3 |
| Macroplacoid 2 | 18 | 4.0–8.7 | 11.4–16.5 | 6.1 | 12.8 | 1.1 | 1.1 |
| Macroplacoid row | 18 | 10.4–18.8 | 29.9–36.9 | 15.4 | 32.4 | 2.1 | 1.6 |
| Claw 1 heights | |||||||
| External base | 18 | 6.9–16.1 | 16.6–31.6 | 10.5 | 22.2 | 2.2 | 3.6 |
| External primary branch | 18 | 15.3–32.1 | 41.6–63.1 | 23.4 | 49.2 | 4.6 | 5.7 |
| External secondary branch | 13 | 7.9–16.1 | 21.0–31.0 | 11.5 | 24.2 | 2.0 | 2.7 |
| External base/primary branch (cct) | 18 | 36.4–57.3 | 45.1 | 5.9 | |||
| Internal base | 18 | 6.1–15.6 | 17.5–30.6 | 10.1 | 21.2 | 2.1 | 3.3 |
| Internal primary branch | 18 | 14.5–31.4 | 40.2–60.5 | 22.4 | 47.1 | 4.2 | 5.1 |
| Internal secondary branch | 13 | 6.1–15.3 | 17.5–29.5 | 11.0 | 23.0 | 2.2 | 3.1 |
| Internal base/primary branch (cct) | 18 | 36.5–55.9 | 45.2 | 5.8 | |||
| Claw 2 heights | |||||||
| External base | 13 | 7.6–18.5 | 20.1–36.3 | 12.1 | 25.6 | 2.8 | 3.9 |
| External primary branch | 14 | 16.5–35.5 | 45.7–69.7 | 25.9 | 54.5 | 5.9 | 7.3 |
| External secondary branch | 9 | 11.2–20.0 | 25.9–39.3 | 14.8 | 29.7 | 2.6 | 4.2 |
| External base/primary branch (cct) | 13 | 40.3–54.7 | 47.1 | 4.6 | |||
| Internal base | 17 | 7.0–16.8 | 17.9–33.0 | 11.3 | 23.9 | 2.5 | 3.5 |
| Internal primary branch | 17 | 15.2–34.4 | 42.1–67.6 | 24.8 | 52.3 | 5.5 | 7.2 |
| Internal secondary branch | 14 | 9.4–17.2 | 21.7–33.8 | 13.4 | 27.3 | 2.0 | 3.2 |
| Internal base/primary branch (cct) | 17 | 37.0–55.6 | 46.1 | 5.2 | |||
| Claw 3 heights | |||||||
| External base | 14 | 6.8–19.2 | 19.5–37.7 | 12.1 | 25.4 | 2.9 | 4.9 |
| External primary branch | 14 | 15.7–37.1 | 45.1–72.9 | 26.4 | 55.4 | 5.2 | 7.1 |
| External secondary branch | 10 | 11.6–20.6 | 25.6–40.5 | 14.0 | 28.9 | 2.5 | 4.3 |
| External base/primary branch (cct) | 14 | 38.5–55.7 | 45.7 | 5.8 | |||
| Internal base | 16 | 6.0–18.9 | 17.2–37.1 | 11.7 | 24.8 | 3.2 | 5.0 |
| Internal primary branch | 16 | 15.7–37.0 | 43.8–71.9 | 25.5 | 54.0 | 6.0 | 8.2 |
| Internal secondary branch | 11 | 11.0–19.5 | 24.1–38.3 | 13.7 | 28.0 | 2.4 | 4.0 |
| Internal base/primary branch (cct) | 16 | 38.0–53.5 | 45.9 | 6.2 | |||
| Claw 4 heights | |||||||
| Anterior base | 12 | 9.1–20.1 | 22.8–39.5 | 13.2 | 28.5 | 3.0 | 4.1 |
| Anterior primary branch | 12 | 27.2–49.4 | 62.8–97.1 | 37.3 | 80.6 | 6.7 | 7.8 |
| Anterior secondary branch | 11 | 11.0–24.4 | 30.1–47.9 | 15.9 | 34.4 | 3.8 | 5.0 |
| Anterior base/primary branch (cct) | 12 | 30.6–44.9 | 35.4 | 4.1 | |||
| Posterior base | 13 | 10.3–21.9 | 23.2–43.0 | 15.6 | 33.4 | 3.6 | 5.3 |
| Posterior primary branch | 13 | 27.4–49.9 | 63.3–98.0 | 40.0 | 85.8 | 7.3 | 8.9 |
| Posterior secondary branch | 12 | 12.4–25.3 | 30.4–49.7 | 19.2 | 40.3 | 4.0 | 5.9 |
| Posterior base/primary branch (cct) | 13 | 30.9–46.7 | 38.9 | 4.4 | |||
3.2.5. Eggs
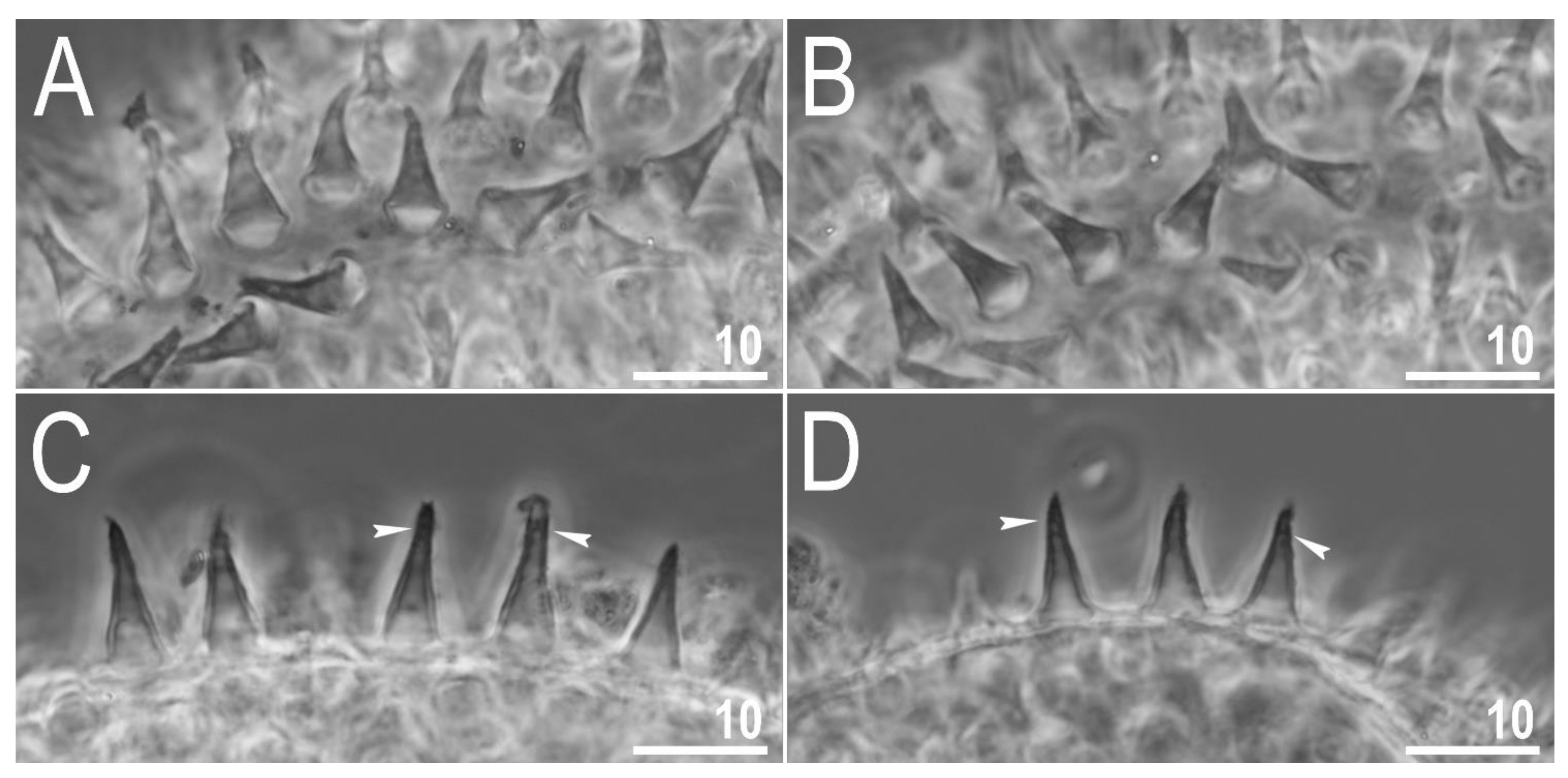
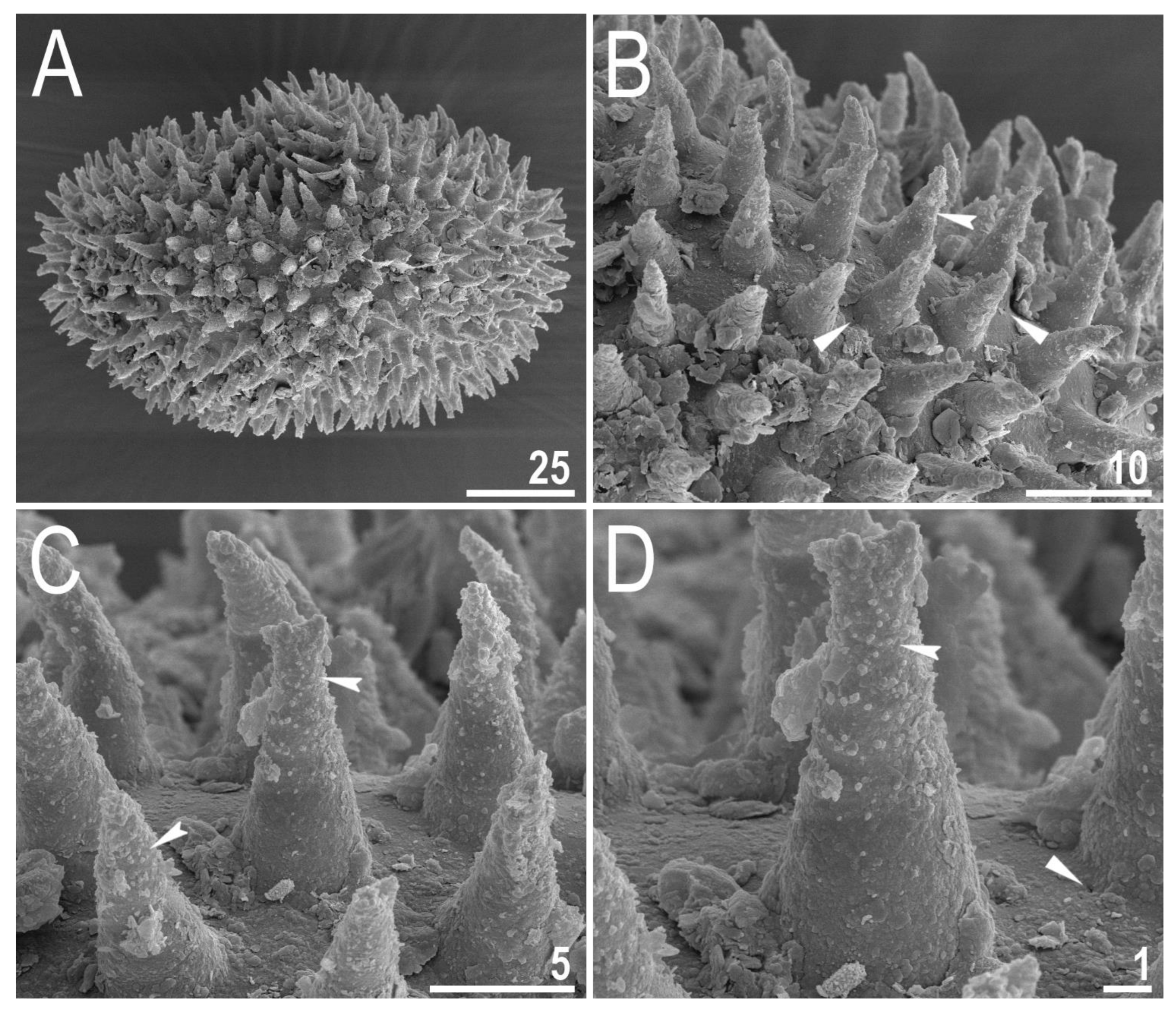
| Character | N | Range | Mean | SD |
|---|---|---|---|---|
| Egg bare diameter | 0 | ? | ? | ? |
| Egg full diameter | 0 | ? | ? | ? |
| Process height | 9 | 9.4–11.9 | 10.4 | 0.8 |
| Process base width | 9 | 4.0–5.5 | 4.7 | 0.5 |
| Process base/height ratio | 9 | 39–50% | 46% | 4% |
| Inter-process distance | 9 | 2.7–4.9 | 3.6 | 0.7 |
| Number of processes on the egg circumference | 0 | ? | ? | ? |
3.2.6. Reproduction
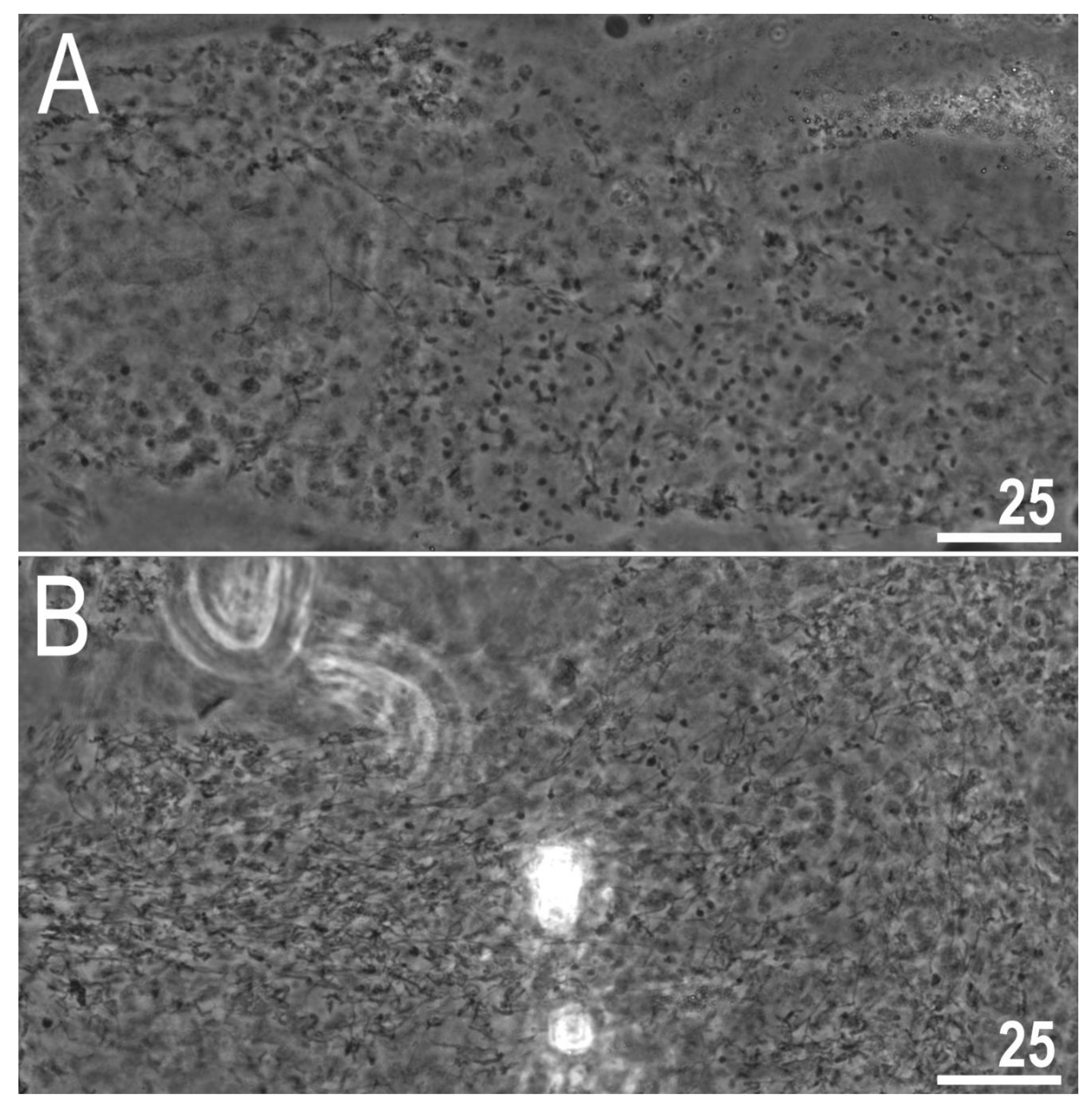
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Nelson, D.R.; Bartels, P.J.; Guil, N. Tardigrade Ecology. In Water Bears: The Biology of Tardigrades; Schill, R.O., Ed.; Springer: Basel, Switzerland, 2019; pp. 163–210. [Google Scholar] [CrossRef]
- Guidetti, R.; Bertolani, R. Tardigrade taxonomy: An updated check list of the taxa and a list of characters for their identification. Zootaxa 2005, 845, 1–46. [Google Scholar] [CrossRef]
- Degma, P.; Guidetti, R. Notes to the current checklist of Tardigrada. Zootaxa 2007, 1579, 41–53. [Google Scholar] [CrossRef]
- Degma, P.; Bertolani, R.; Guidetti, R. Actual Checklist of Tardigrada Species (2009–2021, Ver. 40: 19-07-2021); Universita di Modena e Reggio Emilia: Modena, Italy, 2021. [Google Scholar] [CrossRef]
- Surmacz, B.; Morek, W.; Michalczyk, Ł. What if multiple claw configurations are present in a sample: A case study with the description of Milnesium pseudotardigradum sp. nov. with unique developmental variability. Zool. Stud. 2019, 58, 32. [Google Scholar] [CrossRef]
- Bochnak, M.; Vončina, K.; Kristensen, R.M.; Gąsiorek, P. Continued exploration of Tanzanian rainforests reveals a new echiniscid species (Heterotardigrada). Zool. Stud. 2020, 59, 18. [Google Scholar] [CrossRef]
- Kayastha, P.; Berdi, D.; Mioduchowska, M.; Gawlak, M.; Łukasiewicz, A.; Gołdyn, B.; Jędrzejewski, S.; Kaczmarek, Ł. Description and molecular characterization of Richtersius ziemowiti sp. nov. (Richtersiidae) from Nepal (Asia) with evidence of heterozygous point mutation events in the 28S rRNA. Ann. Zool. 2020, 70, 381–396. [Google Scholar] [CrossRef]
- Tumanov, D.V. Integrative description of Mesobiotus anastasiae sp. nov. (Eutardigrada, Macrobiotoidea) and first record of Lobohalacarus (Chelicerata, Trombidiformes) from the Republic of South Africa. Eur. J. Taxon. 2020, 726, 102–131. [Google Scholar] [CrossRef]
- Tumanov, D.V. Integrative redescription of Hypsibius pallidoides Pilato et al., 2011 (Eutardigrada: Hypsibioidea) with the erection of a new genus and discussion on the phylogeny of Hypsibiidae. Eur. J. Taxon. 2020, 681, 1–37. [Google Scholar] [CrossRef]
- Stec, D.; Dudziak, M.; Michalczyk, Ł. Integrative descriptions of two new Macrobiotidae species (Tardigrada: Eutardigrada: Macrobiotoidea) from French Guiana and Malaysian Borneo. Zool. Stud. 2020, 59, 23. [Google Scholar] [CrossRef]
- Guidetti, R.; Schill, R.O.; Giovannini, I.; Massa, E.; Goldoni, S.E.; Ebel, C.; Förschler, M.I.; Rebecchi, L.; Cesari, M. When DNA sequence data and morphological results fit together: Phylogenetic position of Crenubiotus within Macrobiotoidea (Eutardigrada) with description of Crenubiotus ruhesteini sp. nov. J. Zool. Syst. Evol. Res. 2021, 59, 576–587. [Google Scholar] [CrossRef]
- Vecchi, M.; Stec, D. Integrative descriptions of two new Macrobiotus species (Tardigrada, Eutardigrada, Macrobiotidae) from Mississippi (USA) and Crete (Greece). Zoosyst. Evol. 2021, 97, 281–306. [Google Scholar] [CrossRef]
- Stec, D.; Morek, W.; Gąsiorek, P.; Michalczyk, Ł. Unmasking hidden species diversity within the Ramazzottius oberhaeuseri complex, with an integrative redescription of the nominal species for the family Ramazzottiidae (Tardigrada: Eutardigrada: Parachela). Syst. Biodivers. 2018, 16, 357–376. [Google Scholar] [CrossRef]
- Guidetti, R.; Cesari, M.; Bertolani, R.; Altiero, T.; Rebecchi, L. High diversity in species, reproductive modes and distribution within the Paramacrobiotus richtersi complex (Eutardigrada, Macrobiotidae). Zool. Lett. 2019, 5, 1. [Google Scholar] [CrossRef] [PubMed]
- Cesari, M.; Montanari, M.; Kristensen, R.M.; Bertolani, R.; Guidetti, R.; Rebecchi, L. An integrated study of the biodiversity within the Pseudechiniscus suillus-facettalis group (Heterotardigrada: Echiniscidae). Zool. J. Linn. Soc. 2020, 188, 717–732. [Google Scholar] [CrossRef] [Green Version]
- Grobys, D.; Roszkowska, M.; Gawlak, M.; Kmita, H.; Kepel, A.; Kepel, M.; Parnikoza, I.; Bartylak, T.; Kaczmarek, Ł. High diversity in the Pseudechiniscus suillus-facettalis complex (Heterotardigrada; Echiniscidae) with remarks on the morphology of the genus Pseudechiniscus. Zool. J. Linn. Soc. 2020, 188, 733–752. [Google Scholar] [CrossRef] [Green Version]
- Stec, D.; Krzywański, Ł.; Zawierucha, K.; Michalczyk, Ł. Untangling systematics of the Paramacrobiotus areolatus species complex by an integrative redescription of the nominal species for the group, with multilocus phylogeny and species delineation within the genus Paramacrobiotus. Zool. J. Linn. Soc. 2020, 188, 694–716. [Google Scholar] [CrossRef]
- Stec, D.; Krzywański, Ł.; Arakawa, K.; Michalczyk, Ł. A new redescription of Richtersius coronifer, supported by transcriptome, provides resources for describing concealed species diversity within the monotypic genus Richtersius (Eutardigrada). Zool. Lett. 2020, 6, 2. [Google Scholar] [CrossRef] [PubMed]
- Stec, D.; Vecchi, M.; Dudziak, M.; Bartels, P.J.; Calhim, S.; Michalczyk, Ł. Integrative taxonomy resolves species identities within the Macrobiotus pallarii complex (Eutardigrada: Macrobiotidae). Zool. Lett. 2021, 7, 9. [Google Scholar] [CrossRef]
- Bertolani, R.; Guidetti, R.; Marchioro, T.; Altiero, T.; Rebecchi, L.; Cesari, M. Phylogeny of Eutardigrada: New molecular data and their morphological support lead to the identification of new evolutionary lineages. Mol. Phylogenet. Evol. 2014, 76, 110–126. [Google Scholar] [CrossRef]
- Cesari, M.; Vecchi, M.; Palmer, A.; Bertolani, R.; Pilato, G.; Rebecchi, L.; Guidetti, R. What if the claws are reduced? Morphological and molecular phylogenetic relationships of the genus Haplomacrobiotus May, 1948 (Eutardigrada, Parachela). Zool. J. Linn. Soc. 2016, 178, 819–827. [Google Scholar] [CrossRef]
- Guidetti, R.; Rebecchi, L.; Bertolani, R.; Jönsson, K.I.; Kristensen, R.M.; Cesari, M. Morphological and molecular analyses on Richtersius (Eutardigrada) diversity reveal its new systematic position and lead to the establishment of a new genus and a new family within Macrobiotoidea. Zool. J. Linn. Soc. 2016, 178, 834–845. [Google Scholar] [CrossRef] [Green Version]
- Gąsiorek, P.; Morek, W.; Stec, D.; Michalczyk, Ł. Untangling the Echiniscus Gordian knot: Paraphyly of the “arctomys group” (Heterotardigrada: Echiniscidae). Cladistics 2019, 35, 633–653. [Google Scholar] [CrossRef] [PubMed]
- Gąsiorek, P.; Stec, D.; Morek, W.; Michalczyk, Ł. Deceptive conservatism of claws: Distinct phyletic lineages concealed within Isohypsibioidea (Eutardigrada) revealed by molecular and morphological evidence. Contrib. Zool. 2019, 88, 78–132. [Google Scholar] [CrossRef] [Green Version]
- Gąsiorek, P.; Michalczyk, Ł. Phylogeny of Itaquasconinae in light of the evolution of the flexible pharyngeal tube in Tardigrada. Zool. Scr. 2020, 49, 499–515. [Google Scholar] [CrossRef]
- Morek, W.; Michalczyk, Ł. First extensive multilocus phylogeny of the genus Milnesium Doyère, 1840 (Tardigrada) reveals no congruence between genetic markers and morphological traits. Zool. J. Linn. Soc. 2020, 188, 681–693. [Google Scholar] [CrossRef]
- Stec, D.; Vecchi, M.; Maciejowski, W.; Michalczyk, Ł. Resolving the systematics of Richtersiidae by multilocus phylogeny and an integrative redescription of the nominal species for the genus Crenubiotus (Tardigrada). Sci. Rep. 2020, 10, 19418. [Google Scholar] [CrossRef]
- Morek, W.; Surmacz, B.; López-López, A.; Michalczyk, Ł. “Everything is not everywhere”: Time-calibrated phylogeography of the genus Milnesium (Tardigrada). Mol. Ecol. 2021, 30, 3590–3609. [Google Scholar] [CrossRef]
- Stec, D.; Vecchi, M.; Calhim, S.; Michalczyk, Ł. New multilocus phylogeny reorganises the family Macrobiotidae (Eutardigrada) and unveils complex morphological evolution of the Macrobiotus hufelandi group. Mol. Phylogenet. Evol. 2021, 160, 106987. [Google Scholar] [CrossRef]
- Tumanov, D.V. End of a mystery: Integrative approach reveals the phylogenetic position of an enigmatic Antarctic tardigrade genus Ramajendas (Tardigrada, Eutardigrada). Zool. Scr. 2022. [Google Scholar] [CrossRef]
- Stec, D.; Roszkowska, M.; Kaczmarek, Ł.; Michalczyk, Ł. An integrative description of a population of Mesobiotus radiatus (Pilato, Binda and Catanzaro, 1991) from Kenya. Turk. J. Zool. 2018, 42, 523–540. [Google Scholar] [CrossRef] [Green Version]
- Morek, W.; Blagden, B.; Kristensen, R.M.; Michalczyk, Ł. The analysis of inter- and intrapopulation variability of Milnesium eurystomum Maucci, 1991 reveals high genetic divergence and novel type of ontogenetic variation (Tardigrada: Apochela). Syst. Biodivers. 2020, 18, 614–632. [Google Scholar] [CrossRef]
- Morek, W.; Surmacz, B.; Michalczyk, Ł. Novel integrative data for two Milnesium Doyère, 1840 (Tardigrada: Apochela) species from Central Asia. Zoosyst. Evol. 2020, 96, 499–514. [Google Scholar] [CrossRef]
- Pogwizd, J.; Stec, D. New records of Dactylobiotus parthenogeneticus Bertolani, 1982 provide insight into its genetic variability and geographic distribution. Folia Biol. 2020, 68, 57–72. [Google Scholar] [CrossRef]
- Stec, D.; Kristensen, R.M.; Michalczyk, Ł. An integrative description of Minibiotus ioculator sp. nov. from the Republic of South Africa with notes on Minibiotus pentannulatus Londoño et al., 2017 (Tardigrada: Macrobiotidae). Zool. Anz. 2020, 286, 117–134. [Google Scholar] [CrossRef]
- Kiosya, Y.; Pogwizd, J.; Matsko, Y.; Vecchi, M.; Stec, D. Phylogenetic position of two Macrobiotus species with a revisional note on Macrobiotus sottilei Pilato, Kiosya, Lisi & Sabella, 2012 (Tardigrada: Eutardigrada: Macrobiotidae). Zootaxa 2021, 4933, 113–135. [Google Scholar] [CrossRef]
- Tumanov, D.V. Presence of Notahypsibius pallidoides (Tardigrada: Hypsibiidae) in the fauna of Russia confirmed with the methods of DNA barcoding. Biol. Commun. 2021, 66, 274–280. [Google Scholar] [CrossRef]
- Jørgensen, A.; Kristensen, R.M.; Møbjerg, N. Phylogeny and Integrative Taxonomy of Tardigrada. In Water Bears: The Biology of Tardigrades; Schill, R.O., Ed.; Springer: Basel, Switzerland, 2019; pp. 95–114. [Google Scholar] [CrossRef]
- Gąsiorek, P.; Vončina, K.; Nelson, D.R.; Michalczyk, Ł. The importance of being integrative: A remarkable case of synonymy in the genus Viridiscus (Heterotardigrada: Echiniscidae). Zool. Lett. 2021, 7, 13. [Google Scholar] [CrossRef]
- Maucci, W. Sulla presenza in Italia di Cornechiniscus holmeni (Petersen, 1951), e descrizione di Macrobiotus hyperonyx sp. nov. Boll. Mus. Civ. Stor. Nat. Verona 1982, 9, 175–179. [Google Scholar]
- Maucci, W. A contribution to the knowledge of the North American Tardigrada with emphasis on the fauna of Yellowstone National Park (Wyoming). In Biology of Tardigrada. Selected Symposia and Monographs, U.Z.I., Mucci Modena; Bertolani, R., Ed.; Mucchi Editore: Modena, Italy, 1987; pp. 187–210. [Google Scholar]
- Maucci, W. Tardigradi della Mongolia Esterna, con Descrizione di Macrobiotus mongolicus sp. nov. Boll. Mus. Civ. Stor. Nat. Verona 1988, 14, 339–349. [Google Scholar]
- Tumanov, D.V. Two new species of Macrobiotus (Eutardigrada, Macrobiotidae) from Tien Shan (Kirghizia), with notes on Macrobiotus tenuis group. Zootaxa 2005, 1043, 33–46. [Google Scholar] [CrossRef] [Green Version]
- Tumanov, D.V. Three new species of Macrobiotus (Eutardigrada, Macrobiotidae, tenuis-group) from Tien Shan (Kirghizia) and Spitsbergen. J. Limnol. 2007, 66, 40–48. [Google Scholar] [CrossRef]
- Pilato, G.; Lisi, O. Tenuibiotus, a new genus of Macrobiotidae (Eutardigrada). Zootaxa 2011, 2761, 34–40. [Google Scholar] [CrossRef]
- Zawierucha, K.; Kolicka, M.; Kaczmarek, Ł. Re-description of the Arctic tardigrade Tenuibiotus voronkovi (Tumanov, 2007) (Eutardigrada; Macrobiotidea), with the first molecular data for the genus. Zootaxa 2016, 4196, 498–510. [Google Scholar] [CrossRef] [PubMed]
- Stec, D.; Tumanov, D.V.; Kristensen, R.M. Integrative taxonomy identifies two new tardigrade species (Eutardigrada: Macrobiotidae) from Greenland. Eur. J. Taxon. 2020, 614, 1–40. [Google Scholar] [CrossRef]
- Pilato, G. Macrobiotus willardi a new species of Tardigrada from Canada. Can. J. Zool. 1977, 55, 628–630. [Google Scholar] [CrossRef]
- Stec, D.; Smolak, R.; Kaczmarek, Ł.; Michalczyk, Ł. An integrative description of Macrobiotus paulinae sp. nov. (Tardigrada: Eutardigrada: Macrobiotidae: Hufelandi group) from Kenya. Zootaxa 2015, 4052, 501–526. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Petersen, B. The Tardigrade fauna of Greenland. A faunistic study with some few ecological remarks. Meddelser Grønland København 1951, 150, 5–94. [Google Scholar]
- Doyère, M. Memoire sur les Tardigrades. Ann. Sci. Nat. 1840, 14, 269–362. [Google Scholar]
- Casquet, J.T.; Thebaud, C.; Gillespie, R.G. Chelex without boiling, a rapid and easy technique to obtain stable amplifiable DNA from small amounts of ethanol-stored spiders. Mol. Ecol. Resour. 2012, 12, 136–141. [Google Scholar] [CrossRef]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Stec, D.; Zawierucha, K.; Michalczyk, Ł. An integrative description of Ramazzottius subanomalus (Biserov, 1985) (Tardigrada) from Poland. Zootaxa 2017, 4300, 403–420. [Google Scholar] [CrossRef]
- Gąsiorek, P.; Stec, D.; Zawierucha, Z.; Kristensen, R.M.; Michalczyk, Ł. Revision of Testechiniscus Kristensen, 1987 (Heterotardigrada: Echiniscidae) refutes the polar-temperate distribution of the genus. Zootaxa 2018, 4472, 261–297. [Google Scholar] [CrossRef]
- Mironov, S.V.; Dabert, J.; Dabert, M. A new feather mite species of the genus Proctophyllodes Robin, 1877 (Astigmata: Proctophyllodidae) from the Long-tailed Tit Aegithalos caudatus (Passeriformes: Aegithalidae): Morphological description with DNA barcode data. Zootaxa 2012, 3253, 54–61. [Google Scholar] [CrossRef] [Green Version]
- Astrin, J.; Stüben, P. Phylogeny in cryptic weevils: Molecules, morphology and new genera of western Palaearctic Cryptorhynchinae (Coleoptera: Curculionidae). Invertebr. Syst. 2008, 22, 503–522. [Google Scholar] [CrossRef] [Green Version]
- Gąsiorek, P.; Stec, D.; Morek, W.; Michalczyk, Ł. An integrative redescription of Hypsibius dujardini (Doyère, 1840), the nominal taxon for Hypsibioidea (Tardigrada: Eutardigrada). Zootaxa 2018, 4415, 45–75. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guidetti, R.; Gandolfi, A.; Rossi, V.; Bertolani, R. Phylogenetic analysis of Macrobiotidae (Eutardigrada, Parachela): A combined morphological and molecular approach. Zool. Scr. 2005, 34, 235–244. [Google Scholar] [CrossRef]
- Jørgensen, A.; Faurby, S.; Hansen, J.G.; Møbjerg, N.; Kristensen, R.M. Molecular phylogeny of Arthrotardigrada (Tardigrada). Mol. Phylogenet. Evol. 2010, 54, 1006–1015. [Google Scholar] [CrossRef] [PubMed]
- Stec, D.; Arakawa, K.; Michalczyk, Ł. An integrative description of Macrobiotus shonaicus sp. nov. (Tardigrada: Macrobiotidae) from Japan with notes on its phylogenetic position within the hufelandi group. PLoS ONE 2018, 13, e0192210. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coughlan, K.; Michalczyk, Ł.; Stec, D. Macrobiotus caelestis sp. nov., a new tardigrade species (Macrobiotidae: Hufelandi group) from the Tien Shan mountains (Kyrgyzstan). Ann. Zool. 2019, 69, 499–513. [Google Scholar] [CrossRef]
- Kaczmarek, Ł.; Zawierucha, K.; Buda, J.; Stec, D.; Gawlak, M.; Michalczyk, Ł.; Roszkowska, M. An integrative redescription of the nominal taxon for the Mesobiotus harmsworthi group (Tardigrada: Macrobiotidae) leads to descriptions of two new Mesobiotus species from Arctic. PLoS ONE 2018, 13, e0204756. [Google Scholar] [CrossRef]
- Itang, L.A.M.; Stec, D.; Mapalo, M.A.; Mirano-Bascos, D.; Michalczyk, Ł. An integrative description of Mesobiotus dilimanensis, a new tardigrade species from the Philippines (Eutardigrada: Macrobiotidae: Furciger group). Raffles Bull. Zool. 2020, 68, 19–31. [Google Scholar]
- Guil, N.; Giribet, G. A comprehensive molecular phylogeny of tardigrades—Adding genes and taxa to a poorly resolved phylum-level phylogeny. Cladistics 2012, 28, 21–49. [Google Scholar] [CrossRef]
- Kihm, J.H.; Kim, S.; McInnes, S.J.; Zawierucha, K.; Rho, H.S.; Kang, P.; Park, T.Y.S. Integrative description of a new Dactylobiotus (Eutardigrada: Parachela) from Antarctica that reveals an intraspecific variation in tardigrade egg morphology. Sci. Rep. 2020, 10, 9122. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Guidetti, R.; Rebecchi, L.; Bertolani, R. Cuticule structure and systematics of Macrobiotidae (Tardigrada: Eutardigrada). Acta Zool. 2000, 81, 27–36. [Google Scholar] [CrossRef]
- Bertolani, R.; Kristensen, R.M. New records of Eohypsibius nadjae, Kristensen, 1982, and revision of the taxonomic position of two genera of Eutardigrada (Tardigrada). Biol. Tardigrades 1987, 1, 359–372. [Google Scholar]
- Katoh, K.; Misawa, K.; Kuma, K.; Miyata, T. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef] [Green Version]
- Katoh, K.; Toh, H. Recent developments in the MAFFT multiple sequence alignment program. Brief. Bioinform. 2008, 9, 286–298. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [Green Version]
- Vaidya, G.; Lohman, D.J.; Meier, R. SequenceMatrix: Concatenation software for the fast assembly of multi-gene datasets with character set and codon information. Cladistics 2011, 27, 171–180. [Google Scholar] [CrossRef]
- Lanfear, R.; Frandsen, P.B.; Wright, A.M.; Senfeld, T.; Calcott, B. PartitionFinder 2: New methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol. Biol. Evol. 2016, 34, 772–773. [Google Scholar] [CrossRef] [Green Version]
- Lanfear, R.; Calcott, B.; Ho, S.Y.; Guindon, S. PartitionFinder: Combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol. Biol. Evol. 2012, 29, 1695–1701. [Google Scholar] [CrossRef] [Green Version]
- Ronquist, F.; Huelsenbeck, J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 2003, 19, 1572–1574. [Google Scholar] [CrossRef] [Green Version]
- Rambaut, A.; Suchard, M.A.; Xie, D.; Drummond, A.J. Tracer v1.6. 2014. Available online: http://beast.bio.ed.ac.uk/Tracer (accessed on 13 December 2017).
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef] [PubMed]
- Morek, W.; Stec, D.; Gąsiorek, P.; Schill, R.O.; Kaczmarek, Ł.; Michalczyk, Ł. An experimental test of eutardigrade preparation methods for light microscopy. Zool. J. Linn. Soc. 2016, 178, 785–793. [Google Scholar] [CrossRef] [Green Version]
- Coughlan, K.; Stec, D. Two new species of the Macrobiotus hufelandi complex (Eutardigrada: Macrobiotidae) from Australia and India with notes on their phylogenetic position. Eur. J. Taxon. 2019, 573, 1–38. [Google Scholar] [CrossRef]
- Eibye-Jacobsen, J. Are the supportive structures of the tardigrade pharynx homologous throughout the entire group? J. Zool. Syst. Evol. Res. 2001, 39, 1–11. [Google Scholar] [CrossRef]
- Gąsiorek, P.; Stec, D.; Morek, W.; Zawierucha, K.; Kaczmarek, Ł.; Lachowska-Cierlik, D.; Michalczyk, Ł. An integrative revision of Mesocrista Pilato, 1987 (Tardigrada: Eutardigrada: Hypsibiidae). J. Nat. Hist. 2016, 50, 2803–2828. [Google Scholar] [CrossRef]
- Stec, D.; Gąsiorek, P.; Morek, W.; Kosztyła, P.; Zawierucha, K.; Michno, K.; Kaczmarek, Ł.; Prokop, Z.M.; Michalczyk, Ł. Estimating optimal sample size for tardigrade morphometry. Zool. J. Linn. Soc. 2016, 178, 776–784. [Google Scholar] [CrossRef] [Green Version]
- Michalczyk, Ł.; Kaczmarek, Ł. A description of the new tardigrade Macrobiotus reinhardti (Eutardigrada, Macrobiotidae, harmsworthi group) with some remarks on the oral cavity armature within the genus Macrobiotus Schultze. Zootaxa 2003, 331, 1–24. [Google Scholar] [CrossRef] [Green Version]
- Kaczmarek, Ł.; Michalczyk, Ł. The Macrobiotus hufelandi (Tardigrada) group revisited. Zootaxa 2017, 4363, 101–123. [Google Scholar] [CrossRef]
- Kaczmarek, Ł.; Cytan, J.; Zawierucha, K.; Diduszko, D.; Michalczyk, Ł. Tardigrades from Peru (South America), with descriptions of three new species of Parachela. Zootaxa 2014, 3790, 357–379. [Google Scholar] [CrossRef] [Green Version]
- Pilato, G. Analisi di nuovi caratteri nello studio degli Eutardigradi. Animalia 1981, 8, 51–57. [Google Scholar]
- Michalczyk, Ł.; Kaczmarek, Ł. The Tardigrada Register: A comprehensive online data repository for tardigrade taxonomy. J. Limnol. 2013, 72, 175–181. [Google Scholar] [CrossRef]
- Pilato, G.; Kiosya, Y.; Lisi, O.; Inshina, V.; Biserov, V. Annotated list of Tardigrada records from Ukraine with the description of three new species. Zootaxa 2011, 3123, 1–31. [Google Scholar] [CrossRef]
- Lisi, O.; Londoño, R.; Quiroga, S. Description of a new genus and species (Eutardigrada: Richtersiidae) from Colombia, with comments on the family Richtersiidae. Zootaxa 2020, 4822, 531–550. [Google Scholar] [CrossRef] [PubMed]
- Richters, F. Tardigrada. In Handbuch der Zoologie; Kükenthal, W., Krumbach, T., Eds.; Walter de Gruyter & Co.: Berlin/Leipzig, Germany, 1926; Volume 3, pp. 58–61. [Google Scholar]
- Schuster, R.O.; Nelson, D.R.; Grigarick, A.A.; Christenberry, D. Systematic criteria of the Eutardigrada. Trans. Am. Microsc. Soc. 1980, 99, 284–303. [Google Scholar] [CrossRef]
- Thulin, G. Über die Phylogenie und das System der Tardigraden. Hereditas 1928, 11, 207–266. [Google Scholar] [CrossRef]
- Pilato, G.; Binda, M.G. Richtersius, nuove nome generico in sostituzione di Richtersia Pilato e Binda 1987 (Eutardigrada). Animalia 1989, 16, 147–148. [Google Scholar]
- Maucci, W.; Ramazzotti, G. Adorybiotus gen. nov.: Nouva posizione sistematica per Macrobiotus granulatus Richters, 1903 e per Macrobiotus coronifer Richters, 1903 (Tardigrada, Macrobiotidae). Mem. Ist. Ital. Idrobiol. 1981, 39, 153–159. [Google Scholar]
- Stec, D.; Vončina, K.; Kristensen, R.M.; Michalczyk, Ł. The Macrobiotus ariekammensis species complex provides evidence for parallel evolution of long claws in macrobiotid tardigrades. Zool. J. Linn. Soc. 2022. [Google Scholar] [CrossRef]
- Guil, N.; Guidetti, R.; Machordom, A. Observations on the “tenuis group” (Eutardigrada, Macrobiotidae) and description of a new Macrobiotus species. J. Nat. Hist. 2007, 41, 2741–2755. [Google Scholar] [CrossRef]
| Sample Code | Sample Type | Coordinates | Analyses | ||
|---|---|---|---|---|---|
| PCM | SEM | DNA | |||
| IT.339 | moss | 46°30′29.19″ N | 6A + 0E | 0A + 0E | 1A + 0E |
| 11°49′41″ E | |||||
| IT.341 | moss | 46°30′26.9″ N | 13A + 0E | 0A + 0E | 1A + 0E |
| 11°49′38.4″ E | |||||
| IT.344 | moss + lichen | 46°30′23.23″ N | 18A + 2E | 14A + 1E | 1A + 0E |
| 11°49′31.8″ E | |||||
| IT.345 | moss | 46°30′23.23″ N | 9A + 1E | 0A + 0E | 1A + 0E |
| 11°49′31.8″ E | |||||
| DNA Marker | Primer Name | Primer Direction | Primer Sequence (5′-3′) | Primer Source |
|---|---|---|---|---|
| 18S rRNA | 18S_Tar_Ff1 | forward | AGGCGAAACCGCGAATGGCTC | [54] |
| 18S_Tar_Rr1 | reverse | GCCGCAGGCTCCACTCCTGG | ||
| 28S rRNA | 28S_Eutar_F | forward | ACCCGCTGAACTTAAGCATAT | [55,56] |
| 28SR0990 | reverse | CCTTGGTCCGTGTTTCAAGAC | ||
| ITS-2 | ITS2_Eutar_Ff | forward | CGTAACGTGAATTGCAGGAC | [13] |
| ITS2_Eutar_Rr | reverse | TCCTCCGCTTATTGATATGC | ||
| COI | LCO1490-JJ | forward | CHACWAAYCATAAAGATATYGG | [57] |
| HCO2198-JJ | reverse | AWACTTCVGGRTGVCCAAARAATCA |
| Species | 18S rRNA | 28S rRNA | COI | ITS-2 | Sources |
|---|---|---|---|---|---|
| Hypsibius exemplaris | MG800327 | MG800337 | MG818724 | MG800336 | [58] |
| Ramazzottius subanomalus | MF001997 | MF001998 | MF001999 | MG432819 | [54] |
| Bertolanius volubilis | HQ604918 | – | AY598769 | – | [20,59] |
| Bertolanius nebulosus | GQ849023 | – | – | – | [60] |
| Eohypsibius nadjae | HQ604921 | – | – | – | [20] |
| Minibiotus ioculator | MT023998 | MT024041 | MT023412 | MT024000 | [35] |
| Minibiotus pentannulatus | MT023999 | MT024042 | MT023413 | MT024001 | [35] |
| Tenuibiotus voronkovi | KX810045 | KX810049 | KX810042 | KX810046 | [46] |
| Tenuibiotus zandrae | MN443040 | MN443035 | MN444827 | MN443038 | [47] |
| Paramacrobiotus areolatus | MH664931 | MH664948 | MH675998 | MH666080 | [17] |
| Paramacrobiotus fairbanksi | MH664941 | MH664950 | MH676011 | MH666090 | [17] |
| Macrobiotus shonaicus | MG757132 | MG757133 | MG757136 | MG757134 | [61] |
| Macrobiotus caelestis | MK737073 | MK737071 | MK737922 | MK737072 | [62] |
| Xerobiotus pseudohufelandi | HQ604989 | – | AY598776 | – | [20,59] |
| Mesobiotus harmsworthi | MH197146 | MH197264 | MH195150 | MH197154 | [63] |
| Mesobiotus dilimanensis | MN257048 | MN257049 | MN257047 | MN257050 | [64] |
| Richtersius coronifer NO.385 | MH681760 | MH681757 | MH676053 | MH681763 | [18] |
| Richtersius aff. coronifer GR.008 | MK211386 | MK211384 | MK214323–5 | MK211380–1 | [18] |
| Richtersius aff. coronifer IT.120 | MH681761 | MH681758 | MH676054 | MH681764 | [18] |
| Richtersius aff. coronifer IT.317 | MK211387 | MK211385 | MK214326–8 | MK211382–3 | [18] |
| Richtersius aff. coronifer PL.247 | MH681762 | MH681759 | MH676055 | MH681765 | [18] |
| Richtersius ziemowiti | MT241891 | MT241895 | MT246504 | MT241896 | [7] |
| Diaforobiotus islandicus IS.042 | MT812470 | MT812461 | MT808072 | MT812597 | [27] |
| Diaforobiotus sp. NO.386 | MT812471 | MT812463 | MT808074 | MT812598 | [27] |
| Diaforobiotus sp. ID.517 | MT812472 | MT812462 | MT808073 | MT812599 | [27] |
| Diaforobiotus hyperonyx IT.339 | OM179853 | OM179860 | OM151287 | OM179866 | This study |
| Diaforobiotus hyperonyx IT.341 | OM179855 | OM179861 | OM151288 | OM179868 | This study |
| Diaforobiotus hyperonyx IT.344 | OM179852 | OM179859 | OM151286 | OM179867 | This study |
| Diaforobiotus hyperonyx IT.345 | OM179854 | OM179862 | OM151289 | OM179869 | This study |
| Murrayon dianae | FJ435737 | FJ435762 | FJ435801 | – | [65] |
| Murrayon cf. pullari IT.338 | MT812477 | MT812465 | MT808080 | MT812603 | [27] |
| Murrayon pullari | GQ849026 | – | – | – | [60] |
| Dactylobiotus parthenogeneticus FR.149 | MT373694 | MT373700 | MT373804 | MT374191 | [34] |
| Dactylobiotus parthenogeneticus GB.003 | MT373693 | MT373699 | MT373803 | MT374190 | [34] |
| Dactylobiotus parthenogeneticus PL.317 | MT373695 | MT373701 | MT373805–6 | MT374192 | [34] |
| Dactylobiotus selenicus FI.073 | MT812476 | MT812466 | MT808076 | MT812602 | [27] |
| Dactylobiotus ambiguus | GQ925676–7 | – | – | – | Chen et al. (unpublished) |
| Dactylobiotus ovimutans | MT136805 | – | MT132333 | – | [66] |
| Dactylobiotus octavi | GQ849025 | – | – | – | |
| Crenubiotus sp. GB.108 | MT812473 | MT812467 | MT808077–8 | MT812604–5 | [27] |
| Crenubiotus crenulatus NO.429 | MT812474 | MT812463 | MT808079 | MT812606 | [27] |
| Crenubiotus ruhesteini | MW074384–5, MW074387 | – | MW074336–8 | MW074367–8, MW074370 | [11] |
| Crenubiotus sp. GL.001.01 | OM179850 | OM179857 | OM151284 | OM179864 | This study |
| Crenubiotus sp. GL.001.02 | OM179851 | OM179858 | OM151285 | OM179865 | This study |
| Adorybiotus granulatus | HQ604961–2 | – | – | – | [20] |
| Adorybiotus cf. granulatus JP.008 | MT812475 | MT812464 | MT808075 | MT812600–1 | [27] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Stec, D.; Morek, W. Reaching the Monophyly: Re-Evaluation of the Enigmatic Species Tenuibiotus hyperonyx (Maucci, 1983) and the Genus Tenuibiotus (Eutardigrada). Animals 2022, 12, 404. https://doi.org/10.3390/ani12030404
Stec D, Morek W. Reaching the Monophyly: Re-Evaluation of the Enigmatic Species Tenuibiotus hyperonyx (Maucci, 1983) and the Genus Tenuibiotus (Eutardigrada). Animals. 2022; 12(3):404. https://doi.org/10.3390/ani12030404
Chicago/Turabian StyleStec, Daniel, and Witold Morek. 2022. "Reaching the Monophyly: Re-Evaluation of the Enigmatic Species Tenuibiotus hyperonyx (Maucci, 1983) and the Genus Tenuibiotus (Eutardigrada)" Animals 12, no. 3: 404. https://doi.org/10.3390/ani12030404
APA StyleStec, D., & Morek, W. (2022). Reaching the Monophyly: Re-Evaluation of the Enigmatic Species Tenuibiotus hyperonyx (Maucci, 1983) and the Genus Tenuibiotus (Eutardigrada). Animals, 12(3), 404. https://doi.org/10.3390/ani12030404







