Diversity of Coronaviruses with Particular Attention to the Interspecies Transmission of SARS-CoV-2
Abstract
Simple Summary
Abstract
1. Introduction
2. Diversity of Coronaviruses in Domestic Animals
2.1. Avian Coronaviruses
2.2. Coronaviruses in Pigs
2.3. Coronaviruses in Dogs
2.4. Coronaviruses in Cats
2.5. Coronaviruses in Cattle
2.6. Coronaviruses in Equines
2.7. Coronaviruses in Humans
3. Diversity of Coronavirus Receptors
3.1. Amino Peptidase Receptors
3.2. Carcinoembryonic Antigen-Related Cell Adhesion Molecule 1
3.3. Dipeptidyl Peptidase 4 (DPP4)
3.4. Sialic Acids or Sialosides, Acidic Carbohydrates
3.5. Angiotensin-Converting Enzyme 2 (ACE2)
4. Interaction between SARS-CoV-2 and ACE2 Receptor
5. Interspecies Transmission of Coronaviruses
5.1. SARS-CoV-2 in Dogs and Cats
5.2. SARS-CoV-2 in Mink
5.3. SARS-CoV-2 in Rabbits
5.4. SARS-CoV-2 in Other Animals
| Risk Level | Animals | Experimental | Natural | Remarks | References |
|---|---|---|---|---|---|
| Low | Dog (Canis lupus familiaris) | + | + | No symptoms | [129] |
| Cattle (Bos taurus) | + | - | No symptoms | [163] | |
| Common marmosets (Callithrix jacchus) | + | + | No symptoms | [164] | |
| Tree shrew (Tupaia belangeri) | + | + | No symptoms | [95] | |
| High | Cat (Felis catus) | + | + | mild symptoms | [155,165] |
| Malayan tiger (Panthera tigris subsp. jacksoni) | + | + | Symptoms | [63] | |
| Lion (Panthera leo) | - | + | Symptoms | [151] | |
| Puma (Puma concolor) | - | + | Symptoms | [165] | |
| American mink (Neovison vison) | - | + | Symptoms | [128,148] | |
| Egyptian fruit bats (Rousettus aegyptiacus) | + | - | No symptoms | [154] | |
| Ferret (Mustela putorius furo) | + | - | Very mild | [155,157,166] | |
| Detected | Rabbits (Oryctolagus cuniculus) | + | - | No symptoms | [149] |
| Raccoon dogs (Nyctereutes procyonoides) | + | - | No symptoms | [167] | |
| North American raccoons (Procyon lotor) | + | - | No symptoms | [168] | |
| Striped skunks (Mephitis mephitis) | + | - | No symptoms | [168] | |
| White Chinese geese (Anser cygnoides) | + | - | No symptoms | [169] | |
| Nonsusceptible | Japanese quail (Coturnix japonica) | + | - | No symptoms | [170] |
| White Chinese geese (Anser cygnoides) | + | - | No symptoms | [170] | |
| Turkeys (Meleagris gallopavo) | + | - | No symptoms | [170] | |
| Pekin duck (Anas platyrhinchos domesticus) | + | - | No symptoms | [170] | |
| Duck (Anatidae) | + | - | No symptoms | [155] | |
| Equine (Equus caballus) | + | - | No symptoms | [171] |
6. Control Measures and Implementation of One Health Strategy
7. Conclusions and Recommendations
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Brown, P.A.; Touzain, F.; Briand, F.X.; Gouilh, A.M.; Courtillon, C.; Allée, C.; Lemaitre, E.; De Boisséson, C.; Blanchard, Y.; Eterradossi, N. First complete genome sequence of European turkey coronavirus suggests complex recombination history related with US turkey and guinea fowl coronaviruses. J. Gen. Virol. 2016, 97, 110–120. [Google Scholar] [CrossRef] [PubMed]
- Rahman, M.; Chowdhury, E.H.; Parvin, R. Small-scale poultry production in Bangladesh: Challenges and impact of COVID-19 on sustainability. Ger. J. Vet. Res. 2021, 1, 19–27. [Google Scholar] [CrossRef]
- Shehata, A.A.; Parvin, R.; Nagy, A.; Wang, Y.; Azhar, T.M.; Attia, Y.A.; Azhar, E.I.; Paul, A.K.; Rahmatullah, M. An overview of the ongoing challenges in SARS-CoV-2 global control. Ger. J. Microbiol. 2021, 1, 1–18. [Google Scholar] [CrossRef]
- Sauter, D.; Kirchhoff, F. Key Viral Adaptations Preceding the AIDS Pandemic. Cell Host Microbe 2019, 25, 27–38. [Google Scholar] [CrossRef]
- Andersen, K.G.; Rambaut, A.; Lipkin, W.I.; Holmes, E.C.; Garry, R.F. The proximal origin of SARS-CoV-2. Nat. Med. 2020, 26, 450–452. [Google Scholar] [CrossRef]
- Perlman, S. Another decade, another coronavirus. N. Engl. J. Med. 2020, 382, 760–762. [Google Scholar] [CrossRef] [PubMed]
- Leiser, O.P.; Hobbs, E.C.; Sims, A.C.; Korch, G.W.; Taylor, K.L. Beyond the list: Bioagent-agnostic signatures could enable a more flexible and resilient biodefense posture than an approach based on priority agent lists alone. Pathogens 2021, 10, 1497. [Google Scholar] [CrossRef]
- Rothe, C.; Schunk, M.; Sothmann, P.; Bretzel, G.; Froeschl, G.; Wallrauch, C.; Zimmer, T.; Thiel, V.; Janke, C.; Guggemos, W.; et al. Transmission of 2019-nCoV infection from an asymptomatic contact in Germany. N. Engl. J. Med. 2020, 382, 970–971. [Google Scholar] [CrossRef]
- Zou, L.; Ruan, F.; Huang, M.; Liang, L.; Huang, H.; Hong, Z.; Yu, J.; Kang, M.; Song, Y.; Xia, J.; et al. SARS-CoV-2 viral load in upper respiratory specimens of infected patients. N. Engl. J. Med. 2020, 382, 1177–1179. [Google Scholar] [CrossRef]
- ICTV Code. The international code of virus classification and nomenclature. 2020. Available online: https://talk.ictvonline.org/taxonomy/ (accessed on 15 December 2021).
- Schalk, A.F.; Hawn, M.C. An apparently new respiratory disease of baby chicks. J. Am. Vet. Med. Assoc. 1931, 78, 413–423. [Google Scholar]
- Adams, N.R.; Ball, R.A.; Annis, C.L.; Hofstad, M.S. Ultrastructural changes in the intestines of turkey poults and embryos affected with transmissible enteritis. J. Comp. Pathol. 1972, 82, 187–192. [Google Scholar] [CrossRef]
- Spackman, D.; Cameron, I.R. Isolation of infectious bronchitis virus from pheasants. Vet. Rec. 1983, 113, 354–355. [Google Scholar] [CrossRef]
- Liais, E.; Croville, G.; Mariette, J.; Delverdier, M.; Lucas, M.-N.; Klopp, C.; Lluch, J.; Donnadieu, C.; Guy, J.S.; Corrand, L.; et al. Novel avian coronavirus and fulminating disease in guinea fowl, France. Emerg. Infect. Dis. 2014, 20, 105–108. [Google Scholar] [CrossRef]
- Barr, D.A.; Reece, R.L.; O’Rourke, D.; Button, C.; Faragher, J.T. Isolation of infectious bronchitis virus from a flock of racing pigeons. Aust. Vet. J. 1988, 65, 228. [Google Scholar] [CrossRef]
- Lee, K.M. Propagation of transmissible gastroenteritis virus in tissue culture. Ann. N. Y. Acad. Sci. 1956, 66, 191–195. [Google Scholar] [CrossRef]
- Pensaert, M.B.; de Bouck, P. A new coronavirus-like particle associated with diarrhea in swine. Arch. Virol. 1978, 58, 243–247. [Google Scholar] [CrossRef]
- Pensaert, M.; Callebaut, P.; Vergote, J. Isolation of a porcine respiratory, non-enteric coronavirus related to transmissible gastroenteritis. Vet. Q. 1986, 8, 257–261. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Tian, X.; Qin, P.; Wang, B.; Zhao, P.; Yang, Y.-L.; Wang, L.; Wang, D.; Song, Y.; Zhang, X.; et al. Discovery of a novel swine enteric alphacoronavirus (SeACoV) in southern China. Vet. Microbiol. 2017, 211, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Greig, A.S.; Mitchell, D.; Corner, A.H.; Bannister, G.L.; Meads, E.B.; Julian, R.J. A hemagglutinating virus producing encephalomyelitis in baby pigs. Can. J. Comp. Med. Vet. Sci. 1962, 26, 49–56. [Google Scholar] [PubMed]
- Woo, P.C.Y.; Lau, S.K.P.; Lam, C.S.F.; Lau, C.C.Y.; Tsang, A.K.L.; Lau, J.H.N.; Bai, R.; Teng, J.L.; Tsang, C.C.; Wang, M.; et al. Discovery of seven novel mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavirus. J. Virol. 2012, 86, 3995–4008. [Google Scholar] [CrossRef]
- Binn, L.N.; Lazar, E.C.; Keenan, K.P.; Huxsoll, D.L.; Marchwicki, R.H.; Strano, A.J. Recovery and characterization of a coronavirus from military dogs with diarrhea. Proc. Annu. Meet. U. S. Anim. Health Assoc. 1974, 78, 359–366. [Google Scholar] [PubMed]
- Erles, K.; Toomey, C.; Brooks, H.W.; Brownlie, J. Detection of a group 2 coronavirus in dogs with canine infectious respiratory disease. Virology 2003, 310, 216–223. [Google Scholar] [CrossRef]
- Pedersen, N.C. Morphologic and physical characteristics of feline infectious peritonitis virus and its growth in autochthonous peritoneal cell cultures. Am. J. Vet. Res. 1976, 37, 567–572. [Google Scholar]
- Mebus, C.A.; Stair, E.L.; Rhodes, M.B.; Twiehaus, M.J. Neonatal calf diarrhea: Propagation, attenuation, and characteristics of a coronavirus-like agent. Am. J. Vet. Res. 1973, 34, 145–150. [Google Scholar] [PubMed]
- Guy, J.S.; Breslin, J.J.; Breuhaus, B.; Vivrette, S.; Smith, L.G. Characterization of a coronavirus isolated from a diarrheic foal. J. Clin. Microbiol. 2000, 38, 4523–4526. [Google Scholar] [CrossRef]
- Bucknall, R.A.; King, L.M.; Kapikian, A.Z.; Chanock, R.M. Studies with human coronaviruses. II. Some properties of strains 229E and OC43. Proc. Soc. Exp. Biol. Med. 1972, 139, 722–727. [Google Scholar] [CrossRef]
- Van der Hoek, L.; Pyrc, K.; Jebbink, M.F.; Vermeulen-Oost, W.; Berkhout, R.J.M.; Wolthers, K.C.; Wertheim-van Dillen, P.M.; Kaandorp, J.; Spaargaren, J.; Berkhout, B. Identification of a new human coronavirus. Nat. Med. 2004, 10, 368–373. [Google Scholar] [CrossRef]
- Brucková, M.; McIntosh, K.; Kapikian, A.Z.; Chanock, R.M. The adaptation of two human coronavirus strains (OC38 and OC43) to growth in cell monolayers. Proc. Soc. Exp. Biol. Med. 1970, 135, 431–435. [Google Scholar] [CrossRef]
- Drosten, C.; Günther, S.; Preiser, W.; van der Werf, S.; Brodt, H.-R.; Becker, S.; Rabenau, H.; Panning, M.; Kolesnikova, L.; Fouchier, R.A.; et al. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 2003, 348, 1967–1976. [Google Scholar] [CrossRef]
- Woo, P.C.Y.; Lau, S.K.P.; Chu, C.; Chan, K.; Tsoi, H.; Huang, Y.; Wong, B.H.; Poon, R.W.; Cai, J.J.; Luk, W.K.; et al. Characterization and complete genome sequence of a novel coronavirus, coronavirus HKU1, from patients with pneumonia. J. Virol. 2005, 79, 884–895. [Google Scholar] [CrossRef]
- Ksiazek, T.G.; Erdman, D.; Goldsmith, C.S.; Zaki, S.R.; Peret, T.; Emery, S.; Tong, S.; Urbani, C.; Comer, J.A.; Lim, W.; et al. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 2003, 348, 1953–1966. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Wang, Y.; Li, X.; Ren, L.; Zhao, J.; Hu, Y.; Zhang, L.; Fan, G.; Xu, J.; Gu, X.; et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020, 395, 497–506. [Google Scholar] [CrossRef]
- Chen, G.-Q.; Zhuang, Q.-Y.; Wang, K.-C.; Liu, S.; Shao, J.-Z.; Jiang, W.-M.; Hou, G.Y.; Li, J.P.; Yu, J.M.; Li, Y.P.; et al. Identification and survey of a novel avian coronavirus in ducks. PLoS ONE 2013, 8, e72918. [Google Scholar] [CrossRef]
- De Wit, J.J.; Cook, J.K.A. Spotlight on avian coronaviruses. Avian Pathol. 2020, 49, 313–316. [Google Scholar] [CrossRef] [PubMed]
- Houta, M.H.; Awe, O.O.; Ali, A. Infection with the turkey coronavirus: A recurring problem in turkeys. Ger. J. Vet. Res. 2021, 1, 19–27. [Google Scholar] [CrossRef]
- Jackwood, M.W.; Boynton, T.O.; Hilt, D.A.; McKinley, E.T.; Kissinger, J.C.; Paterson, A.H.; Robertson, J.; Lemke, C.; McCall, A.W.; Williams, S.M.; et al. Emergence of a group 3 coronavirus through recombination. Virology 2010, 398, 98–108. [Google Scholar] [CrossRef] [PubMed]
- Hughes, A.L. Recombinational histories of avian infectious bronchitis virus and turkey coronavirus. Arch. Virol. 2011, 156, 1823–1829. [Google Scholar] [CrossRef] [PubMed]
- Ntafis, V.; Mari, V.; Decaro, N.; Papanastassopoulou, M.; Papaioannou, N.; Mpatziou, R.; Buonavoglia, C.; Xylouri, E. Isolation, tissue distribution and molecular characterization of two recombinant canine coronavirus strains. Vet. Microbiol. 2011, 151, 238–244. [Google Scholar] [CrossRef]
- Le Poder, S. Feline and canine coronaviruses: Common genetic and pathobiological features. Adv. Virol. 2011, 2011, 1–11. [Google Scholar] [CrossRef]
- Priestnall, S.L.; Mitchell, J.A.; Walker, C.A.; Erles, K.; Brownlie, J. New and emerging pathogens in canine infectious respiratory disease. Vet. Pathol. 2014, 51, 492–504. [Google Scholar] [CrossRef]
- Knesl, O.; Allan, F.J.; Shields, S. The seroprevalence of canine respiratory coronavirus and canine influenza virus in dogs in New Zealand. N. Z. Vet. J. 2009, 57, 295–298. [Google Scholar] [CrossRef] [PubMed][Green Version]
- An, D.-J.; Jeoung, H.-Y.; Jeong, W.; Park, J.-Y.; Lee, M.-H.; Park, B.-K. Prevalence of Korean cats with natural feline coronavirus infections. Virol. J. 2011, 8, 455. [Google Scholar] [CrossRef] [PubMed]
- Saif, L.J. Bovine respiratory coronavirus. Vet. Clin. North Am. Food. Anim. Pract. 2010, 26, 349–364. [Google Scholar] [CrossRef] [PubMed]
- Amer, H.M. Bovine-like coronaviruses in domestic and wild ruminants. Anim. Health Res. Rev. 2018, 19, 113–124. [Google Scholar] [CrossRef]
- Saif, L.J. VACCINES FOR COVID-19: Perspectives, prospects, and challenges based on candidate sars, mers, and animal coronavirus vaccines. Eur. Med. J. 2020, 200324. [Google Scholar] [CrossRef]
- Von Borowski, R.G.; Trentin, D.S. Biofilms and coronavirus reservoirs: A perspective review. Appl. Environ. Microbiol. 2021, 87, e0085921. [Google Scholar] [CrossRef] [PubMed]
- Miszczak, F.; Tesson, V.; Kin, N.; Dina, J.; Balasuriya, U.B.; Pronost, S.; Vabret, A. First detection of equine coronavirus (ECoV) in Europe. Vet. Microbiol. 2014, 171, 206–209. [Google Scholar] [CrossRef] [PubMed]
- Giannitti, F.; Diab, S.; Mete, A.; Stanton, J.B.; Fielding, L.; Crossley, B.; Sverlow, K.; Fish, S.; Mapes, S.; Scott, L.; et al. Necrotizing enteritis and hyperammonemic encephalopathy associated with equine coronavirus infection in equids. Vet. Pathol. 2015, 52, 1148–1156. [Google Scholar] [CrossRef]
- Kooijman, L.J.; James, K.; Mapes, S.M.; Theelen, M.J.P.; Pusterla, N. Seroprevalence and risk factors for infection with equine coronavirus in healthy horses in the USA. Vet. J. 2017, 22, 91–94. [Google Scholar] [CrossRef]
- Oue, Y.; Morita, Y.; Kondo, T.; Nemoto, M. Epidemic of equine coronavirus at obihiro racecourse, Hokkaido, Japan in 2012. J. Vet. Med. Sci. 2013, 75, 1261–1265. [Google Scholar] [CrossRef]
- Sanz, M.G.; Kwon, S.Y.; Pusterla, N.; Gold, J.R.; Bain, F.; Evermann, J. Evaluation of equine coronavirus fecal shedding among hospitalized horses. J. Vet. Intern. Med. 2019, 33, 918–922. [Google Scholar] [CrossRef] [PubMed]
- Pusterla, N.; Vin, R.; Leutenegger, C.; Mittel, L.; Divers, T. Enteric coronavirus infection in adult horses. Vet. J. 2018, 231, 13–18. [Google Scholar] [CrossRef]
- Hemida, M.; Chu, D.K.W.; Perera, R.A.; Ko, R.L.W.; So, R.T.Y.; Ng, B.C.Y.; Chan, S.M.S.; Chu, S.; Alnaeem, A.A.; Alhammadi, M.A.; et al. Coronavirus infections in horses in Saudi Arabia and Oman. Transbound. Emerg. Dis. 2017, 64, 2093–2103. [Google Scholar] [CrossRef] [PubMed]
- Schvartz, G.; Tirosh-Levy, S.; Barnum, S.; David, D.; Sol, A.; Pusterla, N.; Steinman, A. Seroprevalence and risk factors for exposure to equine coronavirus in apparently healthy horses in Israel. Animals 2021, 11, 894. [Google Scholar] [CrossRef]
- Wevers, B.A.; van der Hoek, L. Recently discovered human coronaviruses. Clin. Lab. Med. 2009, 29, 715–724. [Google Scholar] [CrossRef]
- Hu, B.; Ge, X.; Wang, L.-F.; Shi, Z. Bat origin of human coronaviruses. Virol. J 2015, 12, 221. [Google Scholar] [CrossRef] [PubMed]
- Millet, J.K.; Whittaker, G.R. Physiological and molecular triggers for SARS-CoV membrane fusion and entry into host cells. Virology 2018, 517, 3–8. [Google Scholar] [CrossRef] [PubMed]
- Hulswit, R.J.G.; De Haan, C.A.M.; Bosch, B.J. Coronavirus spike protein and tropism changes. Adv. Virus Res. 2016, 96, 29–57. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Li, W.; Farzan, M.; Harrison, S.C. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science 2005, 309, 1864–1868. [Google Scholar] [CrossRef]
- Ou, X.; Guan, H.; Qin, B.; Mu, Z.; Wojdyla, J.A.; Wang, M.; Dominguez, S.R.; Qian, Z.; Cui, S. Crystal structure of the receptor binding domain of the spike glycoprotein of human betacoronavirus HKU1. Nat. Commun. 2017, 8, 15216. [Google Scholar] [CrossRef] [PubMed]
- Qian, Z.; Ou, X.; Góes, L.G.B.; Osborne, C.M.; Castano, A.; Holmes, K.V.; Dominguez, S.R. Identification of the receptor-binding domain of the spike glycoprotein of human betacoronavirus HKU1. J. Virol. 2015, 89, 8816–8827. [Google Scholar] [CrossRef]
- Wang, N.; Shi, X.; Jiang, L.; Zhang, S.; Wang, D.; Tong, P.; Guo, D.; Fu, L.; Cui, Y.; Liu, X.; et al. Structure of MERS-CoV spike receptor-binding domain complexed with human receptor DPP4. Cell Res. 2013, 23, 986–993. [Google Scholar] [CrossRef] [PubMed]
- Li, F. Receptor recognition mechanisms of coronaviruses: A decade of structural studies. J. Virol. 2015, 89, 1954–1964. [Google Scholar] [CrossRef] [PubMed]
- Elaswad, A.; Fawzy, M.; Basiouni, S.; Shehata, A.A. Mutational spectra of SARS-CoV-2 isolated from animals. PeerJ 2020, 8, e10609. [Google Scholar] [CrossRef]
- Yeager, C.L.; Ashmun, R.A.; Williams, R.K.; Cardellichio, C.B.; Shapiro, L.H.; Look, A.T.; Holmes, K.V. Human aminopeptidase N is a receptor for human coronavirus 229E. Nature 1992, 357, 420–422. [Google Scholar] [CrossRef] [PubMed]
- Delmas, B.; Gelfi, J.; L’Haridon, R.; Vogel, L.K.; Sjöström, H.; Norén, O.; Laude, H. Aminopeptidase N is a major receptor for the entero-pathogenic coronavirus TGEV. Nature 1992, 357, 417–420. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Moore, M.J.; Vasilieva, N.; Sui, J.; Wong, S.K.; Berne, M.A.; Somasundaran, M.; Sullivan, J.L.; Luzuriaga, K.; Greenough, T.C.; et al. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature 2003, 426, 450–454. [Google Scholar] [CrossRef]
- Tresnan, D.B.; Levis, R.; Holmes, K.V. Feline aminopeptidase N serves as a receptor for feline, canine, porcine, and human coronaviruses in serogroup I. J. Virol. 1996, 70, 8669–8674. [Google Scholar] [CrossRef]
- Benbacer, L.; Kut, E.; Besnardeau, L.; Laude, H.; Delmas, B. Interspecies aminopeptidase-N chimeras reveal species-specific receptor recognition by canine coronavirus, feline infectious peritonitis virus, and transmissible gastroenteritis virus. J. Virol. 1997, 71, 734–737. [Google Scholar] [CrossRef]
- Wu, K.; Li, W.; Peng, G.; Li, F. Crystal structure of NL63 respiratory coronavirus receptor-binding domain complexed with its human receptor. Proc. Natl. Acad. Sci. USA 2009, 106, 19970–19974. [Google Scholar] [CrossRef]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Krüger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.H.; Nitsche, A.; et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 2020, 181, 271–280. [Google Scholar] [CrossRef] [PubMed]
- Raj, V.S.; Mou, H.; Smits, S.L.; Dekkers, D.H.; Müller, M.A.; Dijkman, R.; Muth, D.; Demmers, J.A.; Zaki, A.; Fouchier, R.A.; et al. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature 2013, 495, 251–254. [Google Scholar] [CrossRef]
- Huang, X.C.; Dong, W.J.; Milewska, A.; Golda, A.; Qi, Y.H.; Zhu, Q.K.; Marasco, W.A.; Baric, R.S.; Sims, A.C.; Pyrc, K.; et al. Human coronavirus HKU1 spike protein uses o-acetylated sialic acid as an attachment receptor determinant and employs hemagglutinin-esterase protein as a receptor-destroying enzyme. J. Virol. 2015, 89, 7202–7213. [Google Scholar] [CrossRef] [PubMed]
- Schultze, B.; Wahn, K.; Klenk, H.-D.; Herrler, G. Isolated HE-protein from hemagglutinating encephalomyelitis virus and bovine coronavirus has receptor-destroying and receptor-binding activity. Virology 1991, 180, 221–228. [Google Scholar] [CrossRef]
- Krempl, C.; Schultze, B.; Herrler, G. Analysis of cellular receptors for human coronavirus OC43. Adv. Exp. Med. Biol. 1995, 380, 371–374. [Google Scholar] [CrossRef] [PubMed]
- Williams, R.K.; Jiang, G.S.; Holmes, K.V. Receptor for mouse hepatitis virus is a member of the carcinoembryonic antigen family of glycoproteins. Proc. Natl. Acad. Sci. USA 1991, 88, 5533–5536. [Google Scholar] [CrossRef]
- Schultze, B.; Enjuanes, L.; Cavanagh, D.; Herrler, G. N-acetylneuraminic acid plays a critical role for the haemagglutinating activity of avian infectious bronchitis virus and porcine transmissible gastroenteritis virus. Adv. Exp. Med. Biol. 1993, 342, 305–310. [Google Scholar] [CrossRef] [PubMed]
- Wickramasinghe, I.N.A.; de Vries, R.; Weerts, E.A.W.S.; Van Beurden, S.J.; Peng, W.; McBride, R.; Ducatez, M.; Guy, J.; Brown, P.; Eterradossi, N.; et al. Novel receptor specificity of avian Gammacoronaviruses that cause enteritis. J. Virol. 2015, 89, 8783–8792. [Google Scholar] [CrossRef] [PubMed]
- Tresnan, D.B.; Holmes, K.V. Feline aminopeptidase N is a receptor for all group I coronaviruses. Adv. Exp. Med. Biol. 1998, 440, 69–75. [Google Scholar] [CrossRef]
- Mina-Osorio, P. The moonlighting enzyme CD13: Old and new functions to target. Trends Mol. Med. 2008, 14, 361–371. [Google Scholar] [CrossRef]
- Mesel-Lemoine, M.; Millet, J.K.; Vidalain, P.-O.; Law, H.K.-W.; Vabret, A.; Lorin, V.; Escriou, N.; Albert, M.L.; Nal, B.; Tangy, F. A human coronavirus responsible for the common cold massively kills dendritic cells but not monocytes. J. Virol. 2012, 86, 7577–7587. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Lin, Y.-L.; Peng, G.; Li, F. Structural basis for multifunctional roles of mammalian aminopeptidase N. Proc. Natl. Acad. Sci. USA 2012, 109, 17966–17971. [Google Scholar] [CrossRef] [PubMed]
- Zhou, P.; Yang, X.-L.; Wang, X.-G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.R.; Zhu, Y.; Li, B.; Huang, C.L.; et al. Addendum: A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020, 588, E6. [Google Scholar] [CrossRef]
- Dveksler, G.S.; Pensiero, M.N.; Cardellichio, C.B.; Williams, R.K.; Jiang, G.S.; Holmes, K.V.; Dieffenbach, C.W. Cloning of the mouse hepatitis virus (MHV) receptor: Expression in human and hamster cell lines confers susceptibility to MHV. J. Virol. 1991, 65, 6881–6891. [Google Scholar] [CrossRef]
- Meyerholz, D.K.; Lambertz, A.M.; McCray, P.B. Dipeptidyl Peptidase 4 Distribution in the human respiratory tract: Implications for the middle east respiratory syndrome. Am. J. Pathol. 2016, 186, 78–86. [Google Scholar] [CrossRef]
- Neu, U.; Bauer, J.; Stehle, T. Viruses and sialic acids: Rules of engagement. Curr. Opin. Struct. Biol. 2011, 21, 610–618. [Google Scholar] [CrossRef]
- Stencel-Baerenwald, J.E.; Reiss, K.; Reiter, D.M.; Stehle, T.; Dermody, T.S. The sweet spot: Defining virus-sialic acid interactions. Nat. Rev. Microbiol. 2014, 12, 739–749. [Google Scholar] [CrossRef] [PubMed]
- Sizov, I.; Mitov, B.; Kharalambiev, K.E. Participation of coronaviruses in the etiology of respiratory diseases in calves. Vet. Med. Nauki 1985, 22, 25–30. [Google Scholar]
- Vlasak, R.; Luytjes, W.; Spaan, W.; Palese, P. Human and bovine coronaviruses recognize sialic acid-containing receptors similar to those of influenza C viruses. Proc. Natl. Acad. Sci. USA 1988, 85, 4526–4529. [Google Scholar] [CrossRef]
- Baker, A.N.; Richards, S.-J.; Guy, C.S.; Congdon, T.R.; Hasan, M.; Zwetsloot, A.J.; Gallo, A.; Lewandowski, J.R.; Stansfeld, P.J.; Straube, A.; et al. The SARS-COV-2 spike protein binds sialic acids and enables rapid detection in a lateral flow point of care diagnostic device. ACS Cent. Sci. 2020, 6, 2046–2052. [Google Scholar] [CrossRef]
- Wan, Y.; Shang, J.; Graham, R.; Baric, R.S.; Li, F. Receptor recognition by the novel coronavirus from Wuhan: An analysis based on decade-long structural studies of SARS coronavirus. J. Virol. 2020, 94, e00127-20. [Google Scholar] [CrossRef]
- Crackower, M.A.; Sarao, R.; Oudit, G.Y.; Yagil, C.; Kozieradzki, I.; Scanga, S.E.; Oliveira-dos-Santos, A.J.; da Costa, J.; Zhang, L.; Pei, Y.; et al. Angiotensin-converting enzyme 2 is an essential regulator of heart function. Nature 2002, 417, 822–828. [Google Scholar] [CrossRef] [PubMed]
- Danilczyk, U.; Sarao, R.; Remy, C.; Benabbas, C.; Stange, G.; Richter, A.; Arya, S.; Pospisilik, J.A.; Singer, D.; Camargo, S.M.; et al. Essential role for collectrin in renal amino acid transport. Nature 2006, 444, 1088–1091. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Zhao, Z.; Wang, Y.; Zhou, Y.; Ma, Y.; Zuo, W. Single-cell RNA expression profiling of ACE2, the receptor of SARS-CoV-2. Am. J. Respir. Crit. Care Med. 2020, 202, 756–759. [Google Scholar] [CrossRef] [PubMed]
- Imai, Y.; Kuba, K.; Rao, S.; Huan, Y.; Guo, F.; Guan, B.; Yang, P.; Sarao, R.; Wada, T.; Leong-Poi, H.; et al. Angiotensin-converting enzyme 2 protects from severe acute lung failure. Nature 2005, 436, 112–116. [Google Scholar] [CrossRef]
- Yu, L.; Yuan, K.; Phuong, H.T.A.; Park, B.M.; Kim, S.H. Angiotensin-(1–5), an active mediator of renin-angiotensin system, stimulates ANP secretion via Mas receptor. Peptides 2016, 86, 33–41. [Google Scholar] [CrossRef] [PubMed]
- Bader, M. ACE2—das missbrauchte Multitalent. Der. Nephrol. 2020, 15, 375–380. [Google Scholar] [CrossRef]
- Letko, M.; Marzi, A.; Munster, V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat. Microbiol. 2020, 5, 562–569. [Google Scholar] [CrossRef]
- Ou, X.; Liu, Y.; Lei, X.; Li, P.; Mi, D.; Ren, L.; Guo, L.; Guo, R.; Chen, T.; Hu, J.; et al. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat. Commun. 2020, 11, 1620. [Google Scholar] [CrossRef]
- Bestle, D.; Heindl, M.R.; Limburg, H.; Van, T.V.L.; Pilgram, O.; Moulton, H.; Matrosovich, M.; Abdelwhab, E.M.; Stech, J.; Böttcher-Friebertshäuser, E. TMPRSS2 and furin are both essential for proteolytic activation of SARS-CoV-2 in human airway cells. Life Sci. Alliance 2020, 3, e202000786. [Google Scholar] [CrossRef] [PubMed]
- Matsuyama, S.; Nao, N.; Shirato, K.; Kawase, M.; Saito, S.; Takayama, I.; Nagata, N.; Sekizuka, T.; Katoh, H.; Kato, F.; et al. Enhanced isolation of SARS-CoV-2 by TMPRSS2-expressing cells. Proc. Natl. Acad. Sci. USA 2020, 117, 7001–7003. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, M.; Hofmann-Winkler, H.; Pöhlmann, S. Priming time: How cellular proteases arm coronavirus spike proteins. In Activation of Viruses by Host Proteases; Böttcher-Friebertshäuser, E., Garten, W., Klenk, H.D., Eds.; Springer: Cham, Switzerland, 2018; pp. 71–98. [Google Scholar] [CrossRef]
- Rockwell, N.C.; Krysan, D.J.; Komiyama, T.; Fuller, R.S. Precursor processing by Kex2/Furin proteases. Chem Rev. 2002, 102, 4525–4548. [Google Scholar] [CrossRef] [PubMed]
- Heald-Sargent, T.; Gallagher, T. Ready, Set, Fuse! The coronavirus spike protein and acquisition of fusion competence. Viruses 2012, 4, 557–580. [Google Scholar] [CrossRef] [PubMed]
- Millet, J.K.; Whittaker, G.R. Host cell proteases: Critical determinants of coronavirus tropism and pathogenesis. Virus Res. 2015, 202, 120–134. [Google Scholar] [CrossRef]
- Corman, V.M.; Baldwin, H.J.; Tateno, A.F.; Zerbinati, R.M.; Annan, A.; Owusu, M.; Nkrumah, E.E.; Maganga, G.D.; Oppong, S.; Adu-Sarkodie, Y.; et al. Evidence for an ancestral association of human coronavirus 229E with bats. J. Virol. 2015, 89, 11858–11870. [Google Scholar] [CrossRef]
- Donaldson, E.F.; Haskew, A.N.; Gates, J.E.; Huynh, J.; Moore, C.J.; Frieman, M.B. Metagenomic analysis of the viromes of three North American bat species: Viral diversity among different bat species that share a common habitat. J. Virol. 2010, 84, 13004–13018. [Google Scholar] [CrossRef]
- Ge, X.-Y.; Li, J.-L.; Yang, X.-L.; Chmura, A.A.; Zhu, G.; Epstein, J.H.; Mazet, J.K.; Hu, B.; Zhang, W.; Peng, C.; et al. Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor. Nature 2013, 503, 535–538. [Google Scholar] [CrossRef]
- Cui, J.; Li, F.; Shi, Z.-L. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 2019, 17, 181–192. [Google Scholar] [CrossRef]
- Vlasova, A.N.; Diaz, A.; Damtie, D.; Xiu, L.; Toh, T.-H.; Lee, J.S.; Saif, L.J.; Gray, G.C. Novel Canine coronavirus isolated from a hospitalized pneumonia patient, East Malaysia. Clin. Infect. Dis. 2021, ciab456. [Google Scholar] [CrossRef]
- Su, S.; Wong, G.; Shi, W.; Liu, J.; Lai, A.C.K.; Zhou, J.; Liu, W.; Bi, Y.; Gao, G.F. Epidemiology, Genetic recombination, and pathogenesis of coronaviruses. Trends Microbiol. 2016, 24, 490–502. [Google Scholar] [CrossRef]
- Kuo, L.; Godeke, G.J.; Raamsman, M.J.; Masters, P.S.; Rottier, P.J. Retargeting of coronavirus by substitution of the spike glycoprotein ectodomain: Crossing the host cell species barrier. J. Virol. 2000, 74, 1393–1406. [Google Scholar] [CrossRef] [PubMed]
- Haijema, B.J.; Volders, H.; Rottier, P.J.M. Switching species tropism: An effective way to manipulate the feline coronavirus genome. J. Virol. 2003, 77, 4528–4538. [Google Scholar] [CrossRef] [PubMed]
- Ye, Z.W.; Yuan, S.; Yuen, K.S.; Fung, S.Y.; Chan, C.P.; Jin, D.Y. Zoonotic origins of human coronaviruses. Int. J. Biol. Sci. 2020, 16, 1686–1697. [Google Scholar] [CrossRef]
- Bolles, M.; Donaldson, E.; Baric, R. SARS-CoV and emergent coronaviruses: Viral determinants of interspecies transmission. Curr. Opin. Virol. 2011, 1, 624–634. [Google Scholar] [CrossRef] [PubMed]
- Ren, W.; Qu, X.; Li, W.; Han, Z.; Yu, M.; Zhou, P.; Zhang, S.Y.; Wang, L.F.; Deng, H.; Shi, Z. Difference in receptor usage between severe acute respiratory syndrome (SARS) coronavirus and SARS-like coronavirus of bat origin. J. Virol. 2008, 82, 1899–1907. [Google Scholar] [CrossRef] [PubMed]
- Wu, K.; Peng, G.; Wilken, M.; Geraghty, R.J.; Li, F. Mechanisms of host receptor adaptation by severe acute respiratory syndrome coronavirus. J. Biol. Chem. 2012, 287, 8904–8911. [Google Scholar] [CrossRef] [PubMed]
- Sit, T.H.C.; Brackman, C.J.; Ip, S.M.; Tam, K.W.S.; Law, P.Y.T.; To, E.M.W.; Yu, V.Y.T.; Sims, L.D.; Tsang, D.N.C.; Chu, D.K.W. Infection of dogs with SARS-CoV-2. Nature 2020, 586, 776–778. [Google Scholar] [CrossRef] [PubMed]
- Patterson, E.I.; Elia, G.; Grassi, A.; Giordano, A.; Desario, C.; Medardo, M.; Smith, S.L.; Anderson, E.R.; Prince, T.; Patterson, G.T.; et al. Evidence of exposure to SARS-CoV-2 in cats and dogs from households in Italy. Microbiology 2020, 11, 6231. [Google Scholar] [CrossRef]
- Calvet, G.A.; Pereira, S.A.; Ogrzewalska, M.; Pauvolid-Corrêa, A.; Resende, P.C.; Tassinari, W.S.; Costa, A.P.; Keidel, L.O.; da Rocha, A.S.B.; da Silva, M.F.B.; et al. Investigation of SARS-CoV-2 infection in dogs and cats of humans diagnosed with COVID-19 in Rio de Janeiro, Brazil. PLoS ONE 2021, 16, e0250853. [Google Scholar] [CrossRef]
- Fritz, M.; Rosolen, B.; Krafft, E.; Becquart, P.; Elguero, E.; Vratskikh, O.; Denolly, S.; Boson, B.; Vanhomwegen, J.; Gouilh, M.A.; et al. High prevalence of SARS-CoV-2 antibodies in pets from COVID-19 households. One Health 2020, 11, 100192. [Google Scholar] [CrossRef]
- Jara, L.M.; Ferradas, C.; Schiaffino, F.; Sánchez-Carrión, C.; Martinez, A.; Ulloa, A.; Isasi-Rivas, G.; Montalván, A.; Sarmiento, L.G.; Fernández, M.; et al. Evidence of neutralizing antibodies against SARS-CoV-2 in domestic cats living with owners with a history of COVID-19 in Lima—Peru. One Health 2021, 13, 100318. [Google Scholar] [CrossRef]
- Michelitsch, A.; Hoffmann, D.; Wernike, K.; Beer, M. Occurrence of antibodies against SARS-CoV-2 in thedomestic cat population of Germany. Vaccines 2020, 8, 772. [Google Scholar] [CrossRef]
- Van Aart, A.E.; Velkers, F.C.; Fischer, E.A.; Broens, E.M.; Egberink, H.; Zhao, S.; Engelsma, M.; Hakze-van der Honing, R.W.; Harders, F.; de Rooij, M.M.T.; et al. SARS-CoV-2 infection in cats and dogs in infected mink farms. Transbound Emerg. Dis. 2021, 1-7, tbed.14173. [Google Scholar] [CrossRef]
- Gauntt, J. Texas A&M research uncovers first known COVID-19 UK variant in animals. Available online: https://today.tamu.edu/2021/03/15/texas-am-research-uncovers-first-known-covid-19-uk-variant-in-animals/ (accessed on 15 December 2021).
- Ferasin, L.; Fritz, M.; Ferasin, H.; Becquart, P.; Legros, V.; Leroy, E.M. Myocarditis in naturally infected pets with the British variant of COVID-19. Microbiology 2021, 8, 748896. [Google Scholar] [CrossRef]
- Oreshkova, N.; Molenaar, R.J.; Vreman, S.; Harders, F.; Munnink, B.B.O.; Hakze-van der Honing, R.W.; Gerhards, N.; Tolsma, P.; Bouwstra, R.; Sikkema, R.S.; et al. SARS-CoV-2 infection in farmed minks, the Netherlands, April and May 2020. Euro Surveill. 2020, 25, 210325c. [Google Scholar] [CrossRef]
- De Morais, H.A.; Dos Santos, A.P.; Nascimento, N.C.D.; Kmetiuk, L.B.; Barbosa, D.S.; Brandão, P.E.; Kmetiuk, L.B.; Barbosa, D.S.; Brandão, P.E.; Guimarães, A.M.S.; et al. Natural Infection by SARS-CoV-2 in Companion Animals: A review of case reports and current evidence of their role in the epidemiology of COVID-19. Front Vet. Sci. 2020, 7, 591216. [Google Scholar] [CrossRef] [PubMed]
- Alexander, M.R.; Schoeder, C.T.; Brown, J.A.; Smart, C.D.; Moth, C.; Wikswo, J.P.; Capra, J.A.; Meiler, J.; Chen, W.; Madhur, M.S. Predicting susceptibility to SARS-CoV-2 infection based on structural differences in ACE2 across species. FASEB J. 2020, 34, 15946–15960. [Google Scholar] [CrossRef]
- Sharun, K.; Sircar, S.; Malik, Y.S.; Singh, R.K.; Dhama, K. How close is SARS-CoV-2 to canine and feline coronaviruses? J. Small Anim. Pract. 2020, 61, 523–526. [Google Scholar] [CrossRef] [PubMed]
- Zhai, X.; Sun, J.; Yan, Z.; Zhang, J.; Zhao, J.; Zhao, Z.; Gao, Q.; He, W.T.; Veit, M.; Su, S.; et al. Comparison of severe acute respiratory syndrome coronavirus 2 spike protein binding to ACE2 receptors from human, pets, farm animals, and putative intermediate hosts. J. Virol. 2020, 94, e00831-20. [Google Scholar] [CrossRef]
- Tazerji, S.S.; Duarte, P.M.; Rahimi, P.; Shahabinejad, F.; Dhakal, S.; Malik, Y.S.; Shehata, A.A.; Lama, J.; Klein, J.; Safdar, M.; et al. Transmission of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) to animals: An updated review. J. Transl. Med. 2020, 18, 358. [Google Scholar] [CrossRef] [PubMed]
- Choy, K.W. Specificity and cross-reactivity of a test for anti-SARS-CoV-2 antibodies. Lancet Infect. Dis. 2021, 21, e118. [Google Scholar] [CrossRef]
- Hicks, J.; Klumpp-Thomas, C.; Kalish, H.; Shunmugavel, A.; Mehalko, J.; Denson, J.-P.; Snead, K.R.; Drew, M.; Corbett, K.S.; Graham, B.S.; et al. Serologic cross-reactivity of SARS-CoV-2 with endemic and seasonal Betacoronaviruses. Infect Dis. 2021, 41, 906–913. [Google Scholar] [CrossRef]
- Shrock, E.; Fujimura, E.; Kula, T.; Timms, R.T.; Lee, I.-H.; Leng, Y.; Robinson, M.L.; Sie, B.M.; Li, M.Z.; Chen, Y.; et al. Viral epitope profiling of COVID-19 patients reveals cross-reactivity and correlates of severity. Science 2020, 370. [Google Scholar] [CrossRef] [PubMed]
- Kin, N.; Miszczak, F.; Diancourt, L.; Caro, V.; Moutou, F.; Vabret, A.; Ar Gouilh, M. Comparative molecular epidemiology of two closely related coronaviruses, bovine coronavirus (BCoV) and human coronavirus OC43 (HCoV-OC43), reveals a different evolutionary pattern. Infect. Genet. Evol. 2016, 40, 186–191. [Google Scholar] [CrossRef]
- Csiszar, A.; Jakab, F.; Valencak, T.G.; Lanszki, Z.; Tóth, G.E.; Kemenesi, G.; Tarantini, S.; Fazekas-Pongor, V.; Ungvari, Z. Companion animals likely do not spread COVID-19 but may get infected themselves. GeroScience 2020, 42, 1229–1236. [Google Scholar] [CrossRef]
- Hernández, M.; Abad, D.; Eiros, J.M. Rodríguez-Lázaro, D. Are animals a neglected transmission route of SARS-CoV-2? Pathogens 2020, 9, 480. [Google Scholar] [CrossRef] [PubMed]
- Hobbs, E.C.; Reid, T.J. Animals and SARS-CoV-2: Species susceptibility and viral transmission in experimental and natural conditions, and the potential implications for community transmission. Transbound Emerg. Dis. 2020, 68, 1850–1867. [Google Scholar] [CrossRef]
- Ji, W.; Wang, W.; Zhao, X.; Zai, J.; Li, X. Cross-species transmission of the newly identified coronavirus 2019-nCoV. J. Med. Virol. 2020, 92, 433–440. [Google Scholar] [CrossRef] [PubMed]
- Mazinani, M.; Rude, B.J. The novel zoonotic Coronavirus disease 2019 (COVID-19) pandemic: Health perspective on the outbreak. J. Healthc. Qual. Res. 2021, 36, 47–51. [Google Scholar] [CrossRef] [PubMed]
- Fenollar, F.; Mediannikov, O.; Maurin, M.; Devaux, C.; Colson, P.; Levasseur, A.; Levasseur, A.; Fournier, P.E.; Raoult, D. Mink, SARS-CoV-2, and the human-animal interface. Front Microbiol. 2021, 12, 663815. [Google Scholar] [CrossRef]
- Enserink, M. Coronavirus rips through Dutch mink farms, triggering culls. Science 2020, 368, 1169. [Google Scholar] [CrossRef] [PubMed]
- Hammer, A.S.; Quaade, M.L.; Rasmussen, T.B.; Fonager, J.; Rasmussen, M.; Mundbjerg, K.; Lohse, L.; Strandbygaard, B.; Jørgensen, C.S.; Alfaro-Núñez, A.; et al. SARS-CoV-2 transmission between mink (Neovison vison) and humans, Denmark. Emerg. Infect. Dis. 2021, 27, 547–551. [Google Scholar] [CrossRef]
- Larsen, C.S.; Paludan, S.R. Corona’s new coat: SARS-CoV-2 in Danish minks and implications for travel medicine. Travel Med. Infect. Dis. 2020, 38, 101922. [Google Scholar] [CrossRef] [PubMed]
- Shuai, L.; Zhong, G.; Yuan, Q.; Wen, Z.; Wang, C.; He, X.; Liu, R.; Wang, J.; Zhao, Q.; Liu, Y.; et al. Replication, pathogenicity, and transmission of SARS-CoV-2 in minks. Natl. Sci. Rev. 2021, 8, nwaa291. [Google Scholar] [CrossRef]
- Oude Munnink, B.B.; Sikkema, R.S.; Nieuwenhuijse, D.F.; Molenaar, R.J.; Munger, E.; Molenkamp, R.; van der Spek, A.; Tolsma, P.; Rietveld, A.; Brouwer, M.; et al. Transmission of SARS-CoV-2 on mink farms between humans and mink and back to humans. Science 2021, 371, 172–177. [Google Scholar] [CrossRef]
- Mykytyn, A.Z.; Lamers, M.M.; Okba, N.M.A.; Breugem, T.I.; Schipper, D.; van den Doel, P.B.; van Run, P.; van Amerongen, G.; de Waal, L.; Koopmans, M.P.G.; et al. Susceptibility of rabbits to SARS-CoV-2. Emerg. Microbes. Infect. 2021, 10, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Lamers, M.M.; Mykytyn, A.Z.; I Breugem, T.; Wang, Y.; Wu, D.C.; Riesebosch, S.; van den Doel, P.B.; Schipper, D.; Bestebroer, T.; Wu, N.C.; et al. Human airway cells prevent SARS-CoV-2 multibasic cleavage site cell culture adaptation. eLife 2021, 10, e66815. [Google Scholar] [CrossRef]
- McAloose, D.; Laverack, M.; Wang, L.; Killian, M.L.; Caserta, L.C.; Yuan, F.; Mitchell, P.K.; Queen, K.; Mauldin, M.R.; Cronk, B.D.; et al. From people to panthera: Natural SARS-CoV-2 infection in tigers and lions at the Bronx zoo. MBio 2020, 11. [Google Scholar] [CrossRef] [PubMed]
- Jemeršić, L.; Lojkić, I.; Krešić, N.; Keros, T.; Zelenika, T.; Jurinović, L.; Skok, D.; Bata, I.; Boras, J.; Habrun, B.; et al. Investigating the presence of SARS-CoV-2 in free-living and captive animals. Pathogens 2021, 10, 635. [Google Scholar] [CrossRef]
- Meekins, D.A.; Morozov, I.; Trujillo, J.D.; Gaudreault, N.N.; Bold, D.; Carossino, M.; Artiaga, B.L.; Indran, S.V.; Kwon, T.; Balaraman, V.; et al. Susceptibility of swine cells and domestic pigs to SARS-CoV-2. Emerg. Microbes Infect. 2020, 9, 2278–2288. [Google Scholar] [CrossRef]
- Schlottau, K.; Rissmann, M.; Graaf, A.; Schön, J.; Sehl, J.; Wylezich, C.; Höper, D.; Mettenleiter, T.C.; Balkema-Buschmann, A.; Harder, T.; et al. SARS-CoV-2 in fruit bats, ferrets, pigs, and chickens: An experimental transmission study. Lancet Microbe 2020, 1, e218–e225. [Google Scholar] [CrossRef]
- Shi, J.; Wen, Z.; Zhong, G.; Yang, H.; Wang, C.; Huang, B.; Liu, R.; He, X.; Shuai, L.; Sun, Z.; et al. Susceptibility of ferrets, cats, dogs, and other domesticated animals to SARS-coronavirus-2. Science 2020, 368, 1016–1020. [Google Scholar] [CrossRef] [PubMed]
- Pickering, B.S.; Smith, G.; Pinette, M.M.; Embury-Hyatt, C.; Moffat, E.; Marszal, P.; Lewis, C.E. Susceptibility of domestic swine to experimental infection with severe acute respiratory syndrome coronavirus 2. Emerg. Infect. Dis. 2021, 27, 104–112. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.-I.; Kim, S.-G.; Kim, S.-M.; Kim, E.-H.; Park, S.-J.; Yu, K.-M.; Chang, J.H.; Kim, E.J.; Lee, S.; Casel, M.A.B.; et al. Infection and rapid transmission of SARS-CoV-2 in ferrets. Cell Host Microbe 2020, 27, 704–709. [Google Scholar] [CrossRef] [PubMed]
- Chan, J.F.-W.; Kok, K.-H.; Zhu, Z.; Chu, H.; To, K.K.-W.; Yuan, S.; Yuen, K.Y. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg. Microbes Infect. 2020, 9, 221–236. [Google Scholar] [CrossRef] [PubMed]
- Chandrashekar, A.; Liu, J.; Martinot, A.J.; Mcmahan, K.; Mercado, N.B.; Peter, L.; Tostanoski, L.H.; Yu, J.; Maliga, Z.; Nekorchuk, M.; et al. SARS-CoV-2 infection protects against rechallenge in rhesus macaques. Science 2020, 369, 812–817. [Google Scholar] [CrossRef]
- Yu, P.; Qi, F.; Xu, Y.; Li, F.; Liu, P.; Liu, J.; Bao, L.; Deng, W.; Gao, H.; Xiang, Z.; et al. Age-related rhesus macaque models of COVID-19. Anim. Model Exp. Med 2020, 3, 93–97. [Google Scholar] [CrossRef]
- Bao, L.; Deng, W.; Huang, B.; Gao, H.; Liu, J.; Ren, L.; Wei, Q.; Yu, P.; Xu, Y.; Qi, F.; et al. The pathogenicity of SARS-CoV-2 in hACE2 transgenic mice. Nature 2020, 583, 830–833. [Google Scholar] [CrossRef]
- Damas, J.; Hughes, G.M.; Keough, K.C.; Painter, C.A.; Persky, N.S.; Corbo, M.; Hiller, M.; Koepfli, K.P.; Pfenning, A.R.; Zhao, H.; et al. Broad host range of SARS-CoV-2 predicted by comparative and structural analysis of ACE2 in vertebrates. Proc. Natl. Acad. Sci. USA 2020, 117, 22311–22322. [Google Scholar] [CrossRef]
- Ulrich, L.; Wernike, K.; Hoffmann, D.; Mettenleiter, T.C.; Beer, M. Experimental infection of cattle with SARS-CoV-2. Emerg. Infect. Dis. 2020, 26, 2979–2981. [Google Scholar] [CrossRef]
- Lu, S.; Wang, Y.; Chen, Y.; Wu, B.; Qin, K.; Zhao, J.; Lou, Y.; Tan, W. Discovery of a novel canine respiratory coronavirus support genetic recombination among betacoronavirus1. Virus Res. 2017, 237, 7–13. [Google Scholar] [CrossRef] [PubMed]
- Vale, B.D.; Lopes, A.P.; Fontes, M.D.C.; Silvestre, M.; Cardoso, L.; Coelho, A.C. Bats, pangolins, minks and other animals—villains or victims of SARS-CoV-2? Vet. Res. Commun. 2021, 45, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Richard, M.; Kok, A.; De Meulder, D.; Bestebroer, T.M.; Lamers, M.M.; Okba, N.M.A.; Fentener van Vlissingen, M.; Rockx, B.; Haagmans, B.L.; Koopmans, M.P.G.; et al. SARS-CoV-2 is transmitted via contact and via the air between ferrets. Nat. Commun. 2020, 11, 3496. [Google Scholar] [CrossRef]
- Freuling, C.M.; Breithaupt, A.; Müller, T.; Sehl, J.; Balkema-Buschmann, A.; Rissmann, M.; Klein, A.; Wylezich, C.; Höper, D.; Wernike, K.; et al. Susceptibility of raccoon dogs for experimental SARS-CoV-2 infection. Emerg. Infect. Dis. 2020, 26, 2982–2985. [Google Scholar] [CrossRef] [PubMed]
- Francisco, R.; Hernandez, S.M.; Mead, D.G.; Adcock, K.G.; Burke, S.C.; Nemeth, N.M.; Yabsley, M.J. Experimental susceptibility of North American raccoons (Procyon lotor) and striped skunks (Mephitis mephitis) to SARS-CoV-2. Microbiology 2021. [Google Scholar] [CrossRef] [PubMed]
- Woolsey, C.; Borisevich, V.; Prasad, A.N.; Agans, K.N.; Deer, D.J.; Dobias, N.S.; Heymann, J.C.; Foster, S.L.; Levine, C.B.; Medina, L.; et al. Establishment of an African green monkey model for COVID-19 and protection against re-infection. Nat. Immunol. 2021, 22, 86–98. [Google Scholar] [CrossRef]
- Suarez, D.L.; Pantin-Jackwood, M.J.; Swayne, D.E.; Lee, S.A.; DeBlois, S.M.; Spackman, E. Lack of susceptibility to SARS-CoV-2 and MERS-CoV in poultry. Emerg. Infect. Dis. 2020, 26, 3074–3076. [Google Scholar] [CrossRef]
- Bosco-Lauth, A.M.; Walker, A.; Guilbert, L.; Porter, S.; Hartwig, A.; McVicker, E.; Bielefeldt-Ohmann, H.; Bowen, R.A. Susceptibility of livestock to SARS-CoV-2 infection. Emerg. Microbes Infect. 2021, 10, 2199–2201. [Google Scholar] [CrossRef]
- El Zowalaty, M.; Järhult, J.D. From SARS to COVID-19: A previously unknown SARS- related coronavirus (SARS-CoV-2) of pandemic potential infecting humans—Call for a One Health approach. One Health 2020, 9, 100124. [Google Scholar] [CrossRef]
- Helmy, Y.A.; Fawzy, M.; Elaswad, A.; Sobieh, A.; Kenney, S.P.; Shehata, A.A. The COVID-19 pandemic: A comprehensive review of taxonomy, genetics, epidemiology, diagnosis, treatment, and control. J. Clin. Med. 2020, 9, 1225. [Google Scholar] [CrossRef]
- Fawzy, M.; Hasham, A.; Houta, M.H.; Hasham, M.; Helmy, Y.A. COVID-19: Risk assessment and mitigation measures in healthcare and non-healthcare workplaces. Ger. J. Microbiol. 2021, 1, 19–28. [Google Scholar] [CrossRef]
- Chen, N.; Zhou, M.; Dong, X.; Qu, J.; Gong, F.; Han, Y.; Qiu, Y.; Wang, J.; Liu, Y.; Wei, Y.; et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: A descriptive study. Lancet 2020, 395, 507–513. [Google Scholar] [CrossRef]
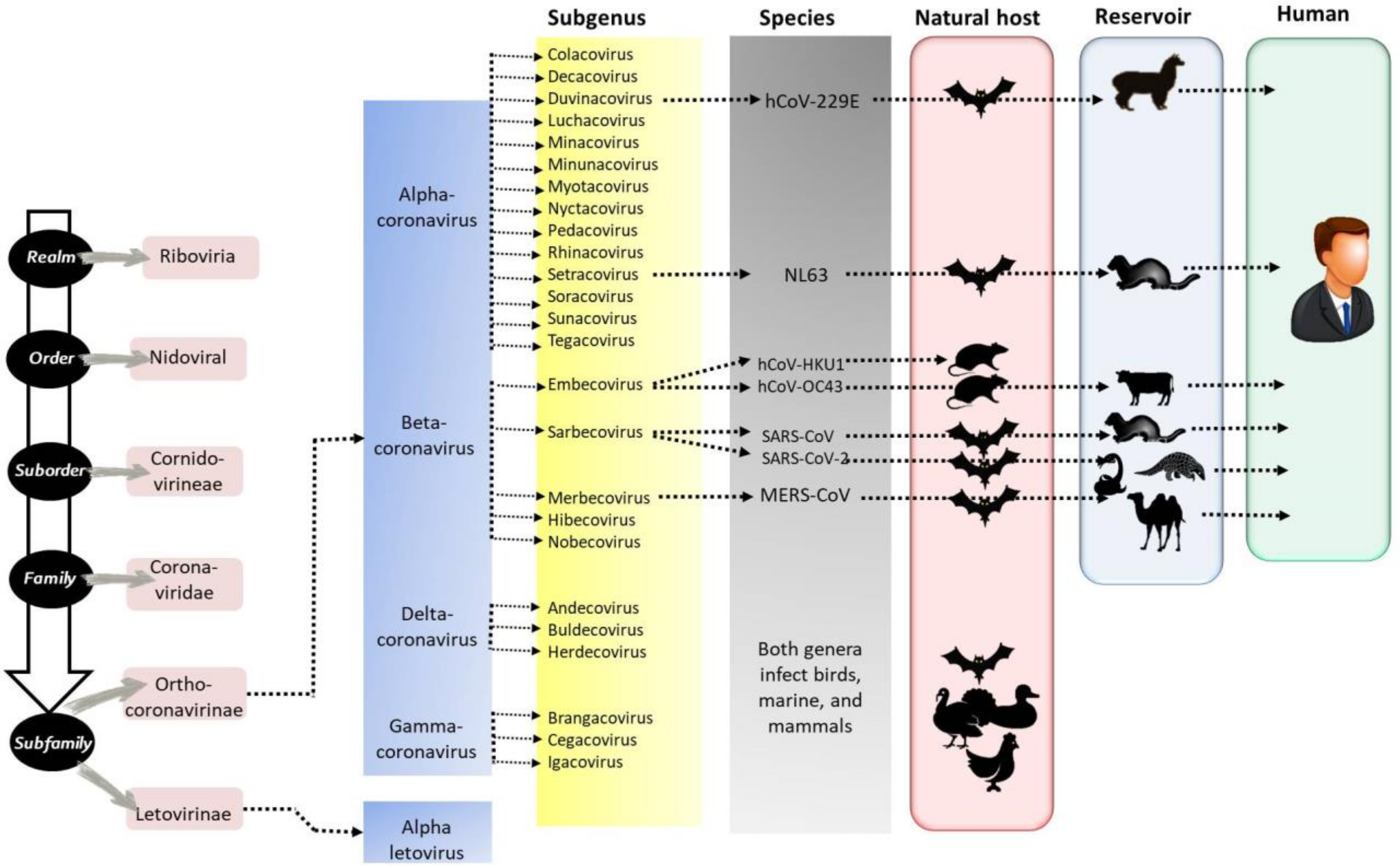
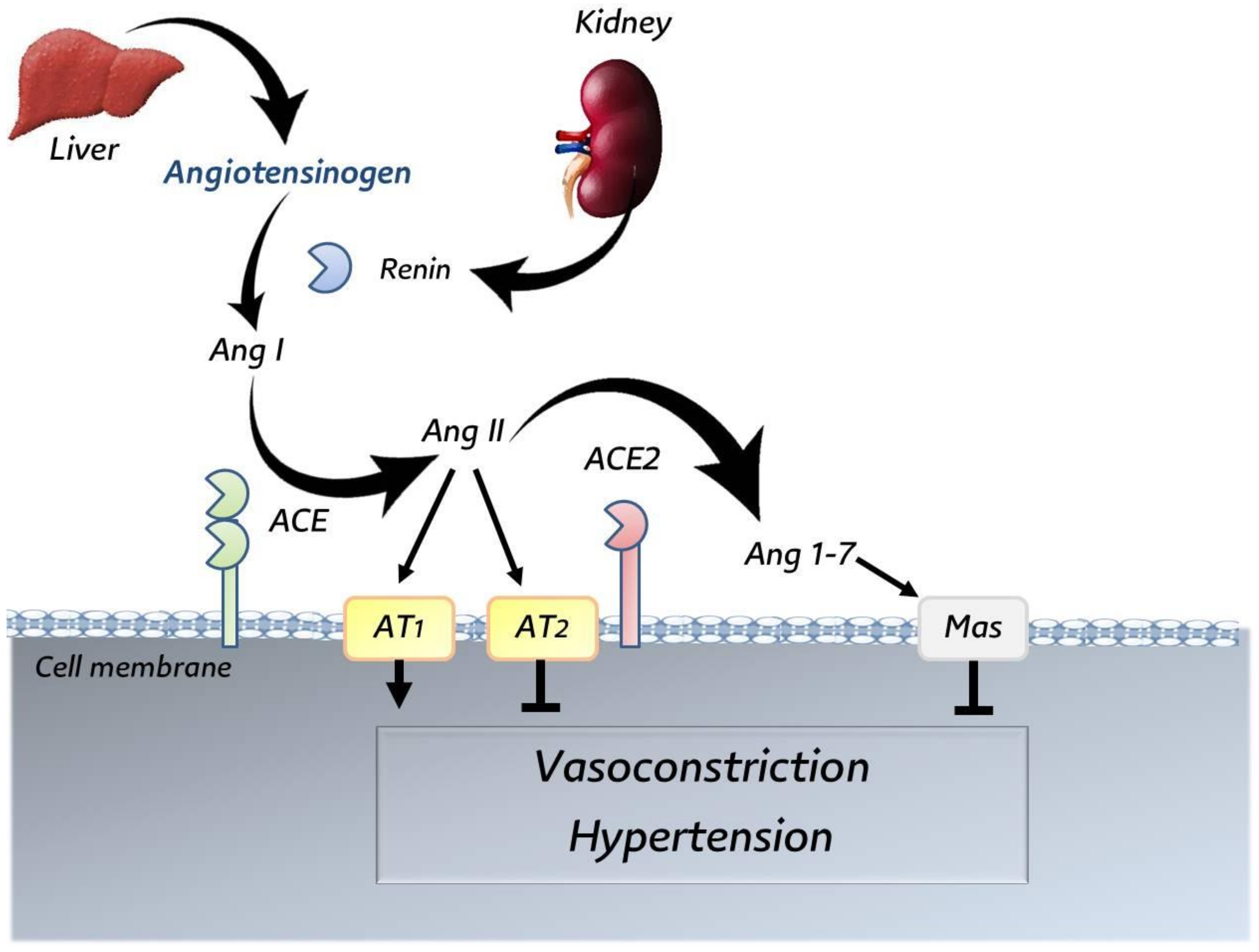
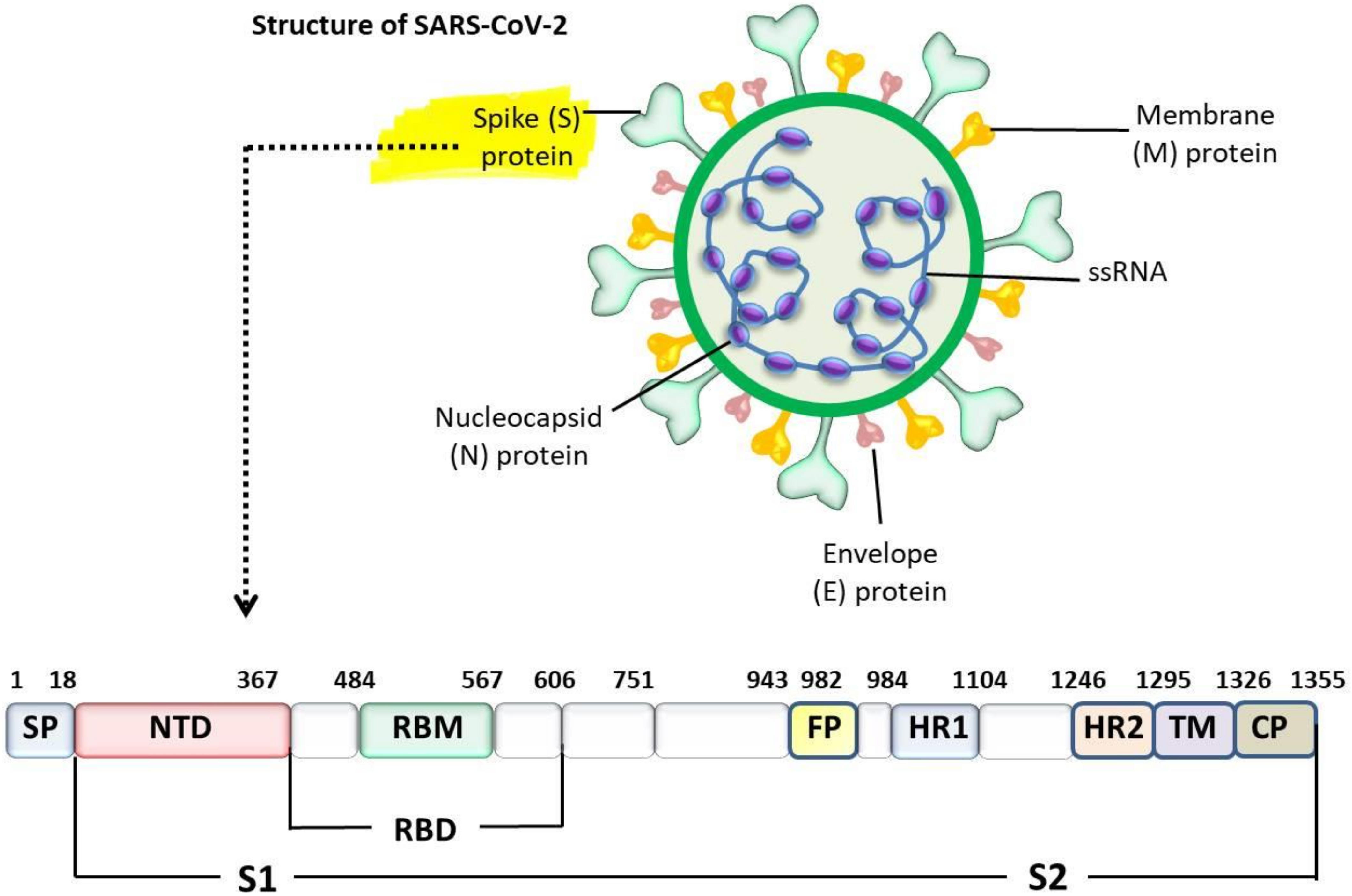
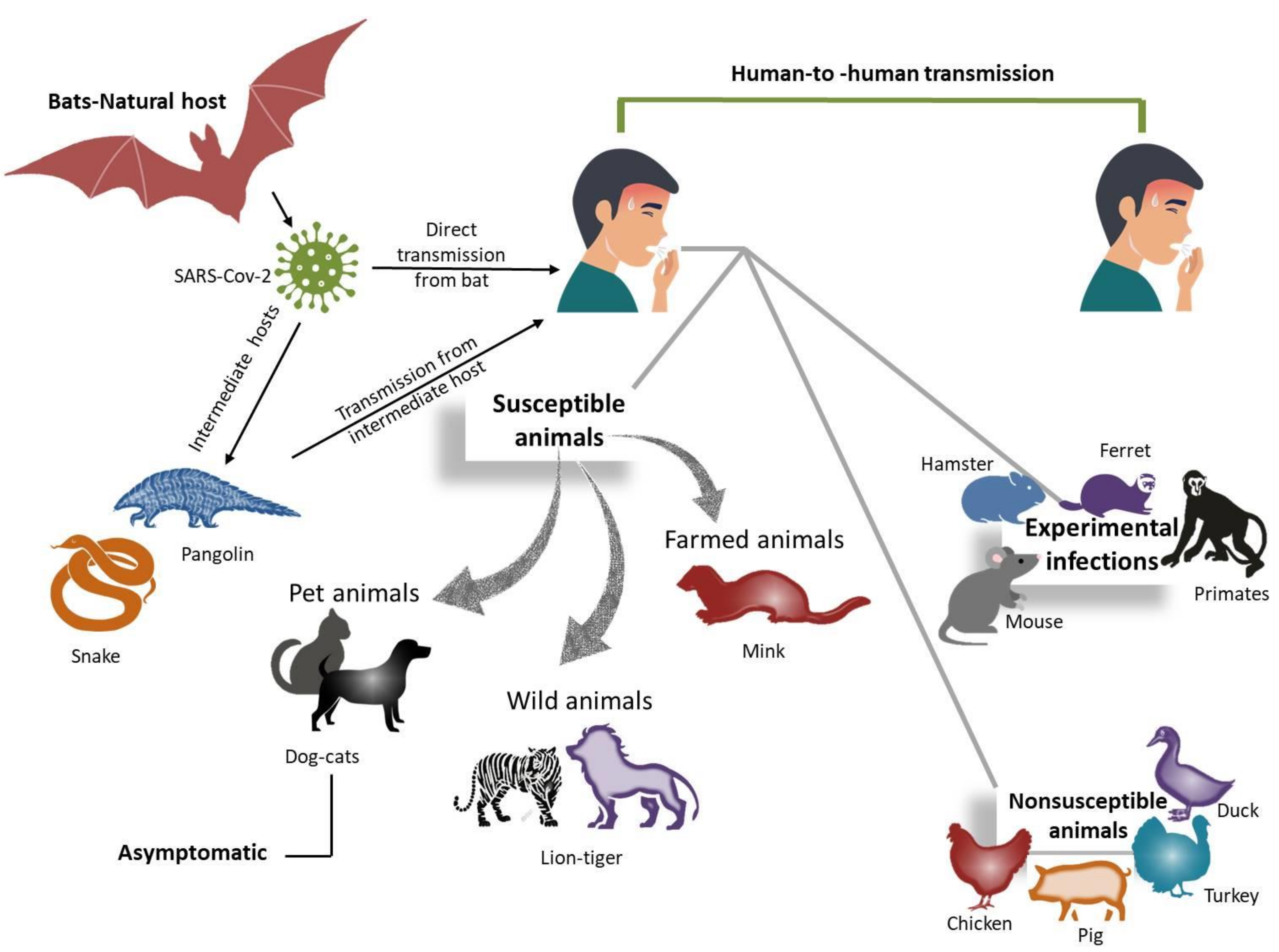
| Host | Virus * | Genus | First Year of Isolation | Country of First Isolation | Tropism | References |
|---|---|---|---|---|---|---|
| Avian species | IBV | β-CoVs | 1930 | USA | Respiratory, urinary, and reproductive | [1,11] |
| TcoV | β-CoVs | 1971 | USA | Enteric | [12] | |
| PhCoV | β-CoVs | 1980 | UK | Respiratory, reproductive, urinary | [13] | |
| GfCoV | β-CoVs | 2011 | France | Fulminating enteritis | [14] | |
| PiCoV | β-CoVs | 1988 | Australia | Enteric | [15] | |
| Pig (Sus scrofa domesticus) | TGEV | α-CoVs | 1946 | USA | Enteric | [16] |
| PEDV | α-CoVs | 1971 | UK | Enteric | [17] | |
| PRCV | α-CoVs | 1986 | Belgium | Respiratory | [18] | |
| SADS-CoV | α-CoVs | 2017 | China | Enteric | [19] | |
| PHEV | β-CoVs | 1962 | Canada | Respiratory, nervous | [20] | |
| PDCoV | δ-CoVs | 2012 | China | Enteric | [21] | |
| Dog (Canis lupus familiaris) | CCoV | α-CoVs | 1971 | Germany | Acute enteritis | [22] |
| CRCoV | β-CoVs | 2003 | UK | Respiratory | [23] | |
| Cat (Felis catus) | FCoV | α-CoVs | 1863 | US | Respiratory, gastrointestinal | [24] |
| Cattle (Bos taurus) | BCoV | β-CoVs | 1973 | USA | Enteric, respiratory | [25] |
| Horse (Equus caballus) | ECoV | β-CoVs | 2007 | USA | Enteric | [26] |
| Human | 229E | α-CoVs | 1966 | USA | Respiratory | [27] |
| NL63 | α-CoVs | 2004 | The Netherlands | Respiratory | [28] | |
| OC43 | β-CoVs | 1967 | USA | Respiratory | [29] | |
| SARS-CoV | β-CoVs | 2002 | China | Respiratory | [30] | |
| HKU1 | β-CoVs | 2005 | China | Respiratory | [31] | |
| MERS-CoV | β-CoVs | 2012 | Middle East | Respiratory | [32] | |
| SARS-CoV-2 | β-CoVs | 2019 | China | Enteric | [33] |
| Receptors | Virus * | Genus | Ref |
|---|---|---|---|
| Amino peptidase (APN, CD13) | Human CoV 229E | α-CoV | [66] |
| TGEV | [67] | ||
| PRCV | [68] | ||
| FCoV | [69] | ||
| CCoV | [70] | ||
| Angiotensin-converting enzyme 2 (ACE2) | Human CoV NL63 | α-CoV | [71] |
| SARS-CoV | β-CoV | [68] | |
| SARS-CoV-2 | β-CoV | [72] | |
| Dipeptidyl peptidase 4 (DPP4) | MERS-CoV | β-CoV | [73] |
| 9-O-acetylated sialic acid | Human CoV HKU1 | β-CoV | [74] |
| PHEV | β-CoV | [75] | |
| Human CoV OC43 | β-CoV | [76] | |
| Carcinoembryonic antigen-related cell adhesion molecule 1 (CEACAM1) | MHV | β-CoV | [77] |
| Leukocyte antigen class I (HLA-1) | BCoV | β-CoV | [68] |
| CRCoV | β-CoV | [72] | |
| 2,3-linked sialylated glycans | IBV | γ-CoV | [78] |
| Nonsialylated type 2 poly-LacNAc | TCoV | γ-CoV | [79] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shehata, A.A.; Attia, Y.A.; Rahman, M.T.; Basiouni, S.; El-Seedi, H.R.; Azhar, E.I.; Khafaga, A.F.; Hafez, H.M. Diversity of Coronaviruses with Particular Attention to the Interspecies Transmission of SARS-CoV-2. Animals 2022, 12, 378. https://doi.org/10.3390/ani12030378
Shehata AA, Attia YA, Rahman MT, Basiouni S, El-Seedi HR, Azhar EI, Khafaga AF, Hafez HM. Diversity of Coronaviruses with Particular Attention to the Interspecies Transmission of SARS-CoV-2. Animals. 2022; 12(3):378. https://doi.org/10.3390/ani12030378
Chicago/Turabian StyleShehata, Awad A., Youssef A. Attia, Md. Tanvir Rahman, Shereen Basiouni, Hesham R. El-Seedi, Esam I. Azhar, Asmaa F. Khafaga, and Hafez M. Hafez. 2022. "Diversity of Coronaviruses with Particular Attention to the Interspecies Transmission of SARS-CoV-2" Animals 12, no. 3: 378. https://doi.org/10.3390/ani12030378
APA StyleShehata, A. A., Attia, Y. A., Rahman, M. T., Basiouni, S., El-Seedi, H. R., Azhar, E. I., Khafaga, A. F., & Hafez, H. M. (2022). Diversity of Coronaviruses with Particular Attention to the Interspecies Transmission of SARS-CoV-2. Animals, 12(3), 378. https://doi.org/10.3390/ani12030378











