Reconstruction of the Major Maternal and Paternal Lineages in the Feral New Zealand Kaimanawa Horses
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. DNA Extraction and SNP Genotyping
2.3. Relatedness Calculation
2.4. Mitochondrial DNA Analysis
2.5. Y-Chromosome Analysis
2.6. Population Structure and Genetic Distance within the KH Population
3. Results
3.1. Relatedness in KH population
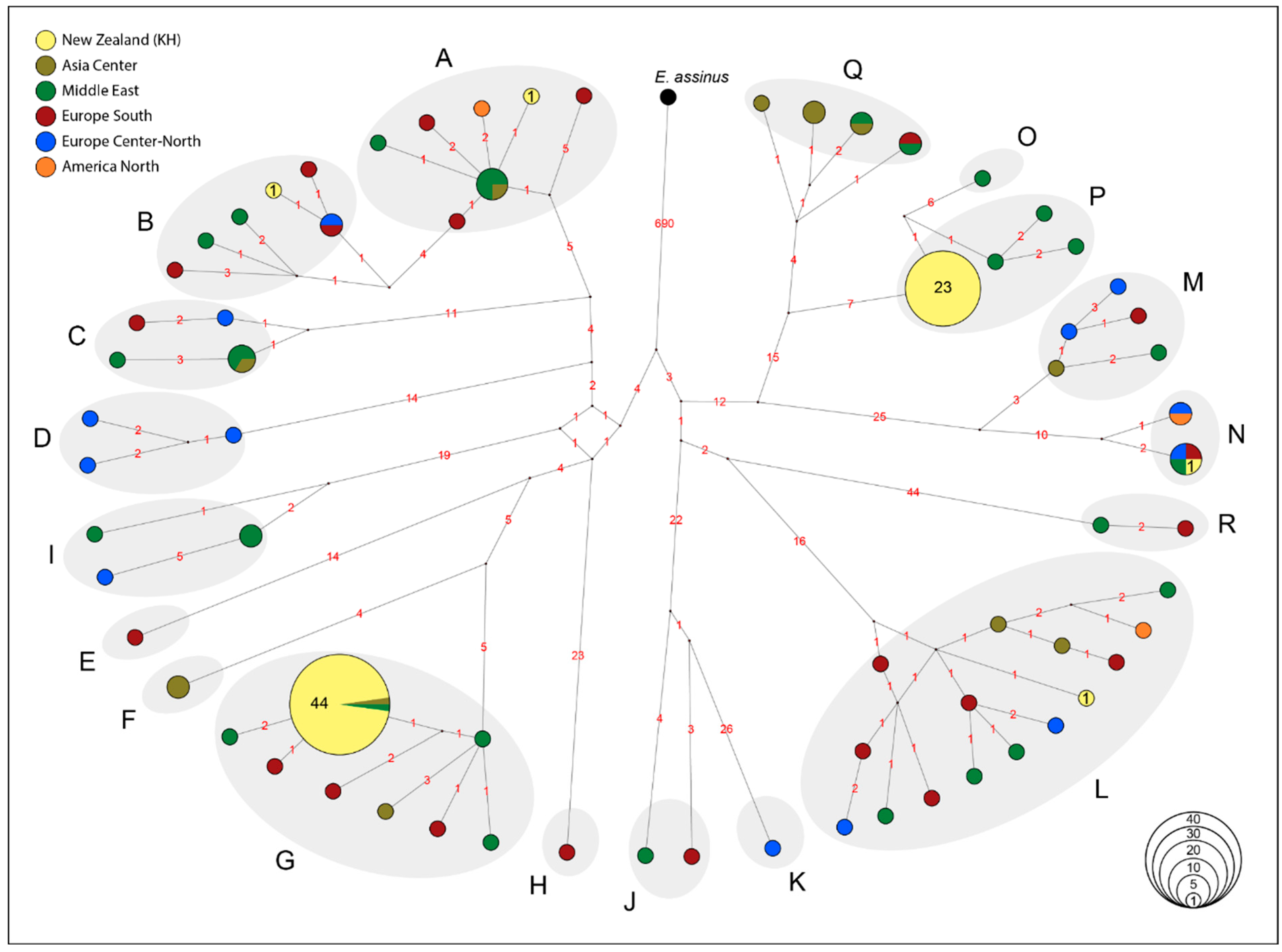
3.2. Mitochondrial DNA Genetic Diversity and Haplogroup Distribution
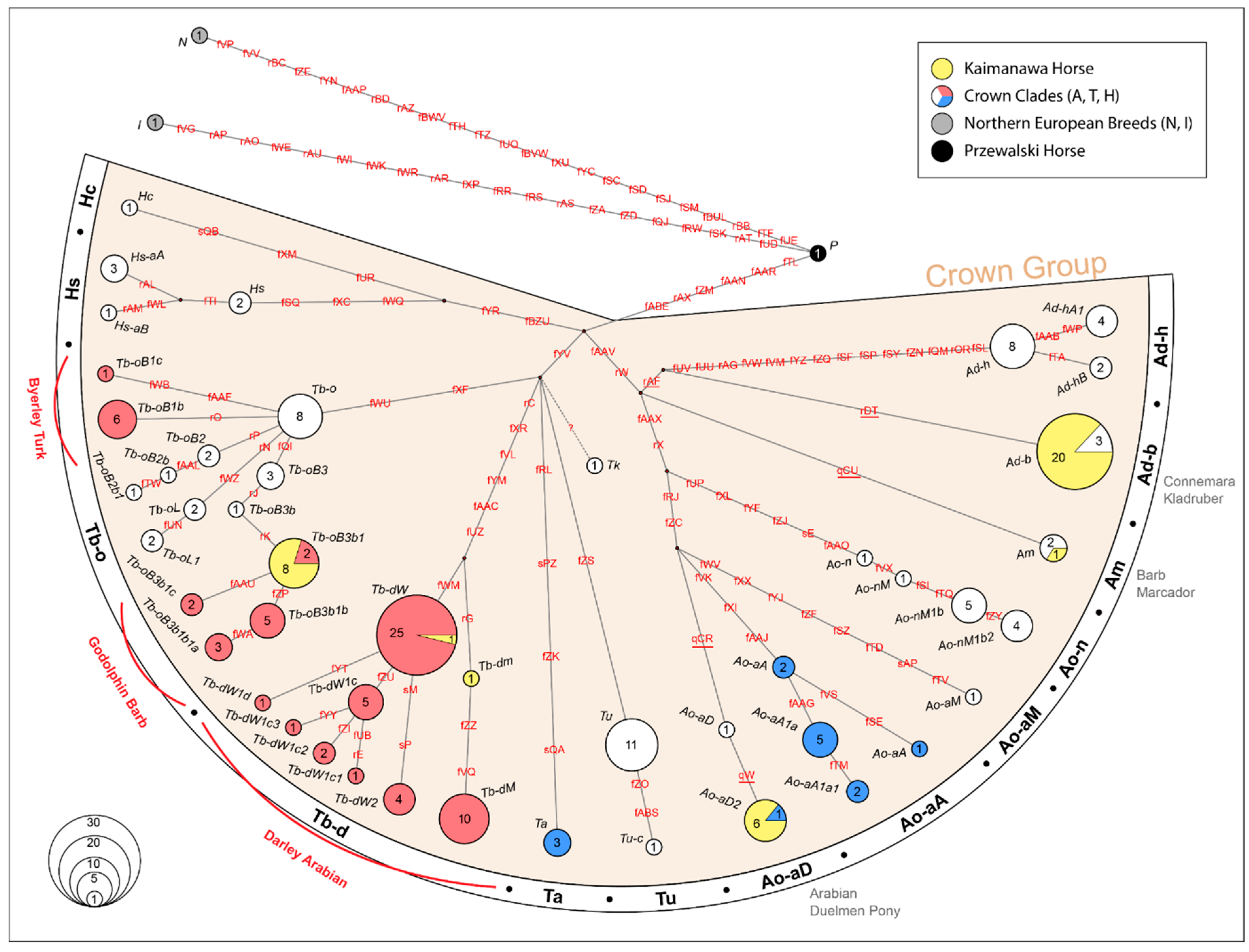
3.3. Y-Chromosome Genetic Diversity and Haplogroup Distribution
3.4. Population Structure and Genetic Distance within the KH Population
4. Discussion
4.1. Genetic Diversity of Maternal Lineages in KHs
4.2. Genetic Diversity of Paternal Lineages in KHs
4.3. Loss of Maternal and Paternal Lineages in the KH Population
4.4. Population Structure within KH Population: Parallel Contribution of European and Middle Eastern Parental Ancestry
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lever, C. Naturalized Mammals of the World; Longman: London, UK, 1985. [Google Scholar]
- Fages, A.; Hanghøj, K.; Khan, N.; Gaunitz, C.; Seguin-Orlando, A.; Leonardi, M.; Constantz, C.M.; Gamba, C.; Al-Rasheid, K.A.; Albizuri, S. Tracking Five Millennia of Horse Management with Extensive Ancient Genome Time Series. Cell 2019, 177, 1419–1435. [Google Scholar] [CrossRef] [PubMed]
- Librado, P.; Gamba, C.; Gaunitz, C.; Der Sarkissian, C.; Pruvost, M.; Albrechtsen, A.; Fages, A.; Khan, N.; Schubert, M.; Jagannathan, V.; et al. Ancient Genomic Changes Associated with Domestication of the Horse. Science 2017, 356, 442–445. [Google Scholar] [CrossRef] [PubMed]
- Cameron, E.; Linklater, W.; Minot, E.; Stafford, K. Population Dynamics 1994–1998, and Management of Kaimanawa Wild Horses. Sci. Conserv. 2001, 171, 1173–2946. [Google Scholar]
- Hendrickson, S.L. A Genome Wide Study of Genetic Adaptation to High Altitude in Feral Andean Horses of the Páramo. BMC Evol. Biol. 2013, 13, 1–13. [Google Scholar] [CrossRef]
- Colpitts, J.; McLoughlin, P.D.; Poissant, J. Runs of Homozygosity in Sable Island Feral Horses Reveal the Genomic Consequences of Inbreeding and Divergence from Domestic Breeds. BMC Genom. 2022, 23, 501. [Google Scholar] [CrossRef] [PubMed]
- Tollett, C.M. Genomic Diversity and Origins of the Feral Horses (Equus ferus caballus) of Sable Island and the Alberta Foothills. Master’s Thesis, University of Saskatchewan, Saskatoon, SK, Canada, 2018. [Google Scholar]
- King, S.R.; Schoenecker, K.A.; Fike, J.A.; Oyler-McCance, S.J. Feral Horse Space Use and Genetic Characteristics from Fecal DNA. J. Wildl. Manag. 2021, 85, 1074–1083. [Google Scholar] [CrossRef]
- Kobayashi, I.; Akita, M.; Takasu, M.; Tozaki, T.; Kakoi, H.; Nakamura, K.; Senju, N.; Matsuyama, R.; Horii, Y. Genetic Characteristics of Feral Misaki Horses Based on Polymorphisms of Microsatellites and Mitochondrial DNA. J. Vet. Med. Sci. 2019, 81, 707–711. [Google Scholar] [CrossRef] [PubMed]
- Ovchinnikov, I.V.; Dahms, T.; Herauf, B.; McCann, B.; Juras, R.; Castaneda, C.; Cothran, E.G. Genetic Diversity and Origin of the Feral Horses in Theodore Roosevelt National Park. PLoS ONE 2018, 13, e0200795. [Google Scholar] [CrossRef]
- Conant, E.; Juras, R.; Cothran, E. A Microsatellite Analysis of Five Colonial Spanish Horse Populations of the Southeastern United States. Anim. Genet. 2012, 43, 53–62. [Google Scholar] [CrossRef]
- Prystupa, J.M.; Hind, P.; Cothran, E.G.; Plante, Y. Maternal Lineages in Native Canadian Equine Populations and Their Relationship to the Nordic and Mountain and Moorland Pony Breeds. J. Hered. 2012, 103, 380–390. [Google Scholar] [CrossRef]
- Wilson, K. Wild Horses of the World; Penguin Random House: Auckland, New Zealand, 2020; ISBN 978-0-14-377291-0. [Google Scholar]
- Fleury, W. Kaimanawa Wild Horse Herd—Draft Management Strategy; Department of Conservation: Wanganui, New Zealand, 1991.
- Boyd, J. Kaimanawa Heritage Horses Welfare SocietyHorse History. Available online: https://kaimanawaheritagehorses.org/horsehistory/ (accessed on 5 November 2022).
- Aitken, V.; Kroef, P.; Pearson, A.; Ricketts, W. Observations of Feral Horses in the Southern Kaimanawas; Report to the Kaimanawa Wild Horse Committee; Massey University: Palmerston North, New Zealand, 1979. [Google Scholar]
- Rogers, G.M. Kaimanawa Feral Horses and Their Environmental Impacts. N. Z. J. Ecol. 1991, 15, 49–64. [Google Scholar]
- Kaimanawa Heritage Horses Welfare Society Immunocontraception. Available online: https://kaimanawaheritagehorses.org/immunocontraception/ (accessed on 12 September 2022).
- Halkett, R.J. A Genetic Analysis of the Kaimanawa Horses and Comparisons with Other Equine Types. Master’s Thesis, Massey University, Palmerston North, New Zealand, 1996. [Google Scholar]
- Achilli, A.; Olivieri, A.; Soares, P.; Lancioni, H.; Kashani, B.H.; Perego, U.A.; Nergadze, S.G.; Carossa, V.; Santagostino, M.; Capomaccio, S. Mitochondrial Genomes from Modern Horses Reveal the Major Haplogroups That Underwent Domestication. Proc. Natl. Acad. Sci. USA 2012, 109, 2449–2454. [Google Scholar] [CrossRef] [PubMed]
- Vilà, C.; Leonard, J.A.; Gotherstrom, A.; Marklund, S.; Sandberg, K.; Lidén, K.; Wayne, R.K.; Ellegren, H. Widespread Origins of Domestic Horse Lineages. Science 2001, 291, 474–477. [Google Scholar] [CrossRef] [PubMed]
- Petersen, J.L.; Mickelson, J.R.; Cothran, E.G.; Andersson, L.S.; Axelsson, J.; Bailey, E.; Bannasch, D.; Binns, M.M.; Borges, A.S.; Brama, P.; et al. Genetic Diversity in the Modern Horse Illustrated from Genome-Wide SNP Data. PLoS ONE 2013, 8, e54997. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.C.; Chow, C.C.; Tellier, L.C.; Vattikuti, S.; Purcell, S.M.; Lee, J.J. Second-Generation PLINK: Rising to the Challenge of Larger and Richer Datasets. GigaScience 2015, 4, s13742-015-0047-8. [Google Scholar] [CrossRef]
- Zhao, S.; Jing, W.; Samuels, D.C.; Sheng, Q.; Shyr, Y.; Guo, Y. Strategies for Processing and Quality Control of Illumina Genotyping Arrays. Brief. Bioinform. 2018, 19, 765–775. [Google Scholar] [CrossRef]
- Guo, Y.; He, J.; Zhao, S.; Wu, H.; Zhong, X.; Sheng, Q.; Samuels, D.C.; Shyr, Y.; Long, J. Illumina Human Exome Genotyping Array Clustering and Quality Control. Nat. Protoc. 2014, 9, 2643–2662. [Google Scholar] [CrossRef]
- Rozas, J.; Ferrer-Mata, A.; Sánchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sánchez-Gracia, A. DnaSP 6: DNA Sequence Polymorphism Analysis of Large Data Sets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Bandelt, H.-J.; Forster, P.; Röhl, A. Median-Joining Networks for Inferring Intraspecific Phylogenies. Mol. Biol. Evol. 1999, 16, 37–48. [Google Scholar] [CrossRef]
- Kong, S.; Sánchez-Pacheco, S.J.; Murphy, R.W. On the Use of Median-Joining Networks in Evolutionary Biology. Cladistics 2016, 32, 691–699. [Google Scholar] [CrossRef] [PubMed]
- Eggert, L.S.; Powell, D.M.; Ballou, J.D.; Malo, A.F.; Turner, A.; Kumer, J.; Zimmerman, C.; Fleischer, R.C.; Maldonado, J.E. Pedigrees and the Study of the Wild Horse Population of Assateague Island National Seashore. J. Wildl. Manag. 2010, 74, 963–973. [Google Scholar] [CrossRef]
- Luís, C.; Bastos-Silveira, C.; Cothran, E.G.; Oom, M. do M. Iberian Origins of New World Horse Breeds. J. Hered. 2006, 97, 107–113. [Google Scholar] [CrossRef] [PubMed]
- Jansen, T.; Forster, P.; Levine, M.A.; Oelke, H.; Hurles, M.; Renfrew, C.; Weber, J.; Olek, K. Mitochondrial DNA and the Origins of the Domestic Horse. Proc. Natl. Acad. Sci. USA 2002, 99, 10905–10910. [Google Scholar] [CrossRef] [PubMed]
- Felkel, S.; Vogl, C.; Rigler, D.; Dobretsberger, V.; Chowdhary, B.P.; Distl, O.; Fries, R.; Jagannathan, V.; Janečka, J.E.; Leeb, T.; et al. The Horse Y Chromosome as an Informative Marker for Tracing Sire Lines. Sci. Rep. 2019, 9, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Remer, V.; Bozlak, E.; Felkel, S.; Radovic, L.; Rigler, D.; Grilz-Seger, G.; Stefaniuk-Szmukier, M.; Bugno-Poniewierska, M.; Brooks, S.; Miller, D.C.; et al. Y-Chromosomal Insights into Breeding History and Sire Line Genealogies of Arabian Horses. Genes 2022, 13, 229. [Google Scholar] [CrossRef]
- He, C.; Holme, J.; Anthony, J. SNP Genotyping: The KASP Assay. In Crop Breeding; Springer: Berlin/Heidelberg, Germany, 2014; pp. 75–86. [Google Scholar]
- Lippold, S.; Xu, H.; Ko, A.; Li, M.; Renaud, G.; Butthof, A.; Schröder, R.; Stoneking, M. Human Paternal and Maternal Demographic Histories: Insights from High-Resolution Y Chromosome and MtDNA Sequences. Investig. Genet. 2014, 5, 1–17. [Google Scholar] [CrossRef]
- Wright, S. The Genetical Structure of Populations. Ann. Eugen. 1949, 15, 323–354. [Google Scholar] [CrossRef]
- Excoffier, L.; Lischer, H.E. Arlequin Suite Ver 3.5: A New Series of Programs to Perform Population Genetics Analyses under Linux and Windows. Mol. Ecol. Resour. 2010, 10, 564–567. [Google Scholar] [CrossRef]
- Tajima, F. Statistical Method for Testing the Neutral Mutation Hypothesis by DNA Polymorphism. Genetics 1989, 123, 585–595. [Google Scholar] [CrossRef]
- Fu, Y.-X.; Li, W.-H. Statistical Tests of Neutrality of Mutations. Genetics 1993, 133, 693–709. [Google Scholar] [CrossRef] [PubMed]
- Kavar, T.; Dovč, P. Domestication of the Horse: Genetic Relationships between Domestic and Wild Horses. Livest. Sci. 2008, 116, 1–14. [Google Scholar] [CrossRef]
- Cieslak, M.; Pruvost, M.; Benecke, N.; Hofreiter, M.; Morales, A.; Reissmann, M.; Ludwig, A. Origin and History of Mitochondrial DNA Lineages in Domestic Horses. PLoS ONE 2010, 5, e15311. [Google Scholar] [CrossRef] [PubMed]
- Gurney, S.; Schneider, S.; Pflugradt, R.; Barrett, E.; Forster, A.C.; Brinkmann, B.; Jansen, T.; Forster, P. Developing Equine MtDNA Profiling for Forensic Application. Int. J. Leg. Med. 2010, 124, 617–622. [Google Scholar] [CrossRef]
- Mühlberger, M. Genetische Untersuchung Zu Paternalen Linien in Kaltblutpferden Und Kleinpferden Europas. Master’s Thesis, Veterinärmedizinischen Universität Wien, Wien, Austria, 2019. [Google Scholar]
- Willmann, R. Das Exmoor-Pferd: Eines Der Ursprünglichsten Halbwilden Pferde Der Welt. Nat. Und Mus. 1999, 129, 389–407. [Google Scholar]
- Allfrey, S. Horses & Foals; WHSmith: London, UK, 1980; ISBN 9780706342130. [Google Scholar]
- Duncan, P.; Boy, V.; Monard, A.-M. The Proximate Mechanisms of Natal Dispersal in Female Horses. Behaviour 1996, 133, 1095–1124. [Google Scholar] [CrossRef]
- Linklater, W.L. Adaptive Explanation in Socio-Ecology: Lessons from the Equidae. Biol. Rev. 2000, 75, 1–20. [Google Scholar] [CrossRef]
- OROZCO-terWENGEL, P.; Corander, J.; Schlötterer, C. Genealogical Lineage Sorting Leads to Significant, but Incorrect Bayesian Multilocus Inference of Population Structure. Mol. Ecol. 2011, 20, 1108–1121. [Google Scholar] [CrossRef]
- Hein, J.; Schierup, M.; Wiuf, C. Gene Genealogies, Variation and Evolution: A Primer in Coalescent Theory; Oxford University Press: New York, NY, USA, 2004. [Google Scholar]
- Jagannathan, V.; Gerber, V.; Rieder, S.; Tetens, J.; Thaller, G.; Drögemüller, C.; Leeb, T. Comprehensive Characterization of Horse Genome Variation by Whole-Genome Sequencing of 88 Horses. Anim. Genet. 2019, 50, 74–77. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Hall, T.; Biosciences, I.; Carlsbad, C. BioEdit: An Important Software for Molecular Biology. GERF Bull. Biosci. 2011, 2, 60–61. [Google Scholar]
- Letunic, I.; Bork, P. Interactive Tree Of Life (ITOL) v5: An Online Tool for Phylogenetic Tree Display and Annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef] [PubMed]

| Genetic Diversity Indices | Kaimanawa Horse | Global Modern Horse [20] | |
|---|---|---|---|
| 1081 SNPs | 1081 SNPs | Complete mtDNA | |
| Number of samples (N) | 71 | 81 | 81 |
| Number of polymorphic (segregating) sites (S) | 126 | 334 | 659 |
| Number of haplotypes (h) | 6 | 66 | 79 |
| Average number of nucleotide differences (k) | 27.3 | 45 | 83 |
| Haplotype diversity (Hd) | 0.518 (0.043) | 0.994 (0.003) | 0.999 (0.002) |
| Nucleotide diversity (π) | 0.025 (0.002) | 0.042 (0.001) | 0.0049 (0.0002) |
| Theta estimator based on the segregating sites (θW) | 0.024 (0.006) | 0.062 (0.016) | 0.008 (0.002) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sharif, M.B.; Fitak, R.R.; Wallner, B.; Orozco-terWengel, P.; Frewin, S.; Fremaux, M.; Mohandesan, E. Reconstruction of the Major Maternal and Paternal Lineages in the Feral New Zealand Kaimanawa Horses. Animals 2022, 12, 3508. https://doi.org/10.3390/ani12243508
Sharif MB, Fitak RR, Wallner B, Orozco-terWengel P, Frewin S, Fremaux M, Mohandesan E. Reconstruction of the Major Maternal and Paternal Lineages in the Feral New Zealand Kaimanawa Horses. Animals. 2022; 12(24):3508. https://doi.org/10.3390/ani12243508
Chicago/Turabian StyleSharif, Muhammad Bilal, Robert Rodgers Fitak, Barbara Wallner, Pablo Orozco-terWengel, Simone Frewin, Michelle Fremaux, and Elmira Mohandesan. 2022. "Reconstruction of the Major Maternal and Paternal Lineages in the Feral New Zealand Kaimanawa Horses" Animals 12, no. 24: 3508. https://doi.org/10.3390/ani12243508
APA StyleSharif, M. B., Fitak, R. R., Wallner, B., Orozco-terWengel, P., Frewin, S., Fremaux, M., & Mohandesan, E. (2022). Reconstruction of the Major Maternal and Paternal Lineages in the Feral New Zealand Kaimanawa Horses. Animals, 12(24), 3508. https://doi.org/10.3390/ani12243508






