Joint Models to Predict Dairy Cow Survival from Sensor Data Recorded during the First Lactation
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Data
2.2. Data Processing
2.3. Algorithm Development
2.4. Algorithm Evaluation
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- van der Heide, E.M.M.; Kamphuis, C.; Veerkamp, R.F.; Athanasiadis, I.N.; Azzopardi, G.; van Pelt, M.L.; Ducro, B.J. Improving Predictive Performance on Survival in Dairy Cattle Using an Ensemble Learning Approach. Comput. Electron. Agric. 2020, 177, 105675. [Google Scholar] [CrossRef]
- van Pelt, M.L.; Meuwissen, T.H.E.; de Jong, G.; Veerkamp, R.F. Genetic Analysis of Longevity in Dutch Dairy Cattle Using Random Regression. J. Dairy Sci. 2015, 98, 4117–4130. [Google Scholar] [CrossRef] [PubMed]
- Adriaens, I.; Friggens, N.C.; Ouweltjes, W.; Scott, H.; Aernouts, B.; Statham, J. Productive Life Span and Resilience Rank Can Be Predicted from On-Farm First-Parity Sensor Time Series but Not Using a Common Equation across Farms. J. Dairy Sci. 2020, 103, 7155–7171. [Google Scholar] [CrossRef]
- Schuster, J.C.; Barkema, H.W.; De Vries, A.; Kelton, D.F.; Orsel, K. Invited Review: Academic and Applied Approach to Evaluating Longevity in Dairy Cows. J. Dairy Sci. 2020, 103, 11008–11024. [Google Scholar] [CrossRef] [PubMed]
- Dallago, G.M.; Wade, K.M.; Cue, R.I.; McClure, J.T.; Lacroix, R.; Pellerin, D.; Vasseur, E. Keeping Dairy Cows for Longer: A Critical Literature Review on Dairy Cow Longevity in High Milk-Producing Countries. Animals 2021, 11, 808. [Google Scholar] [CrossRef] [PubMed]
- Hoffman, J.M.; Valencak, T.G. A Short Life on the Farm: Aging and Longevity in Agricultural, Large-Bodied Mammals. GeroScience 2020, 42, 909–922. [Google Scholar] [CrossRef] [PubMed]
- Bach, A. Associations between Several Aspects of Heifer Development and Dairy Cow Survivability to Second Lactation. J. Dairy Sci. 2011, 94, 1052–1057. [Google Scholar] [CrossRef] [PubMed]
- Cabrera, V.E. Invited Review: Helping Dairy Farmers to Improve Economic Performance Utilizing Data-Driving Decision Support Tools. Animal 2018, 12, 134–144. [Google Scholar] [CrossRef]
- Grandl, F.; Furger, M.; Kreuzer, M.; Zehetmeier, M. Impact of Longevity on Greenhouse Gas Emissions and Profitability of Individual Dairy Cows Analysed with Different System Boundaries. Animal 2019, 13, 198–208. [Google Scholar] [CrossRef]
- Bruijnis, M.R.N.; Meijboom, F.L.B.; Stassen, E.N. Longevity as an Animal Welfare Issue Applied to the Case of Foot Disorders in Dairy Cattle. J. Agric. Environ. Ethics 2013, 26, 191–205. [Google Scholar] [CrossRef]
- Steeneveld, W.; Hogeveen, H. Characterization of Dutch Dairy Farms Using Sensor Systems for Cow Management. J. Dairy Sci. 2015, 98, 709–717. [Google Scholar] [CrossRef] [PubMed]
- Lora, I.; Gottardo, F.; Contiero, B.; Zidi, A.; Magrin, L.; Cassandro, M.; Cozzi, G. A Survey on Sensor Systems Used in Italian Dairy Farms and Comparison between Performances of Similar Herds Equipped or Not Equipped with Sensors. J. Dairy Sci. 2020, 103, 10264–10272. [Google Scholar] [CrossRef] [PubMed]
- Rutten, C.J.; Velthuis, A.G.J.; Steeneveld, W.; Hogeveen, H. Invited Review: Sensors to Support Health Management on Dairy Farms. J. Dairy Sci. 2013, 96, 1928–1952. [Google Scholar] [CrossRef] [PubMed]
- King, M.T.M.; DeVries, T.J. Graduate Student Literature Review: Detecting Health Disorders Using Data from Automatic Milking Systems and Associated Technologies. J. Dairy Sci. 2018, 101, 8605–8614. [Google Scholar] [CrossRef] [PubMed]
- Rizopoulos, D. Joint Models for Longitudinal and Time-to-Event Data: With Applications in R, 1st ed.; Chapman & Hall/CRC: Boca Raton, FL, USA, 2012. [Google Scholar]
- Steensels, M.; Antler, A.; Bahr, C.; Berckmans, D.; Maltz, E.; Halachmi, I. A Decision-Tree Model to Detect Post-Calving Diseases Based on Rumination, Activity, Milk Yield, BW and Voluntary Visits to the Milking Robot. Animal 2016, 10, 1493–1500. [Google Scholar] [CrossRef]
- Ouweltjes, W.; Spoelstra, M.; Ducro, B.; de Haas, Y.; Kamphuis, C. A Data-Driven Prediction of Lifetime Resilience of Dairy Cows Using Commercial Sensor Data Collected during First Lactation. J. Dairy Sci. 2021, 104, 11759–11769. [Google Scholar] [CrossRef]
- Græsbøll, K.; Kirkeby, C.; Nielsen, S.S.; Halasa, T.; Toft, N.; Christiansen, L.E. Models to Estimate Lactation Curves of Milk Yield and Somatic Cell Count in Dairy Cows at the Herd Level for the Use in Simulations and Predictive Models. Front. Vet. Sci. 2016, 3, 115. [Google Scholar] [CrossRef]
- Fitzmaurice, G.; Davidian, M.; Verbeke, G.; Molenberghs, G. Generalized Linear Mixed-Effects Models. In Longitudinal Data Analysis; Chapman & Hall/CRC: Boca Raton, FL, USA, 2008. [Google Scholar]
- Kalbfleisch, J.D.; Prentice, R.L. Relative Risk (Cox) Regression Models. In The Statistical Analysis of Failure Time Data; Wiley and Sons: New York, NY, USA, 2002; pp. 95–147. [Google Scholar]
- van Ravenzwaaij, D.; Cassey, P.; Brown, S.D. A Simple Introduction to Markov Chain Monte–Carlo Sampling. Psychon. Bull. Rev. 2018, 25, 143–154. [Google Scholar] [CrossRef]
- VandeHaar, M.J.; St-Pierre, N. Major Advances in Nutrition: Relevance to the Sustainability of the Dairy Industry. J. Dairy Sci. 2006, 89, 1280–1291. [Google Scholar] [CrossRef]
- Rizopoulos, D.; Papageorgiou, G.; Miranda Afonso, P. JMbayes2: Extended Joint Models for Longitudinal and Time-to-Event Data. Available online: https://cran.r-project.org/web/packages/JMbayes2/JMbayes2.pdf (accessed on 11 July 2022).
- Stone, M. Cross-Validatory Choice and Assessment of Statistical Predictions. J. R. Stat. Soc. Ser. B Methodol. 1974, 36, 111–147. [Google Scholar] [CrossRef]
- Heagerty, P.J.; Zheng, Y. Survival Model Predictive Accuracy and ROC Curves. Biometrics 2005, 61, 92–105. [Google Scholar] [CrossRef] [PubMed]
- Gerds, T.A.; Schumacher, M. Consistent Estimation of the Expected Brier Score in General Survival Models with Right-Censored Event Times. Biom. J. 2006, 48, 1029–1040. [Google Scholar] [CrossRef] [PubMed]
- Goldstein-Greenwood, J. A Brief on Brier Scores. Available online: https://data.library.virginia.edu/a-brief-on-brier-scores/ (accessed on 9 August 2022).
- Levene, H. Robust Tests for Equality of Variances. In Contributions to Probability and Statistics: Essays in Honor of Harold Hotelling; Stanford University Press: Redwood City, CA, USA, 1960; pp. 278–292. [Google Scholar]
- van der Heide, E.M.M.; Veerkamp, R.F.; van Pelt, M.L.; Kamphuis, C.; Athanasiadis, I.; Ducro, B.J. Comparing Regression, Naive Bayes, and Random Forest Methods in the Prediction of Individual Survival to Second Lactation in Holstein Cattle. J. Dairy Sci. 2019, 102, 9409–9421. [Google Scholar] [CrossRef] [PubMed]
- Olechnowicz, J.; Kneblewski, P.; Jaśkowski, J.M.; Włodarek, J. Effect of Selected Factors on Longevity in Cattle: A Review. J. Anim. Plant Sci. 2016, 26, 1533–1541. [Google Scholar]
- Poppe, M.; Veerkamp, R.F.; van Pelt, M.L.; Mulder, H.A. Exploration of Variance, Autocorrelation, and Skewness of Deviations from Lactation Curves as Resilience Indicators for Breeding. J. Dairy Sci. 2020, 103, 1667–1684. [Google Scholar] [CrossRef]
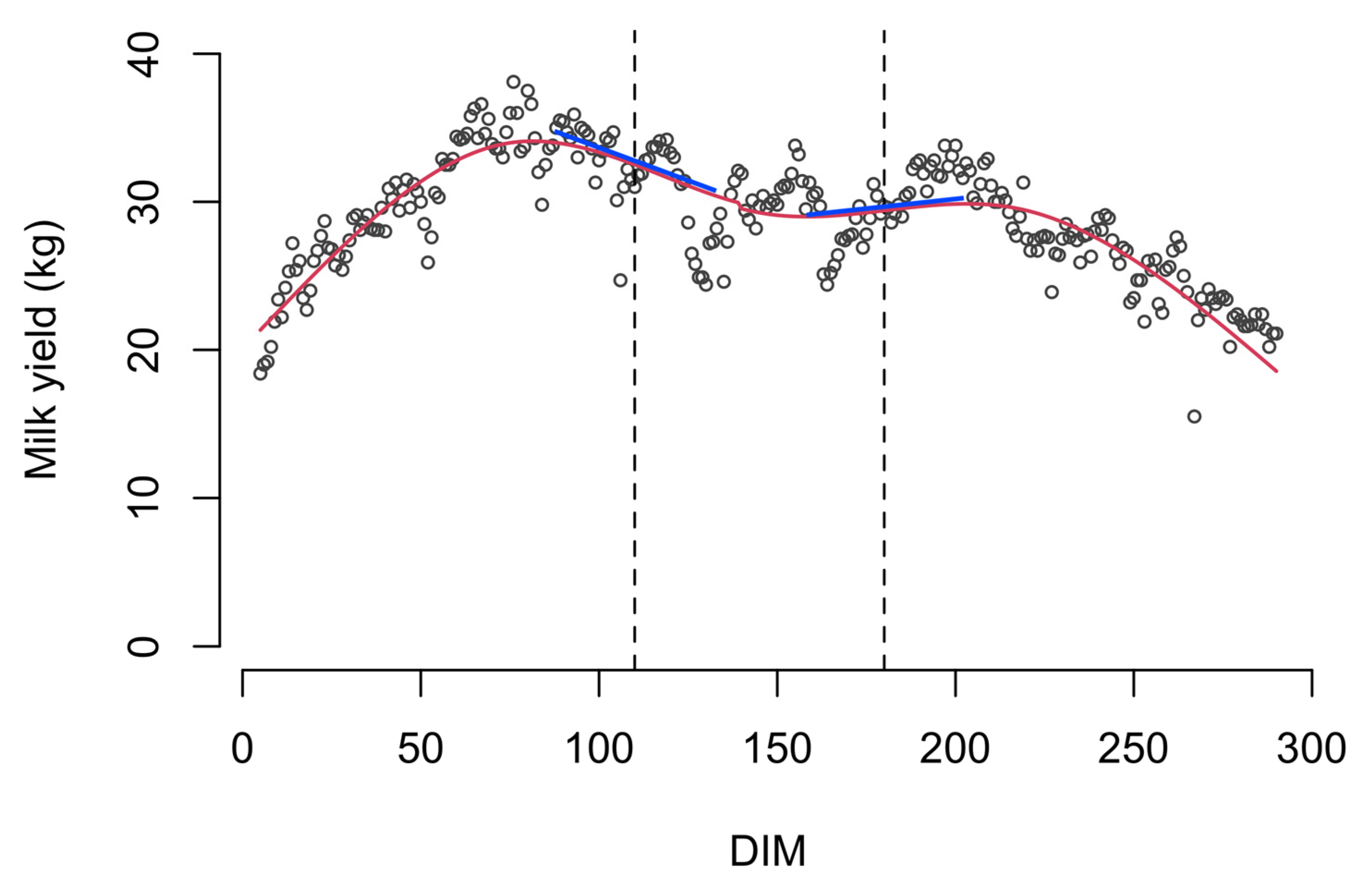
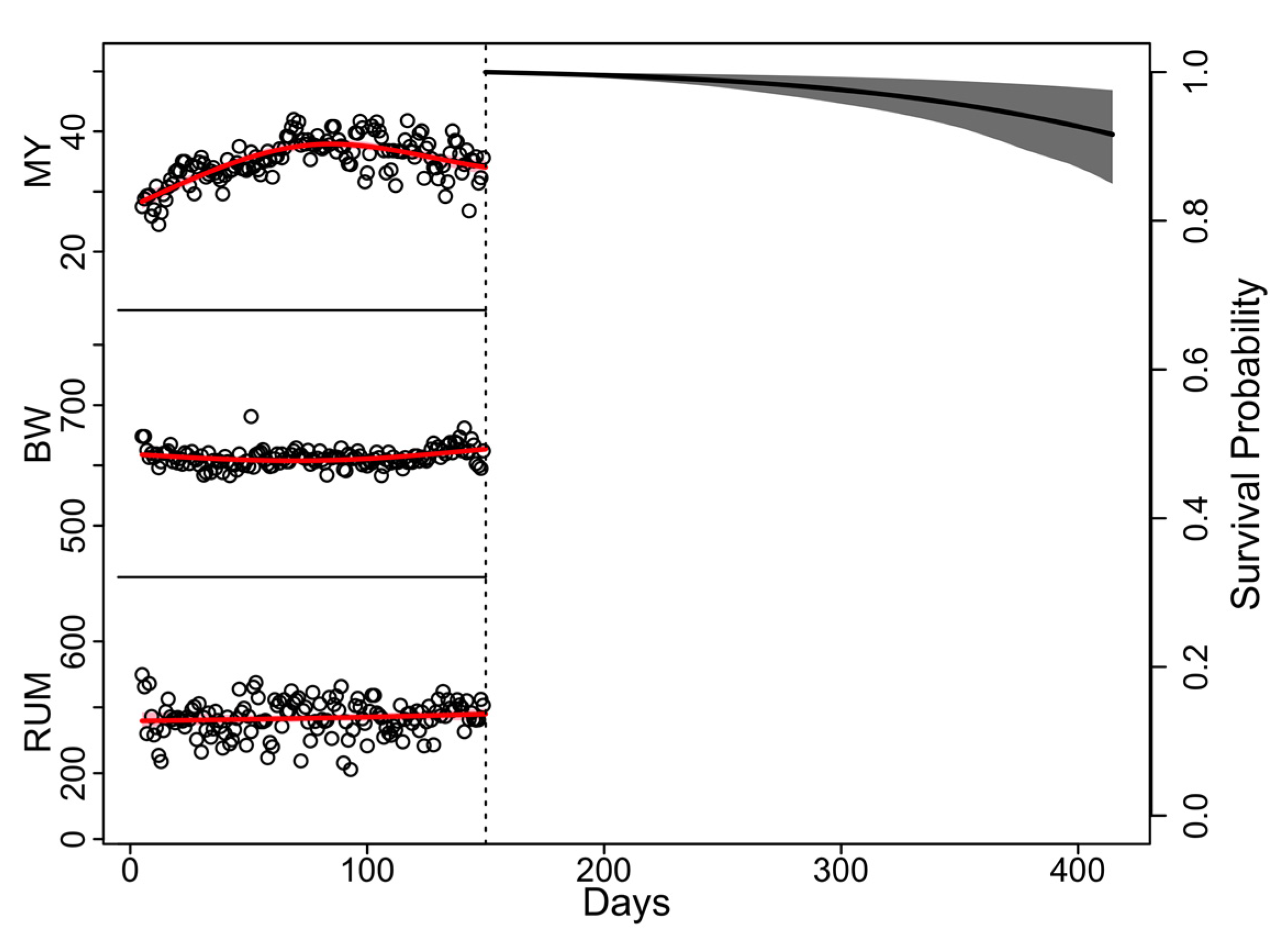
| Farm | Time Period | Cows (n) | 1 | 2 | ||
|---|---|---|---|---|---|---|
| Italian | 2014–2020 | 98 | 414 | 12% | 828 | 41% |
| Belgian 1 | 2014–2020 | 169 | 422 | 18% | 843 | 34% |
| Belgian 2 | 2013–2019 | 182 | 397 | 9% | 793 | 21% |
| British 1 | 2013–2019 | 266 | 384 | 9% | 768 | 24% |
| British 2 | 2013–2019 | 101 | 402 | 11% | 805 | 26% |
| British 3 | 2013–2019 | 226 | 400 | 6% | 799 | 17% |
| Farm | MY 1 | BW 2 | RUM 3 | |||
|---|---|---|---|---|---|---|
| Mean | SD | Mean | SD | Mean | SD | |
| Italian | 32.8 | 6.60 | 595 | 62.1 | 458 | 94.2 |
| Belgian 1 | 27.5 | 6.33 | 530 | 65.9 | 470 | 98.0 |
| Belgian 2 | 32.4 | 6.57 | 548 | 109 | 487 | 126 |
| British 1 | 31.9 | 8.01 | 634 | 65.1 | 491 | 102 |
| British 2 | 24.2 | 6.11 | 567 | 60.0 | 500 | 120 |
| British 3 | 33.9 | 6.84 | 578 | 59.0 | 484 | 125 |
| Survival Outcome | ||||||
| Parameters | Coeff. 1 | p2 | ||||
| (AFC 3 medium) | 0.997 | * | ||||
| (AFC high) | 1.35 | * | ||||
| (slope MY 4) | 0.005 | n.s. | ||||
| (slope BW 5) | 0.178 | n.s. | ||||
| (slope RUM 6) | −0.633 | ** | ||||
| Longitudinal Outcomes | ||||||
| Parameters | ||||||
| Fixed | coeff. | p | coeff. | p | coeff. | p |
| (intercept) | 28.0 | *** | 469 | *** | 472 | *** |
| ( 1) 7 | 6.42 | *** | 95.2 | *** | −18.6 | ** |
| ( 2) | 4.34 | *** | 68.9 | *** | 34.6 | *** |
| ( 3) | 13.8 | *** | 86.3 | *** | - | - |
| ( 4) | −13.5 | *** | 99.7 | *** | - | - |
| (AFC medium) | −0.268 | n.s. | −3.64 | n.s. | −21.3 | *** |
| (AFC high) | 1.24 | * | 50.9 | *** | −24.8 | *** |
| (SEAS 8 warm) | −0.021 | n.s. | 0.918 | n.s. | −9.03 | * |
| Random | SD 9 | SD | SD | |||
| (intercept) | 4.61 | 48.3 | 129 | |||
| ( 1) | 7.56 | 43.3 | 191 | |||
| ( 2) | 6.64 | 44.8 | 106 | |||
| ( 3) | 11.4 | 74.5 | - | |||
| ( 4) | 10.1 | 45.4 | - | |||
| DIM 1 | Farm | AUC 2 | PE 3 | ||
|---|---|---|---|---|---|
| 4 | 5 | ||||
| 60 | Italian | 0.558 | 0.505 | 0.098 † | 0.263 |
| Belgian 1 | 0.580 * | 0.497 | 0.091 † | 0.228 † | |
| Belgian 2 | - | 0.451 | - | 0.146 † | |
| British 1 | 0.526 | 0.519 | 0.061 † | 0.202 † | |
| British 2 | - | 0.498 | - | 0.196 † | |
| British 3 | 0.476 | 0.508 | 0.037 † | 0.164 † | |
| 150 | Italian | 0.605 | 0.556 | 0.096 † | 0.256 |
| Belgian 1 | 0.578 | 0.513 | 0.085 † | 0.225 † | |
| Belgian 2 | - | 0.475 | - | 0.143 † | |
| British 1 | 0.562 | 0.526 * | 0.060 † | 0.202 † | |
| British 2 | - | 0.520 | - | 0.194 † | |
| British 3 | 0.535 | 0.514 | 0.036 † | 0.164 † | |
| 240 | Italian | 0.616 | 0.566 * | 0.096 † | 0.259 |
| Belgian 1 | 0.597 * | 0.533 | 0.083 † | 0.229 | |
| Belgian 2 | - | 0.507 | - | 0.143 † | |
| British 1 | 0.577 | 0.539 | 0.060 † | 0.200 † | |
| British 2 | - | 0.593 * | - | 0.189 † | |
| British 3 | 0.763 * | 0.559 * | 0.035 † | 0.158 † | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ranzato, G.; Adriaens, I.; Lora, I.; Aernouts, B.; Statham, J.; Azzolina, D.; Meuwissen, D.; Prosepe, I.; Zidi, A.; Cozzi, G. Joint Models to Predict Dairy Cow Survival from Sensor Data Recorded during the First Lactation. Animals 2022, 12, 3494. https://doi.org/10.3390/ani12243494
Ranzato G, Adriaens I, Lora I, Aernouts B, Statham J, Azzolina D, Meuwissen D, Prosepe I, Zidi A, Cozzi G. Joint Models to Predict Dairy Cow Survival from Sensor Data Recorded during the First Lactation. Animals. 2022; 12(24):3494. https://doi.org/10.3390/ani12243494
Chicago/Turabian StyleRanzato, Giovanna, Ines Adriaens, Isabella Lora, Ben Aernouts, Jonathan Statham, Danila Azzolina, Dyan Meuwissen, Ilaria Prosepe, Ali Zidi, and Giulio Cozzi. 2022. "Joint Models to Predict Dairy Cow Survival from Sensor Data Recorded during the First Lactation" Animals 12, no. 24: 3494. https://doi.org/10.3390/ani12243494
APA StyleRanzato, G., Adriaens, I., Lora, I., Aernouts, B., Statham, J., Azzolina, D., Meuwissen, D., Prosepe, I., Zidi, A., & Cozzi, G. (2022). Joint Models to Predict Dairy Cow Survival from Sensor Data Recorded during the First Lactation. Animals, 12(24), 3494. https://doi.org/10.3390/ani12243494






