Extensive Adaptive Variation in Gene Expression within and between Closely Related Horseshoe Bats (Chiroptera, Rhinolophus) Revealed by Three Organs
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection and Data Acquisition
2.2. Expression Variation between Taxa and among Individuals within Taxon
2.3. Identifying Expression Variation Forced by Natural Selection, Especially Related to Phenotypic Divergence
2.4. Functional Enrichment Analysis
3. Results
3.1. Differences in Gene Expression within and between Taxa
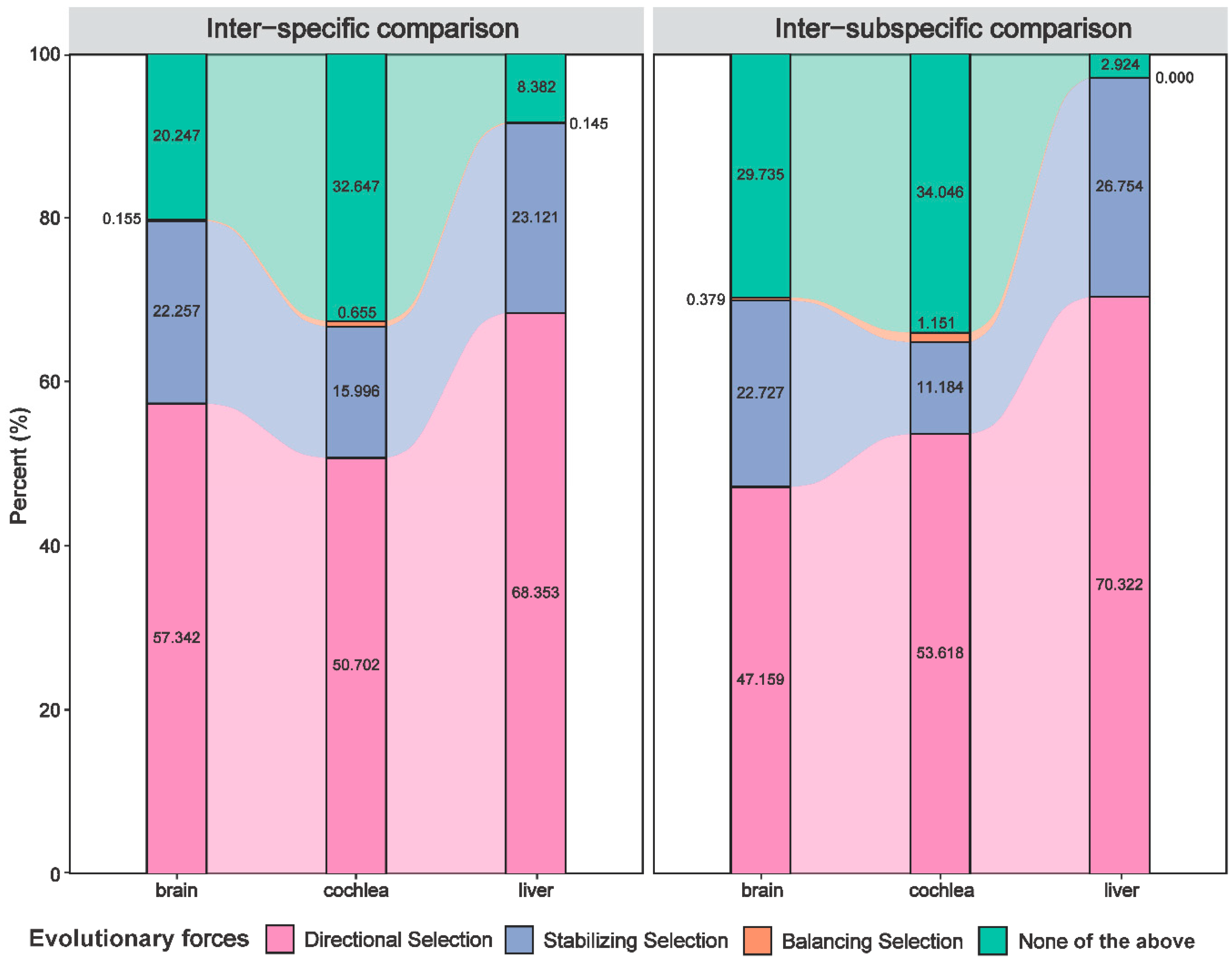
3.2. Expression Variation Extensively Governed by Natural Selection
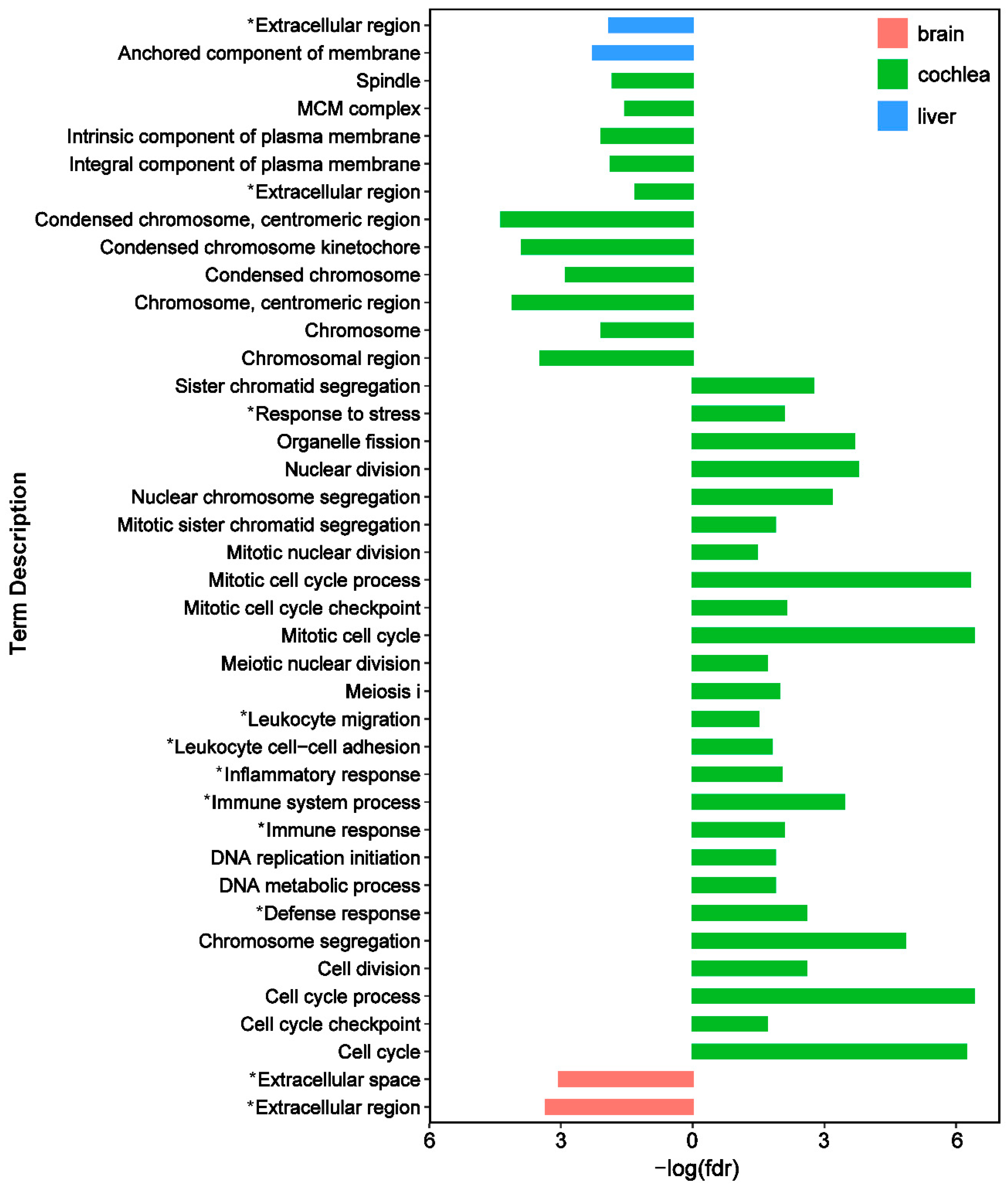
3.3. Functional Patterns of Adaptive Expression Variation
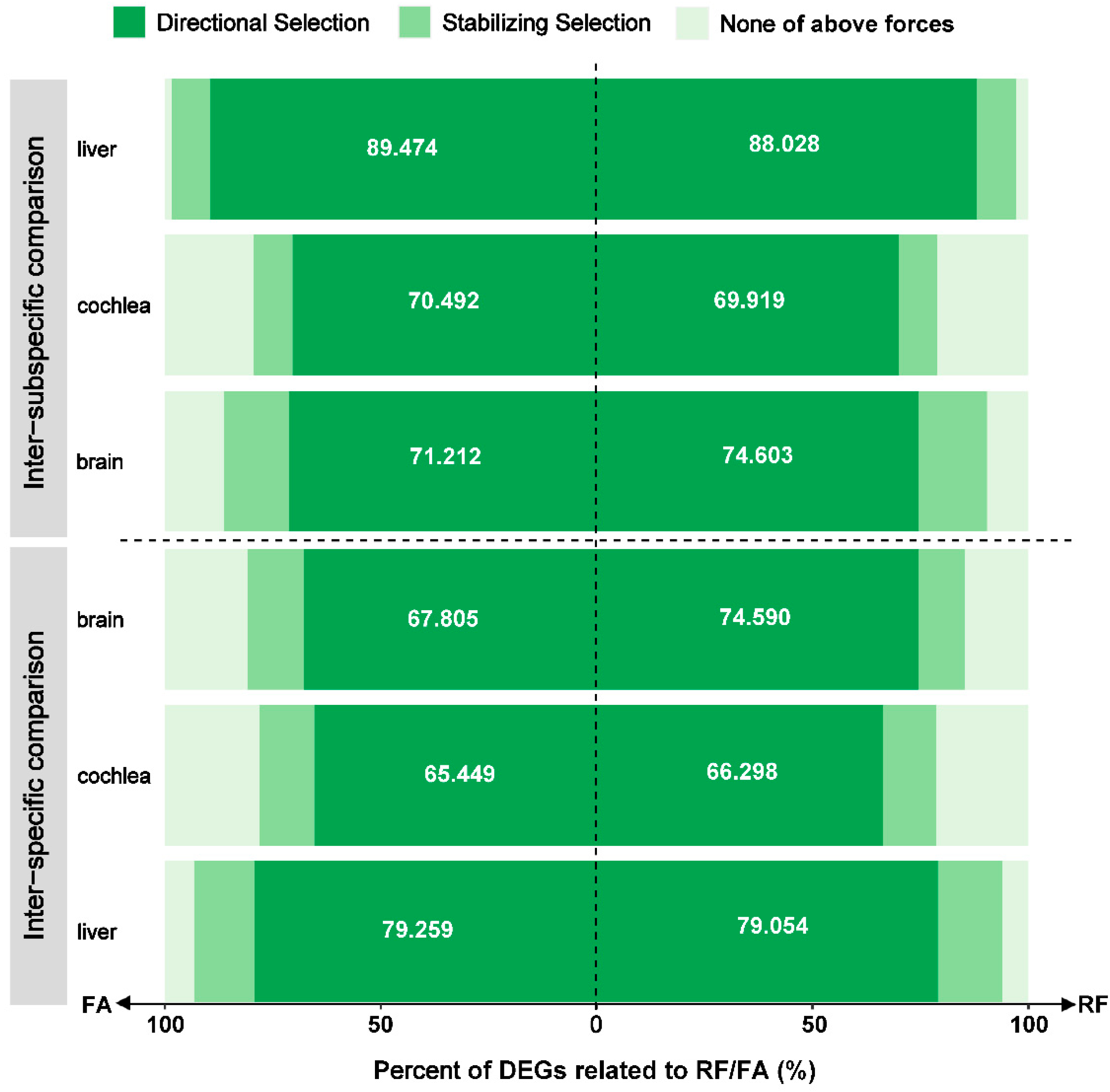
3.4. Adaptive Expression Variation Related to Phenotypes
4. Discussion
4.1. Extensive Adaptive Variation in Inter-Specific and Inter-Subspecific Gene Expression
4.2. Organ-Specificity of Expression Variation Governed by Natural Selection
4.3. Functional Patterns of Adaptive Expression Variation at Two Different Classification Categories
4.4. Trait-Related Expression Variation Forced by Natural Selection
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fushan, A.A.; Turanov, A.A.; Lee, S.G.; Kim, E.B.; Lobanov, A.V.; Yim, S.H.; Buffenstein, R.; Lee, S.R.; Chang, K.T.; Rhee, H.; et al. Gene expression defines natural changes in mammalian lifespan. Aging Cell 2015, 14, 352–365. [Google Scholar] [CrossRef] [PubMed]
- Jarosaw, B.; Mehmet, S.; Anna, L.; Meike, T. Early gene expression divergence between allopatric populations of the house mouse (Mus musculus domesticus). Ecol. Evol. 2013, 3, 558–568. [Google Scholar]
- Pavey, S.A.; Collin, H.; Nosil, P.; Rogers, S.M. The role of gene expression in ecological speciation. Ann. N. Y. Acad. Sci. 2010, 1206, 110–129. [Google Scholar] [CrossRef]
- Wolf, J.B.; Bayer, T.; Haubold, B.; Schilhabel, M.; Rosenstiel, P.; Tautz, D. Nucleotide divergence vs. gene expression differentiation: Comparative transcriptome sequencing in natural isolates from the carrion crow and its hybrid zone with the hooded crow. Mol. Ecol. 2010, 19 (Suppl. 1), 162–175. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Liu, R.; Huang, L.; Zheng, X.M.; Liu, P.L.; Du, Y.S.; Cai, Z.; Zhou, L.; Wei, X.H.; Zhang, F.M.; et al. Widespread and adaptive alterations in genome-wide gene expression associated with ecological divergence of two Oryza species. Mol. Biol. Evol. 2016, 33, 62–78. [Google Scholar] [CrossRef]
- Gilad, Y.; Oshlack, A.; Rifkin, S.A. Natural selection on gene expression. Trends Genet. 2006, 22, 456–461. [Google Scholar] [CrossRef] [PubMed]
- Khaitovich, P.; Weiss, G.; Lachmann, M.; Hellmann, I.; Enard, W.; Muetzel, B.; Wirkner, U.; Ansorge, W.; Paabo, S. A neutral model of transcriptome evolution. PLoS Biol. 2004, 2, E132. [Google Scholar] [CrossRef]
- Uebbing, S.; Kunstner, A.; Makinen, H.; Backstrom, N.; Bolivar, P.; Burri, R.; Dutoit, L.; Mugal, C.F.; Nater, A.; Aken, B.; et al. Divergence in gene expression within and between two closely related flycatcher species. Mol. Ecol. 2016, 25, 2015–2028. [Google Scholar] [CrossRef]
- Fraser, H.B.; Moses, A.M.; Schadt, E.E. Evidence for widespread adaptive evolution of gene expression in budding yeast. Proc. Natl. Acad. Sci. USA 2010, 107, 2977–2982. [Google Scholar] [CrossRef]
- Yang, J.-R.; Maclean, C.; Park, C.; Zhao, H.; Zhang, J. Intra- and inter-specific variations of gene expression levels in yeast are largely neutral. Mol. Biol. Evol. 2016, 34, 2125–2139. [Google Scholar] [CrossRef]
- Khaitovich, P.; Enard, W.; Lachmann, M.; Paabo, S. Evolution of primate gene expression. Nat. Rev. Genet. 2006, 7, 693–702. [Google Scholar] [CrossRef] [PubMed]
- Whitehead, A.; Crawford, D.L. Neutral and adaptive variation in gene expression. Proc. Natl. Acad. Sci. USA 2006, 103, 5425–5430. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Gao, B.; Wang, H.; Han, N.; Shao, S.; Wu, S.; Song, G.; Zhang, Y.E.; Zhu, X.; Lu, X.; et al. Evolution of beak morphology in the Ground Tit revealed by comparative transcriptomics. Front. Zool. 2017, 14, 58. [Google Scholar] [CrossRef]
- Lencer, E.S.; Warren, W.C.; Harrison, R.; McCune, A.R. The Cyprinodon variegatus genome reveals gene expression changes underlying differences in skull morphology among closely related species. BMC Genom. 2017, 18, 424. [Google Scholar] [CrossRef] [PubMed]
- Haas, F.; Brodin, A. The crow Corvus corone hybrid zone in southern Denmark and northern Germany. Ibis 2005, 147, 649–656. [Google Scholar] [CrossRef]
- Haas, F.; Pointer, M.A.; Saino, N.; Brodin, A.; Mundy, N.I.; Hansson, B. An analysis of population genetic differentiation and genotype–phenotype association across the hybrid zone of carrion and hooded crows using microsatellites and MC1R. Mol. Ecol. 2009, 18, 294–305. [Google Scholar] [CrossRef]
- Zhao, H.B.; Wang, H.; Liu, T.; Liu, S.; Jin, L.R.; Huang, X.B.; Dai, W.T.; Sun, K.P.; Feng, J. Gene expression vs. sequence divergence: Comparative transcriptome sequencing among natural Rhinolophus ferrumequinum populations with different acoustic phenotypes. Front. Zool. 2019, 16, 37. [Google Scholar] [CrossRef]
- He, T.; Lamont, B.B.; Enright, N.J.; D’Agui, H.M.; Stock, W. Environmental drivers and genomic architecture of trait differentiation in fire-adapted Banksia attenuata ecotypes. J. Integr. Plant. Biol. 2019, 61, 417–432. [Google Scholar] [CrossRef]
- Liu, T.; Sun, K.P.; Csorba, G.; Zhang, K.K.; Zhang, L.; Zhao, H.B.; Jin, L.R.; Thong, V.D.; Xiao, Y.H.; Feng, J. Species delimitation and evolutionary reconstruction within an integrative taxonomic framework: A case study on Rhinolophus macrotis complex (Chiroptera: Rhinolophidae). Mol. Phylogen. Evol. 2019, 139, 106544. [Google Scholar] [CrossRef]
- Li, J.; Sun, K.P.; Dai, W.T.; Leng, H.X.; Feng, J. Divergence in interspecific and intersubspecific gene expression between two closely related horseshoe bats (Rhinolophus). J. Mammal. 2022, 363, 2813–2820. [Google Scholar] [CrossRef]
- Sun, K.; Kimball, R.T.; Liu, T.; Wei, X.; Jin, L.; Jiang, T.; Lin, A.; Feng, J. The complex evolutionary history of big-eared horseshoe bats (Rhinolophus macrotis complex): Insights from genetic, morphological and acoustic data. Sci. Rep. 2016, 6, 35417. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Zhang, K.K.; Dai, W.T.; Jin, L.R.; Sun, K.P.; Feng, J. Evolutionary insights into Rhinolophus episcopus (Chiroptera, Rhinolophidae) in China: Isolation by distance, environment, or sensory system? J. Zool. Syst. Evol. Res. 2020, 59, 249–310. [Google Scholar]
- Puechmaille, S.J.; Gouilh, M.A.; Piyapan, P.; Yokubol, M.; Mie, K.M.; Bates, P.J.; Satasook, C.; Nwe, T.; Bu, S.S.; Mackie, I.J.; et al. The evolution of sensory divergence in the context of limited gene flow in the bumblebee bat. Nat. Commun. 2011, 2, 573. [Google Scholar] [CrossRef] [PubMed]
- Davies, K.T.; Maryanto, I.; Rossiter, S.J. Evolutionary origins of ultrasonic hearing and laryngeal echolocation in bats inferred from morphological analyses of the inner ear. Front. Zool. 2013, 10, 2. [Google Scholar] [CrossRef] [PubMed]
- Vannatta, J.M.; Carver, B.D. Sexual size dimorphism and geographic variation in forearm length of Rafinesque’s Big-eared Bat (Corynorhinus rafinesquii) and Southeastern Myotis (Myotis austroriparius). Mammalia 2021, 86, 280–286. [Google Scholar] [CrossRef]
- Luo, B.; Leiser-Miller, L.; Santana, S.E.; Zhang, L.; Liu, T.; Xiao, Y.H.; Liu, Y.; Feng, J. Echolocation call divergence in bats: A. comparative analysis. Behav. Ecol. Sociobiol. 2019, 73, 154. [Google Scholar] [CrossRef]
- Sun, K.P.; Luo, L.; Kimball, R.T.; Wei, X.W.; Jin, L.R.; Jiang, T.L.; Feng, J. Geographic variation in the acoustic traits of greater horseshoe bats: Testing the importance of drift and ecological selection in evolutionary processes. PLoS ONE 2013, 8, e70368. [Google Scholar] [CrossRef]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; van Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef]
- Hart, T.; Komori, H.K.; LaMere, S.; Podshivalova, K.; Salomon, D.R. Finding the active genes in deep RNA-seq gene expression studies. BMC Genom. 2013, 14, 778. [Google Scholar] [CrossRef]
- Blekhman, R.; Oshlack, A.; Chabot, A.E.; Smyth, G.K.; Gilad, Y. Gene regulation in primates evolves under tissue-specific selection pressures. PLoS Genet. 2008, 4, e1000271. [Google Scholar] [CrossRef]
- Chapman, M.G.; Underwood, A.J. Ecological patterns in multivariate assemblages: Information and interpretation of negative values in ANOSIM tests. Mar. Ecol. Prog. Ser. 1999, 180, 257–265. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Alexander, J.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, 607–613. [Google Scholar] [CrossRef] [PubMed]
- Enard, W.; Khaitovich, P.; Klose, J.; Zollner, S.; Heissig, F.; Giavalisco, P.; Nieselt-Struwe, K.; Muchmore, E.; Varki, A.; Ravid, R.; et al. Intra- and interspecific variation in primate gene expression patterns. Science 2002, 296, 340–343. [Google Scholar] [CrossRef]
- Fraser, H.B. Genome-wide approaches to the study of adaptive gene expression evolution: Systematic studies of evolutionary adaptations involving gene expression will allow many fundamental questions in evolutionary biology to be addressed. Bioessays 2011, 33, 469–477. [Google Scholar] [CrossRef]
- Brawand, D.; Soumillon, M.; Necsulea, A.; Julien, P.; Csardi, G.; Harrigan, P.; Weier, M.; Liechti, A.; Aximu-Petri, A.; Kircher, M.; et al. The evolution of gene expression levels in mammalian organs. Nature 2011, 478, 343–348. [Google Scholar] [CrossRef]
- Chan, E.T.; Quon, G.T.; Chua, G.; Babak, T.; Trochesset, M.; Zirngibl, R.A.; Aubin, J.; Ratcliffe, M.J.; Wilde, A.; Brudno, M.; et al. Conservation of core gene expression in vertebrate tissues. J. Biol. 2009, 8, 33. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Zhu, T.; Xue, H.; Fang, N.; Zhang, J.; Zhang, L.; Pang, J.; Teeling, E.C.; Zhang, S. Prenatal development supports a single origin of laryngeal echolocation in bats. Nat. Ecol. Evol. 2017, 1, 21. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Xie, S.B.; Wu, C.H.; Hu, Y.; Zhang, Q.; Li, S.; Fan, Y.G.; Leng, R.X.; Pan, H.F.; Xiong, H.B.; et al. Coagulation cascade and complement system in systemic lupus erythematosus. Oncotarget 2018, 9, 14862–14881. [Google Scholar] [CrossRef]
- Dave, D.D.; Jha, B.K. Analytically depicting the calcium diffusion for Alzheimer’s affected cell. Int. J. Biomath. 2018, 11, 13. [Google Scholar] [CrossRef]
- DeCoster, M.A. Calcium dynamics in the central nervous system. Adv. Neuroimmunol. 1995, 5, 233–239. [Google Scholar] [CrossRef]
- Burdette, S.C.; Lippard, S.J. Meeting of the minds: Metalloneurochemistry. Proc. Natl. Acad. Sci. USA 2003, 100, 3605–3610. [Google Scholar] [CrossRef] [PubMed]
- Marambaud, P.; Dreses-Werringloer, U.; Vingtdeux, V. Calcium signaling in neurodegeneration. Mol. Neurodegener. 2009, 4, 20. [Google Scholar] [CrossRef] [PubMed]
- Hrynkow, S.H.; Morest, D.K.; Brumwell, C.; Rutishauser, U. Spatio-temporal diversity in the microenvironments for neural cell adhesion molecule, neural cell adhesion molecule polysialic acid, and L1-cell adhesion molecule expression by sensory neurons and their targets during cochleo-vestibular innervation. Neuroscience 1998, 87, 401–422. [Google Scholar] [CrossRef] [PubMed]
- Ricard-Blum, S.; Perez, S. Glycosaminoglycan interaction networks and databases. Curr. Opin. Struct. Biol. 2022, 74, 102355. [Google Scholar] [CrossRef] [PubMed]
- Blaner, W.S.; Brun, P.J.; Calderon, R.M.; Golczak, M. Retinol-binding protein 2 (RBP2): Biology and pathobiology. Crit. Rev. Biochem. Mol. Biol. 2020, 55, 197–218. [Google Scholar] [CrossRef] [PubMed]
- Kuroiwa, S.; Kikuchi, T.; Yoshimura, N. A novel compound heterozygous mutation in the RDH5 gene in a patient with fundus albipunctatus. Am. J. Ophthalmol. 2000, 130, 672–675. [Google Scholar] [CrossRef]
- Zhao, H.; Rossiter, S.J.; Teeling, E.C.; Li, C.; Cotton, J.A.; Zhang, S. The evolution of color vision in nocturnal mammals. Proc. Natl. Acad. Sci. USA 2009, 106, 8980–8985. [Google Scholar] [CrossRef]
- Galvan, I.; Vargas-Mena, J.C.; Herrera, B.R. Tent-roosting may have driven the evolution of yellow skin coloration in Stenodermatinae bats. J. Zool. Syst. Evol. Res. 2020, 58, 519–527. [Google Scholar] [CrossRef]
- Simões, B.F.; Foley, N.M.; Hughes, G.M.; Zhao, H.; Shuyi, Z.; Teeling, E.C. As blind as a bat? Opsin phylogenetics illuminates the evolution of color vision in bats. Mol. Biol. Evol. 2018, 36, 54–68. [Google Scholar] [CrossRef]
- Danilovich, S.; Yovel, Y. Integrating vision and echolocation for navigation and perception in bats. Sci. Adv. 2019, 5, eaaw6503. [Google Scholar] [CrossRef]
- Qin, H.; Hu, C.; Zhao, X.; Tian, M.; Zhu, B. Usefulness of candidate mRNAs and miRNAs as biomarkers for mild cognitive impairment and Alzheimer’s disease. Int. J. Neurosci. 2021, 10, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Lin, P.B.C.; Tsai, A.P.Y.; Nho, K.; Lamb, B.T.; Oblak, A.L. INPP5D regulates the amyloid pathology in Alzheimer’s disease. Alzheimers Dement. 2021, 17, e058724. [Google Scholar] [CrossRef]
- Wang, H.; Zhao, H.B.; Sun, K.P.; Huang, X.B.; Feng, J. Evolutionary basis of high-frequency hearing in the cochleae of echolocators revealed by comparative genomics. Genome Biol. Evol. 2019, 12, 3740–3753. [Google Scholar] [CrossRef] [PubMed]
- Delmonte, O.M.; Bergerson, J.R.E.; Kawai, T.; Kuehn, H.S.; McDermott, D.H.; Cortese, I.; Zimmermann, M.T.; Dobbs, A.K.; Bosticardo, M.; Fink, D.; et al. SASH3 variants cause a novel form of X-linked combined immunodeficiency with immune dysregulation. Blood 2021, 138, 1019–1033. [Google Scholar] [CrossRef] [PubMed]
- Udayanga, K.G.; Nakamura, Y.; Nakahashi-Oda, C.; Shibuya, A. Immunoreceptor CD300A on mast cells and dendritic cells regulates neutrophil recruitment in a murine model of sepsis. Int. Immunol. 2016, 28, 611–615. [Google Scholar] [CrossRef] [PubMed]
- Chien, Y.H.; Hwu, W.L.; Ariga, T. Gene symbol: WAS. Disease: Wiskott-Aldrich syndrome. Hum. Genet. 2004, 115, 532. [Google Scholar] [PubMed]
- Castro, C.N.; Rosenzwajg, M.; Carapito, R.; Shahrooei, M.; Konantz, M.; Khan, A.; Miao, Z.; Gross, M.; Tranchant, T.; Radosavljevic, M.; et al. NCKAP1L defects lead to a novel syndrome combining immunodeficiency, lymphoproliferation, and hyperinflammation. J. Exp. Med. 2020, 217, e20192275. [Google Scholar] [CrossRef]
- Skokowa, J.; Klimiankou, M.; Klimenkova, O.; Lan, D.; Gupta, K.; Hussein, K.; Carrizosa, E.; Kusnetsova, I.; Li, Z.; Sustmann, C.; et al. Interactions among HCLS1, HAX1 and LEF-1 proteins are essential for G-CSF-triggered granulopoiesis. Nat. Med. 2012, 18, 1550–1559. [Google Scholar] [CrossRef]
- Achieng, A.O.; Guyah, B.; Cheng, Q.; Ong’echa, J.M.; Ouma, C.; Lambert, C.G.; Perkins, D.J. Molecular basis of reduced LAIR1 expression in childhood severe malarial anaemia: Implications for leukocyte inhibitory signalling. EBioMedicine 2019, 45, 278–289. [Google Scholar] [CrossRef]
- He, L.; Wang, B.; Li, Y.; Zhu, L.; Li, P.; Zou, F.; Bin, L. The solute carrier transporter SLC15A3 participates in antiviral innate immune responses against Herpes Simplex Virus-1. J. Immunol. Res. 2018, 2018, 5214187. [Google Scholar] [CrossRef] [PubMed]
- Cortes-Hernandez, J.; Fossati-Jimack, L.; Petry, F.; Loos, M.; Izui, S.; Walport, M.J.; Cook, H.T.; Botto, M. Restoration of C1q levels by bone marrow transplantation attenuates autoimmune disease associated with C1q deficiency in mice. Eur. J. Immunol. 2004, 34, 3713–3722. [Google Scholar] [CrossRef] [PubMed]
- Leman, J.C.; Weddle, C.B.; Gershman, S.N.; Kerr, A.M.; Ower, G.D.; St John, J.M.; Vogel, L.A.; Sakaluk, S.K. Lovesick: Immunological costs of mating to male sagebrush crickets. J. Evol. Biol. 2009, 22, 163–171. [Google Scholar] [CrossRef] [PubMed]
- Matthews, L.P.; Parks, S.E. An overview of North Atlantic right whale acoustic behavior, hearing capabilities, and responses to sound. Mar. Pollut. Bull. 2021, 173, 113043. [Google Scholar] [CrossRef] [PubMed]
- Shibkov, A.A.; Movchan, V.N.; Sobolevskii, S.A. Acoustic signaling of the water shrew, Neomys fodiens (Insectivora, Soricidae), in conflict interruptions. Zool. Zhurnal 2001, 80, 454–458. [Google Scholar]
- Goutte, S.; Dubois, A.; Howard, S.D.; Marquez, R.; Rowley, J.J.L.; Dehling, J.M.; Grandcolas, P.; Xiong, R.C.; Legendre, F. How the environment shapes animal signals: A test of the acoustic adaptation hypothesis in frogs. J. Evol. Biol. 2018, 31, 148–158. [Google Scholar] [CrossRef] [PubMed]
- Hughes, A.C.; Satasook, C.; Bates, P.J.J.; Soisook, P.; Sritongchuay, T.; Jones, G.; Bumrungsri, S. Echolocation call analysis and presence-only modelling as conservation monitoring tools for rhinolophoid bats in Thailand. Acta Chiropt. 2010, 12, 311–327. [Google Scholar] [CrossRef]
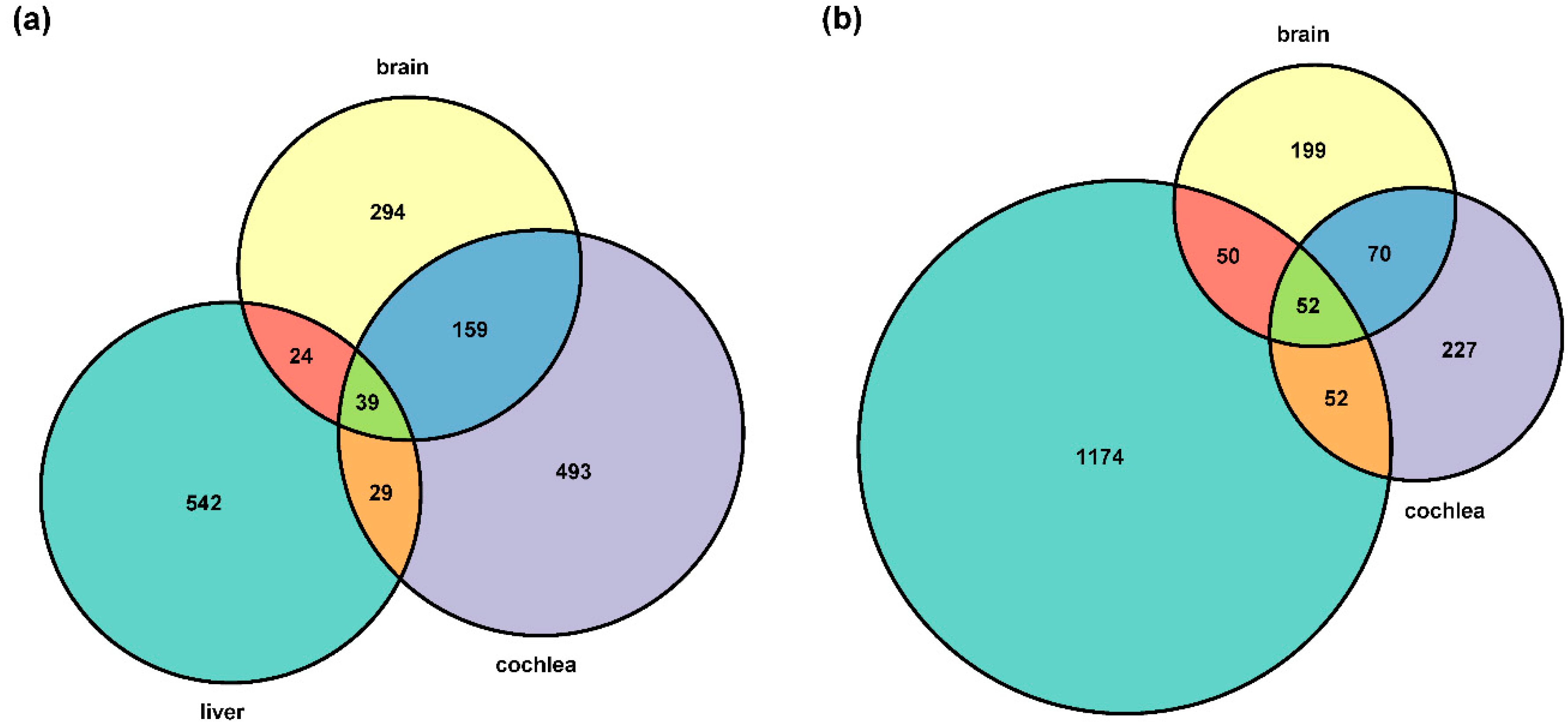
| Comparison | Organ | Number of DEGs | |||
|---|---|---|---|---|---|
| Directional Selection | Stabilizing Selection | Balancing Selection | In Total | ||
| Inter-specific comparison | brain | 371 | 144 | 1 | 516 |
| cochlea | 542 | 171 | 7 | 720 | |
| liver | 473 | 160 | 1 | 634 | |
| Inter-subspecific comparison | brain | 249 | 120 | 2 | 371 |
| cochlea | 326 | 68 | 7 | 401 | |
| liver | 962 | 366 | 0 | 1328 | |
| Comparison | Organ | Number of DEGs Related to RF/FA | |||
|---|---|---|---|---|---|
| Directional Selection | Stabilizing Selection | Balancing Selection | In Total | ||
| Inter-specific comparison | brain | 91/139 | 13/27 | 0/0 | 104/166 |
| cochlea | 240/233 | 45/45 | 0/0 | 285/278 | |
| liver | 117/107 | 22/19 | 0/0 | 139/126 | |
| Inter-subspecific comparison | brain | 47/47 | 10/10 | 0/0 | 57/57 |
| cochlea | 86/86 | 11/11 | 0/0 | 97/97 | |
| liver | 125/170 | 13/17 | 0/0 | 138/187 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, J.; Sun, K.; Dai, W.; Leng, H.; Li, A.; Feng, J. Extensive Adaptive Variation in Gene Expression within and between Closely Related Horseshoe Bats (Chiroptera, Rhinolophus) Revealed by Three Organs. Animals 2022, 12, 3432. https://doi.org/10.3390/ani12233432
Li J, Sun K, Dai W, Leng H, Li A, Feng J. Extensive Adaptive Variation in Gene Expression within and between Closely Related Horseshoe Bats (Chiroptera, Rhinolophus) Revealed by Three Organs. Animals. 2022; 12(23):3432. https://doi.org/10.3390/ani12233432
Chicago/Turabian StyleLi, Jun, Keping Sun, Wentao Dai, Haixia Leng, Aoqiang Li, and Jiang Feng. 2022. "Extensive Adaptive Variation in Gene Expression within and between Closely Related Horseshoe Bats (Chiroptera, Rhinolophus) Revealed by Three Organs" Animals 12, no. 23: 3432. https://doi.org/10.3390/ani12233432
APA StyleLi, J., Sun, K., Dai, W., Leng, H., Li, A., & Feng, J. (2022). Extensive Adaptive Variation in Gene Expression within and between Closely Related Horseshoe Bats (Chiroptera, Rhinolophus) Revealed by Three Organs. Animals, 12(23), 3432. https://doi.org/10.3390/ani12233432






