Assessing Molecular Diversity in Native and Introduced Populations of Red Wood Ant Formica paralugubris
Simple Summary
Abstract
1. Introduction
2. Materials and Method
2.1. Sampling Site
2.2. AFLP Fingerprinting
2.3. AFLP Analysis
2.3.1. Genetic Diversity
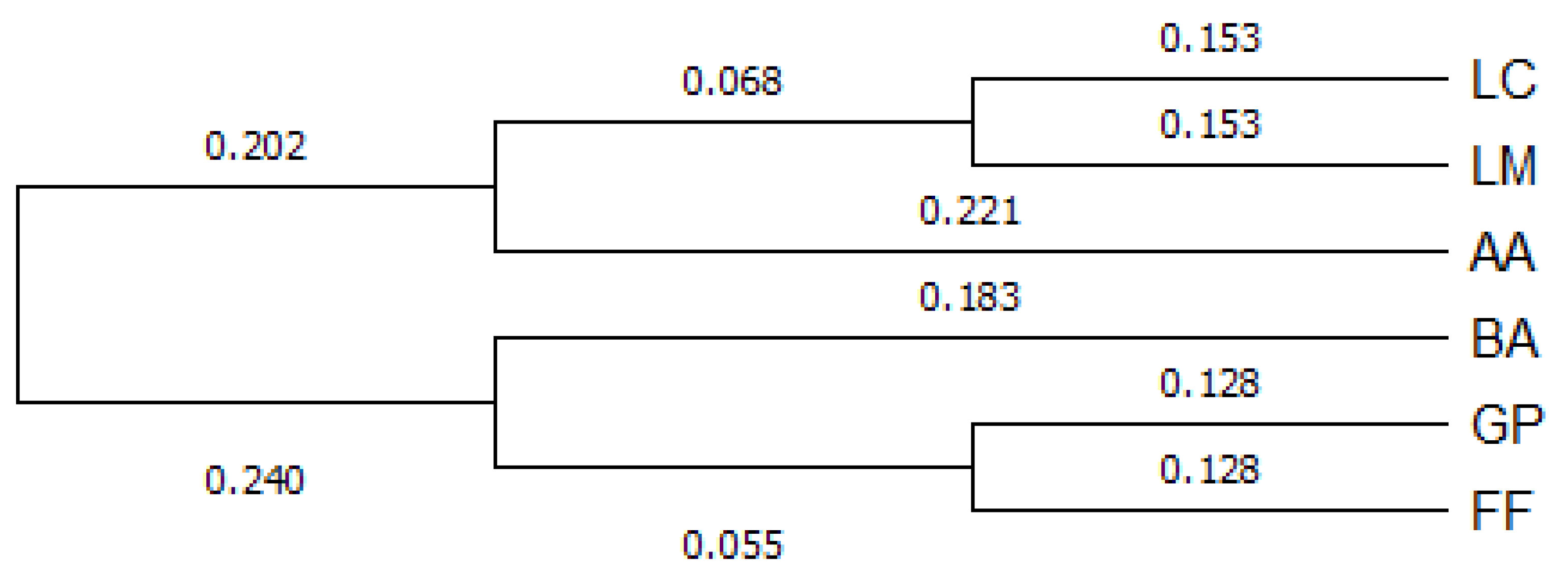
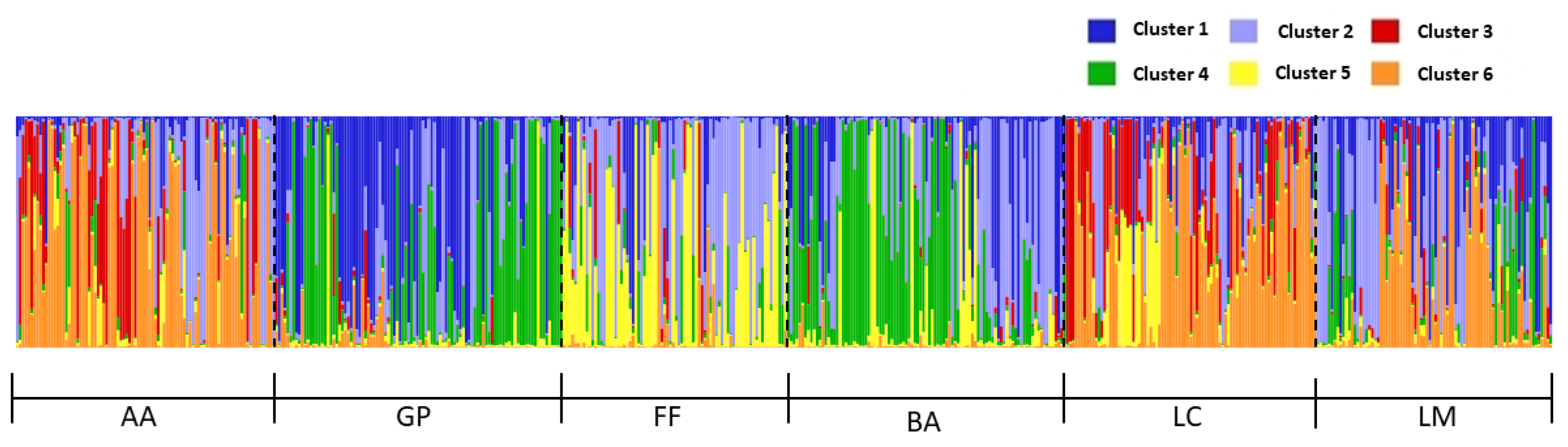
2.3.2. Genetic Distance and Structure
3. Results
4. Discussion and Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Stockan, J.; Robinson, E.; Trager, J.; Yao, I.; Seifert, B. Introducing wood ants: Evolution, phylogeny, identification and distribution. In Wood Ant Ecology and Conservation (Ecology, Biodiversity and Conservation), 1st ed.; Stockan, J., Robinson, E., Eds.; Cambridge University Press: Cambridge, UK, 2016; pp. 1–36. [Google Scholar] [CrossRef]
- Risch, A.C.; Ellis, S.; Wiswell, H. Where and why? Wood ant population ecology. In Wood Ant Ecology and Conservation (Ecology, Biodiversity and Conservation), 1st ed.; Stockan, J., Robinson, E., Eds.; Cambridge University Press: Cambridge, UK, 2016; pp. 81–105. [Google Scholar] [CrossRef]
- Punttila, P.; Niemelä, P.; Karhu, K. The impact of wood ants (Hymenoptera: Formicidae) on the structure of invertebrate community on mountain birch (Betula pubescens ssp. czerepanovii). Ann. Zool. Fenn. 2004, 41, 429–446. [Google Scholar]
- Ohashi, M.; Kilpeläinen, J.; Finér, L.; Risch, A.C.; Domisch, T.; Neuvonen, S.; Niemelä, P. The effect of red wood ant (Formica rufa group) mounds on root biomass, density, and nutrient concentrations in boreal managed forests. Eur. J. For. Res. 2007, 12, 113–119. [Google Scholar] [CrossRef]
- Di Nuzzo, L.; Masoni, A.; Frizzi, F.; Bianchi, E.; Castellani, M.B.; Balzani, P.; Benesperi, R. Red wood ants shape epiphytic lichen assemblages in montane silver fir forests. iForest 2022, 15, 71–76. [Google Scholar] [CrossRef]
- Frouz, J.; Jílková, V.; Sorvari, J. Contribution of wood ants to nutrient cycling and ecosystem function. In Wood Ant Ecology and Conservation (Ecology, Biodiversity and Conservation), 1st ed.; Stockan, J., Robinson, E., Eds.; Cambridge University Press: Cambridge, UK, 2016; pp. 81–105. [Google Scholar] [CrossRef]
- Balzani, P.; Masoni, A.; Venturi, S.; Frizzi, F.; Bambi, M.; Fani, R.; Santini, G. CO2 biogeochemical investigation and microbial characterization of red wood ant mounds in a Southern Europe montane forest. Soil Biol. Biochem. 2022, 166, 108536. [Google Scholar] [CrossRef]
- Mabelis, A.A.; Korczyńska, J. Long-term impact of agriculture on the survival of wood ants of the Formica rufa group (Formicidae). J. Insect Conserv. 2016, 20, 621–628. [Google Scholar] [CrossRef]
- Balzani, P.; Dekoninck, W.; Feldhaar, H.; Freitag, A.; Frizzi, F.; Frouz, J.; Santini, G. Challenges and a call to action for protecting European red wood ants. Cons. Biol. 2022, e13959. [Google Scholar] [CrossRef]
- Gosswald, K. Die Rote Waldameise im Dienste der Waldhygiene: Forstwirtschaftliche Bedeutung, Nutzung, Lebensweise, Zucht, Wermehrung und Schutt; Metta Kinau Verlag: Lüneburg, Germany, 1951; p. 160. [Google Scholar]
- Ronchetti, G.; Mazzoldi, P.; Groppali, R. Venticinque Anni di Osservazioni Sui Trapianti di Formica Lugubris Zett. Dalla Alpi Alle Foreste Demaniali Casentinesi (Italia centrale): (Hymen. Formicidae); Università di Pavia: Pavia, Italy, 1986; p. 121. [Google Scholar]
- Ronchetti, G.; Groppali, R. Quarantacinque Anni di Protezione Forestale con Formica lugubris Zett. (Hymenoptera Formicidae). L’esperienza di Monte d’Alpe (Appennino Ligure in Provincia di Pavia); Istituto di Entomologia dell’Università di Pavia: Pavia, Italy, 1995; p. 271. [Google Scholar]
- Frizzi, F.; Masoni, A.; Quilghini, G.; Ciampelli, P.; Santini, G. Chronicle of an impact foretold: The fate and effect of the introduced Formica paralugubris ant. Biol. Inv. 2018, 20, 3575–3589. [Google Scholar] [CrossRef]
- Seifert, B. The supercolonial European wood ant Formica paralugubris Seifert, 1996 (Hymenoptera: Formicidae) introduced to Canada and its predicted role in Nearctic forests. Myrmecol. News 2016, 22, 11–20. [Google Scholar]
- Masoni, A.; Frizzi, F.; Natali, C.; Bernasconi, C.; Ciofi, C.; Santini, G. Molecular identification of imported red wood ant populations in the Campigna biogenetic nature Reserve (Foreste Casentinesi national Park, Italy). Cons. Gen. Res. 2019, 11, 231–236. [Google Scholar] [CrossRef]
- Seifert, B. Formica paralugubris nov. spec.—A sympatric sibling species of Formica lugubris from the western Alps (Insecta: Hymenoptera: Formicoidea: Formicidae). Reichenbachia 1996, 35, 193–201. [Google Scholar]
- Bernasconi, C.; Maeder, A.; Freitag, A.; Cherix, D. Formica paralugubris (Hymenoptera, Formicidae) in the Italian Alps from new data and old data revisited. Myrmecol. News 2006, 8, 251–256. [Google Scholar]
- Cherix, D. Note preliminaire sur la structure, la phenologie et le regime alimentaire d’une super-colonie de Formica lugubris Zett. Insect. Soc. 1980, 27, 226–236. [Google Scholar] [CrossRef]
- Chapuisat, M.; Goudet, J.; Keller, L. Microsatellites reveal high population viscosity and limited dispersal in the ant Formica paralugubris. Evolution 1997, 51, 475–482. [Google Scholar] [CrossRef] [PubMed]
- Holzer, B.; Meunier, J.; Keller, L.; Chapuisat, M. Stay or drift? Queen acceptance in the ant Formica paralugubris. Insect. Soc. 2008, 55, 392–396. [Google Scholar] [CrossRef][Green Version]
- Cherix, D. Red wood ants. Ethol. Ecol. Evol. 1991, 3, 165. [Google Scholar] [CrossRef]
- Chapuisat, M.; Keller, L. Extended family structure in the ant Formica paralugubris: The role of the breeding system. Behav. Ecol. Soc. 1999, 46, 405–412. [Google Scholar] [CrossRef]
- Blears, M.J.; De Grandis, S.A.; Lee, H.; Trevors, J.T. Amplified fragment length polymorphism (AFLP): A review of the procedure and its applications. JIMB 1998, 21, 99–114. [Google Scholar] [CrossRef]
- Bernasconi, C.; Pamilio, P.; Cherix, D. Molecular markers allow sibling species identification in red wood ants (Formica rufa group). Syst. Entomol. 2010, 35, 243–249. [Google Scholar] [CrossRef]
- Balzani, P.; Vizzini, S.; Frizzi, F.; Masoni, A.; Lessard, J.P.; Bernasconi, C.; Santini, G. Plasticity in the trophic niche of an invasive ant explains establishment success and long-term coexistence. Oikos 2021, 130, 691–696. [Google Scholar] [CrossRef]
- Lienhard, A.; Schäffer, S. Extracting the invisible: Obtaining high quality DNA is a challenging task in small arthropods. PeerJ 2019, 7, e6753. [Google Scholar] [CrossRef]
- Coppi, A.; Cecchi, L.; Mengoni, A.; Phustahija, F.; Tomović, G.; Selvi, F. Low genetic diversity and contrasting patterns of differentiation in the two monotypic genera Halacsya and Paramoltkia (Boraginaceae) endemic to the Balkan serpentines. Flora 2014, 209, 5–14. [Google Scholar] [CrossRef]
- Foll, M.; Fischer, M.C.; Heckel, G.; Excoffier, L. Estimating population structure from AFLP amplification intensity. Mol. Ecol. 2010, 19, 4638–4647. [Google Scholar] [CrossRef] [PubMed]
- Burr, T.L. Quasi-equilibrium theory for the distribution of rare alleles in a subdivided population: Justification and implications. Theor. Popul. Biol. 2000, 57, 297–306. [Google Scholar] [CrossRef] [PubMed]
- Yang, A.H.; Dick, C.W.; Yao, X.H.; Huang, H.W. Impacts of biogeographic history and marginal population genetics on species range limits: A case study of Liriodendron chinense. Sci. Rep. 2016, 6, 25632. [Google Scholar] [CrossRef]
- Nei, M. Molecular Evolutionary Genetics; Columbia University Press: New York, NY, USA, 1987; p. 512. [Google Scholar]
- Lewontin, R.C. Evolutionary biology: The apportionment of human diversity. In Evolutionary Biology; Dobzhansky, T., Hecht, M.K., Steere, W.C., Eds.; Springer: New York, NY, USA, 1995; pp. 381–398. [Google Scholar] [CrossRef]
- Yeh, F.; Yang, R.; Boyle, T.; Ye, Z.; Mao, J. POPGEN Ver. 1.32. The User-Friendly Software for Population Genetic Analysis; University of Alberta, Molecular Biology and Biotechnology Center: Edmonton, AB, Canada, 1997. [Google Scholar]
- Schneider, S.; Roessli, D.; Excer, L. Arlequin: A Software for Population Genetics Data Analysis; University of Geneva, Genetics and Biometry Laboratory: Geneva, Switzerland, 2000. [Google Scholar]
- Tamura, K.; Nei, M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 1993, 10, 512–526. [Google Scholar] [CrossRef]
- Kumar, S.; Nei, M.; Dudley, J.; Tamura, K. MEGA: A biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief. Bioinform. 2008, 9, 299–306. [Google Scholar] [CrossRef]
- Hubisz, M.J.; Falush, D.; Stephens, M.; Pritchard, J.K. Inferring weak population structure with the assistance of sample group information. Mol. Ecol. Res. 2009, 9, 1322–1332. [Google Scholar] [CrossRef]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef]
- Earl, D.A.; Von Holdt, B.M. STRUCTURE HARVESTER: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 2012, 4, 359–361. [Google Scholar] [CrossRef]
- Menchetti, M.; Talavera, G.; Cini, A.; Salvati, V.; Dincă, V.; Platania, L.; Dapporto, L. Two ways to be endemic. Alps and Apennines are different functional refugia during climatic cycles. Mol. Ecol. 2021, 30, 1297–1310. [Google Scholar] [CrossRef]
- Frizzi, F.; Masoni, A.; Santedicola, M.; Servini, M.; Simoncini, N.; Palmieri, J.; Santini, G. Intraspecific relationships and nest mound shape are affected by habitat features in introduced populations of the red wood ant Formica paralugubris. Insects 2022, 13, 198–204. [Google Scholar] [CrossRef] [PubMed]
- Kulmuni, J.; Nouhaud, P.; Pluckrose, L.; Satokangas, I.; Dhaygude, K.; Butlin, R.K. Instability of natural selection at candidate barrier loci underlying speciation in wood ants. Mol. Ecol. 2020, 29, 3988–3999. [Google Scholar] [CrossRef] [PubMed]
- Maky-Petays, H.; Zakharov, A.; Viljakainen, L.; Corander, J.; Pamilo, P. Genetic changes associated to declining populations of Formica ants in fragmented forest landscape. Mol. Ecol. 2005, 14, 733–742. [Google Scholar] [CrossRef] [PubMed]
- Allendorf, F.W.; England, P.R.; Luikart, G.; Ritchie, P.A.; Ryman, N. Genetic effects of harvest on wild animal populations. Trends Ecol. Evol. 2008, 23, 327–337. [Google Scholar] [CrossRef]
- Groppali, R.; Crudele, G. Le formiche del gruppo Formica rufa trapiantate nel Parco nazionale delle Foreste Casentinesi, Monte Falterona e Campigna. Quad. Stud. Nat. Romagna 2005, 20, 63–73. [Google Scholar]
- Flanagan, S.P.; Jones, A.G. The future of parentage analysis: From microsatellites to SNPs and beyond. Mol. Ecol. 2019, 28, 544–567. [Google Scholar] [CrossRef]
- Gzyl, A.; Augustynowicz, E.; Mosiej, E.; Zawadka, M.; Gniadek, G.; Nowaczek, A.; Slusarczyk, J. Amplified fragment length polymorphism (AFLP) versus randomly amplified polymorphic DNA (RAPD) as new tools for inter-and intra-species differentiation within Bordetella. J. Med. Microbiol. 2005, 54, 333–346. [Google Scholar] [CrossRef]
- Kulmuni, J.; Seifert, B.; Pamilo, P. Segregation distortion causes large-scale differences between male and female genomes in hybrid ants. Proc. Nas. Acad. Sci. USA 2010, 107, 7371–7376. [Google Scholar] [CrossRef]
- Schluns, E.A.; Neumann, P.; Schluns, H.; Hepburn, H.R.; Moritz, R.F.A. Nestmate recognition and genetic variability among individuals from nests of the queenless ponerine ant, Streblognathus aethiopicus Smith (Hymenoptera: Formicidae). Afr. Entomol. 2006, 14, 95–102. [Google Scholar]
- Nouhaud, P.; Beresford, J.; Kulmuni, J. Assembly of a hybrid Formica aquilonia × F. polyctena ant genome from a haploid male. J. Hered. 2022, 9, 353–359. [Google Scholar] [CrossRef]
- Portinha, B.; Avril, A.; Bernasconi, C.; Helanterä, H.; Monaghan, J.; Seifert, B.; Nouhaud, P. Whole-genome analysis of multiple wood ant population pairs supports similar speciation histories, but different degrees of gene flow, across their European ranges. Mol. Ecol. 2022, 31, 3416–3431. [Google Scholar] [CrossRef] [PubMed]
- Smith, C.C.; Weber, J.N.; Mikheyev, A.S.; Roces, F.; Bollazzi, M.; Kellner, K.; Mueller, U.G. Landscape genomics of an obligate mutualism: Concordant and discordant population structures between the leafcutter ant Atta texana and its two main fungal symbiont types. Mol. Ecol. 2019, 28, 2831–2845. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.M.; Vitone, T.R.; Storer, C.G.; Payton, A.C.; Dunn, R.R.; Hulcr, J.; Lucky, A. From pavement to population genomics: Characterizing a long-established non-native ant in North America through citizen science and ddRADseq. Front. Ecol. Evol. 2019, 7, 453. [Google Scholar] [CrossRef]
- Jay, P.; Leroy, M.; Le Poul, Y.; Whibley, A.; Arias, M.; Chouteau, M.; Joron, M. Association mapping of colour variation in a butterfly provides evidence that a supergene locks together a cluster of adaptive loci. Philos. Trans. R. Soc. B 2022, 377, 20210193. [Google Scholar] [CrossRef]
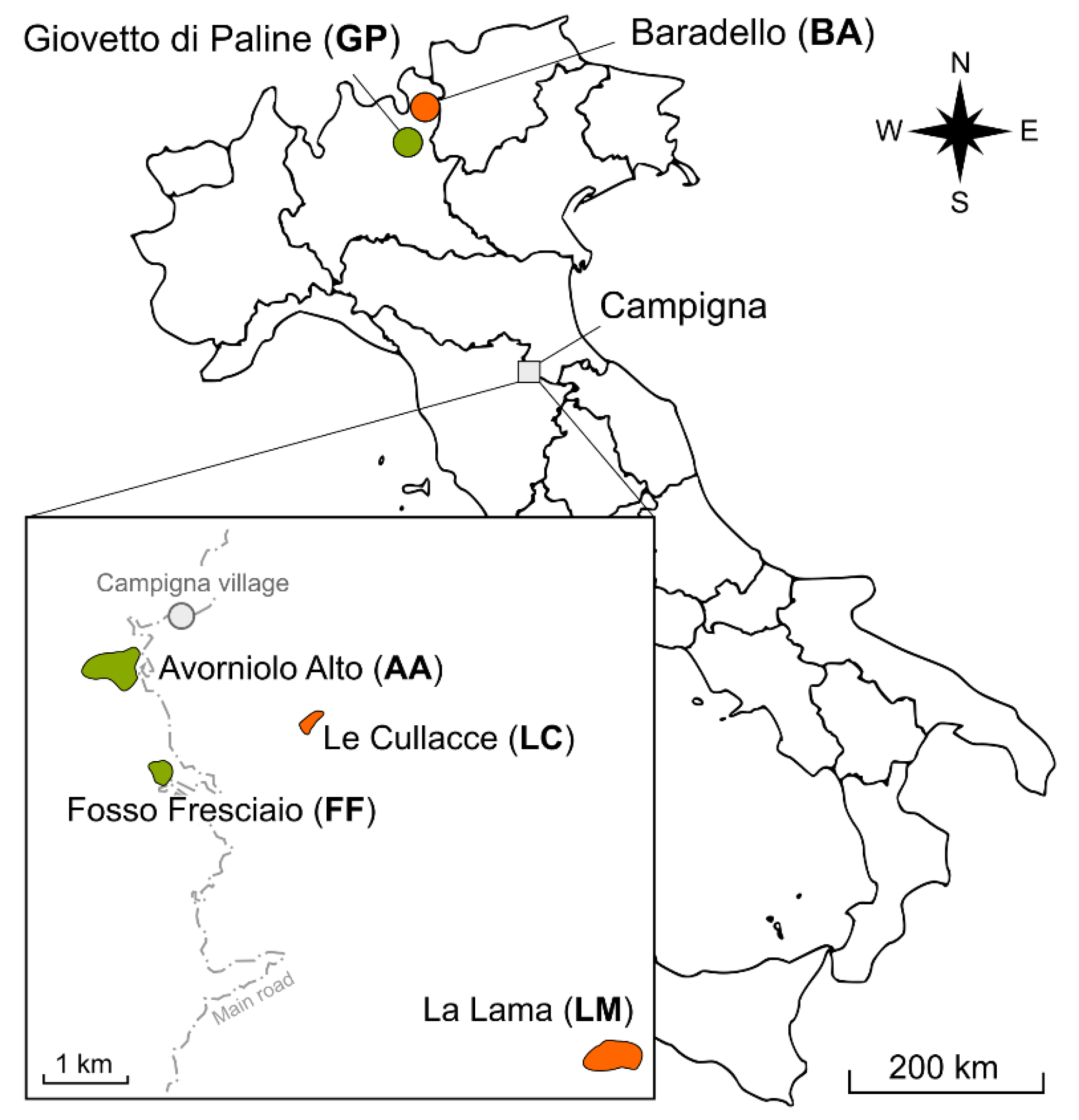
| Population | locality | NN | Nw | NPL | PL(%) | FST-POP | h | I | |
|---|---|---|---|---|---|---|---|---|---|
| GP | Native | Alps, Giovetto Paline | 10 | 100 | 99 | 67.35 | 0.207 ± 0.003 | 0.104 ± 0.007 | 0.160 ± 0.020 |
| AA | Transplanted | Apennine, Avorniolo | 10 | 100 | 115 | 78.23 | 0.157 ± 0.006 | 0.184 ± 0.014 | 0.235 ± 0.027 |
| FF | Transplanted | Apennine, Fosso Fresciaio | 10 | 75 | 100 | 68.03 | 0.208 ± 0.005 | 0.104 ± 0.01 | 0.155 ± 0.023 |
| BA | Native | Alps, Baradello | 9 | 90 | 76 | 51.70 | 0.208 ± 0.002 | 0.102 ± 0.004 | 0.139 ± 0.021 |
| LC | Transplanted | Apennine, Le Cullacce | 9 | 87 | 133 | 90.48 | 0.147 ± 0.006 | 0.205 ± 0.011 | 0.231 ± 0.022 |
| LM | Transplanted | Apennine, La Lama | 10 | 82 | 118 | 80.71 | 0.183 ± 0.006 | 0.191 ± 0.012 | 0.199 ± 0.021 |
| Source of Variation | df | Sum of Squares | Variance Components | % Variation | Fix.Index | p-Values | |
|---|---|---|---|---|---|---|---|
| All population | |||||||
| Among populations | 5 | 728.16 | 1.56 | 12.15 | FCT = 0.121 | <0.001 | |
| Among nests within populations | 43 | 878.42 | 1.02 | 8.05 | FSC = 0.091 | <0.001 | |
| Among workers | 435 | 4465.26 | 10.26 | 79.84 | FST = 0.201 | <0.001 | |
| Total | 483 | 6071.87 | 12.85 | 100 | |||
| Alpine vs. Apennine | |||||||
| Among groups | 1 | 294.21 | 1.13 | 8.6 | FCT = 0.085 | <0.05 | |
| Among nests within groups | 47 | 1312.33 | 1.78 | 13.56 | FSC = 0.148 | <0.05 | |
| Among workers | 435 | 4465.22 | 10.26 | 77.84 | FST = 0.221 | <0.001 | |
| Total | 483 | 6071.81 | 13.17 | 100 | |||
| BA vs. (LC + LM) | |||||||
| Among groups | 1 | 182.21 | 1.23 | 9.23 | FCT = 0.092 | <0.001 | |
| Among nests within groups | 28 | 714.33 | 1.68 | 12.57 | FSC = 0.138 | <0.001 | |
| Among workers | 239 | 2504.91 | 10.48 | 78.20 | FST = 0.217 | <0.001 | |
| Total | 268 | 3401.45 | 100 | ||||
| GP vs. (AA + FF) | |||||||
| Among groups | 1 | 144.39 | 0.94 | 7.65 | FCT = 0.076 | <0.001 | |
| Among nests within groups | 28 | 701.93 | 1.75 | 14.20 | FSC = 0.153 | <0.001 | |
| Among workers | 235 | 2269.92 | 9.65 | 78.15 | FST = 0.218 | <0.001 | |
| Total | 264 | 3116.24 | 12.35 | 100 | |||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Masoni, A.; Coppi, A.; Balzani, P.; Frizzi, F.; Fani, R.; Zaccaroni, M.; Santini, G. Assessing Molecular Diversity in Native and Introduced Populations of Red Wood Ant Formica paralugubris. Animals 2022, 12, 3165. https://doi.org/10.3390/ani12223165
Masoni A, Coppi A, Balzani P, Frizzi F, Fani R, Zaccaroni M, Santini G. Assessing Molecular Diversity in Native and Introduced Populations of Red Wood Ant Formica paralugubris. Animals. 2022; 12(22):3165. https://doi.org/10.3390/ani12223165
Chicago/Turabian StyleMasoni, Alberto, Andrea Coppi, Paride Balzani, Filippo Frizzi, Renato Fani, Marco Zaccaroni, and Giacomo Santini. 2022. "Assessing Molecular Diversity in Native and Introduced Populations of Red Wood Ant Formica paralugubris" Animals 12, no. 22: 3165. https://doi.org/10.3390/ani12223165
APA StyleMasoni, A., Coppi, A., Balzani, P., Frizzi, F., Fani, R., Zaccaroni, M., & Santini, G. (2022). Assessing Molecular Diversity in Native and Introduced Populations of Red Wood Ant Formica paralugubris. Animals, 12(22), 3165. https://doi.org/10.3390/ani12223165









