Potential Involvement of ewsr1-w Gene in Ovarian Development of Chinese Tongue Sole, Cynoglossus semilaevis
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Approval
2.2. Samples Collection
2.3. Gene Cloning of Cs-ewsr1-w and Cs-ewsr1-z
2.4. Characterization of Cs-ewsr1-w and Cs-ewsr1-z
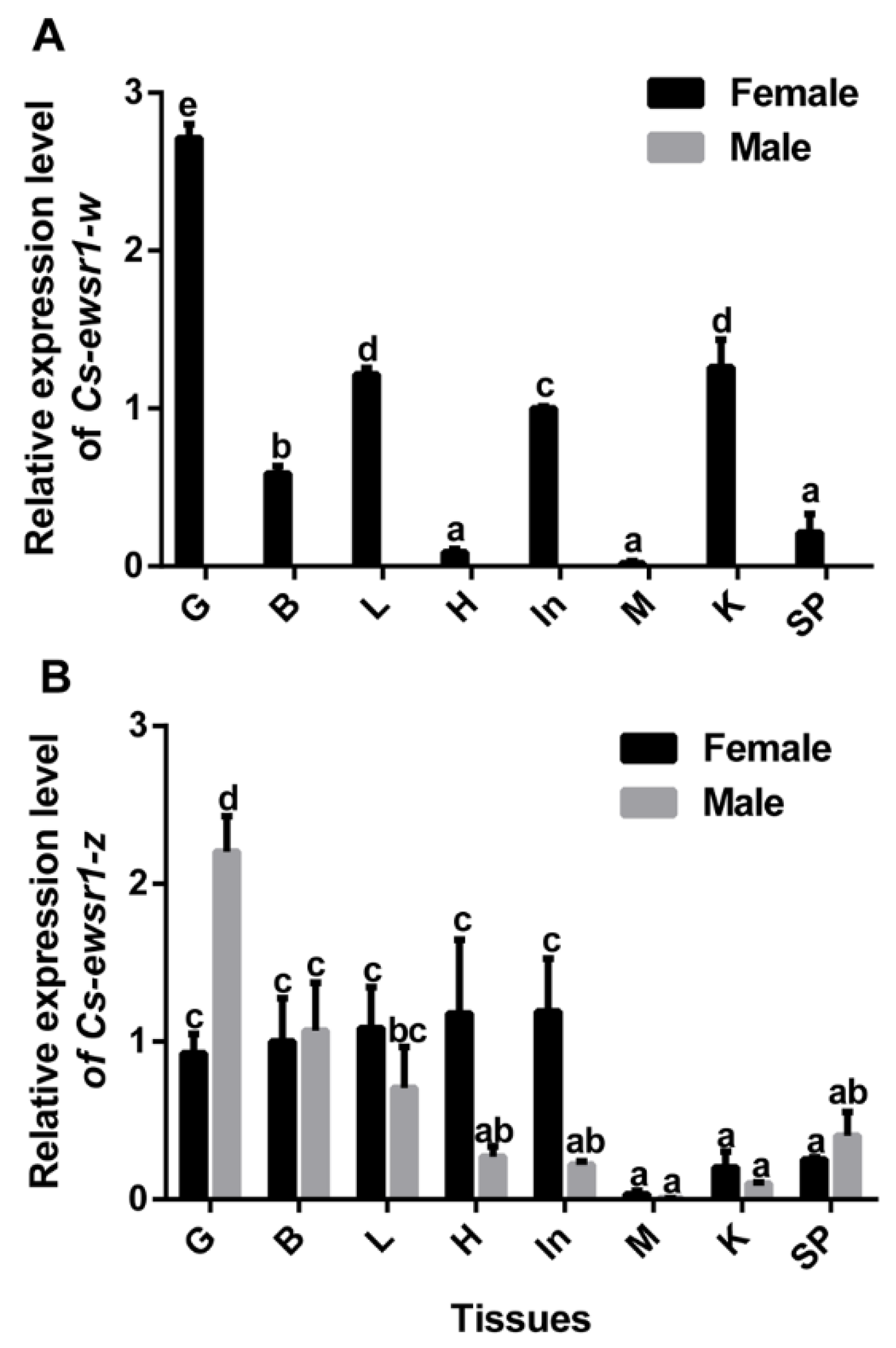
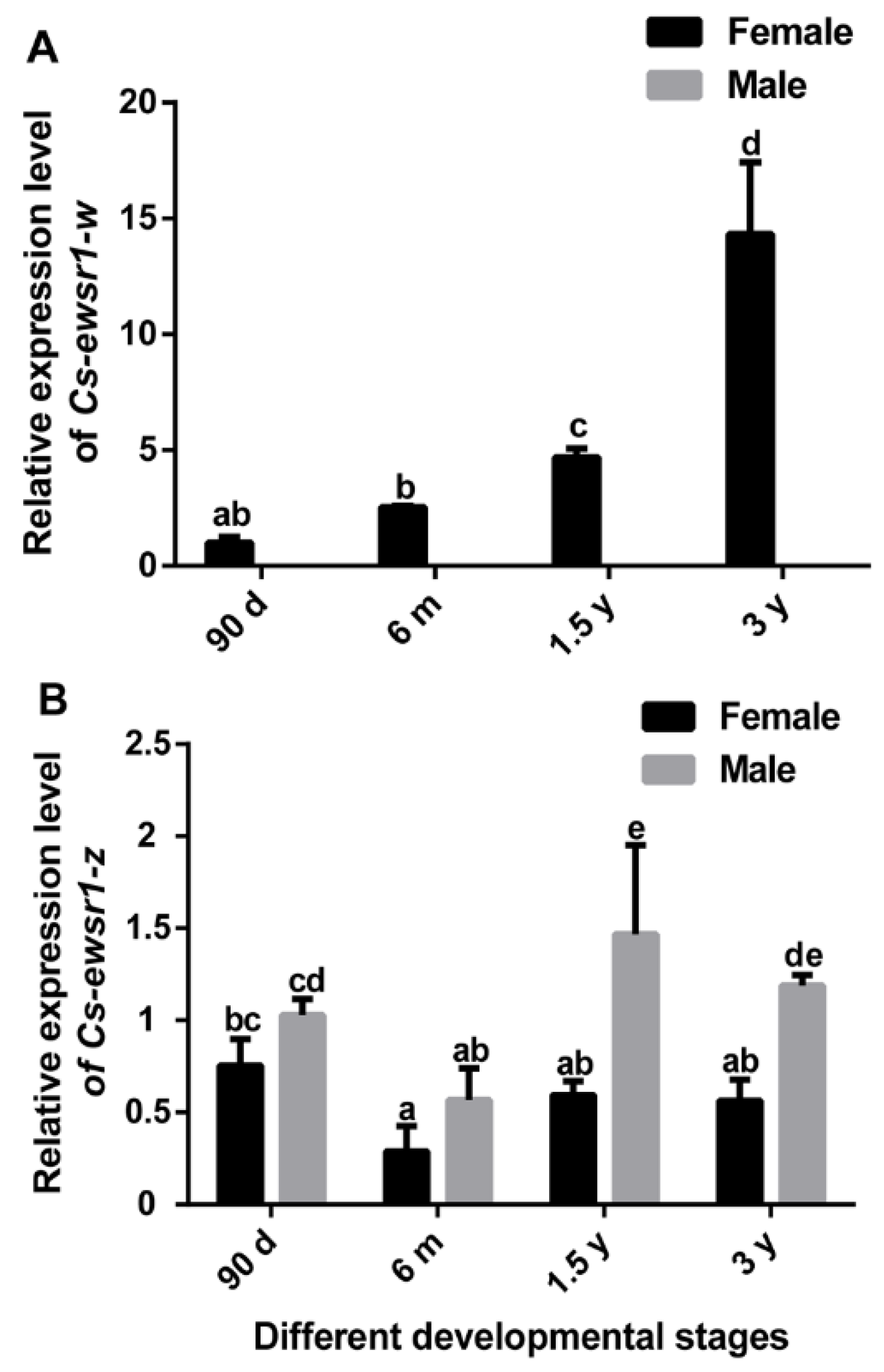
2.5. Gene Expression Patterns of Cs-ewsr1-w and Cs-ewsr1-z in Different Tissues and Stages
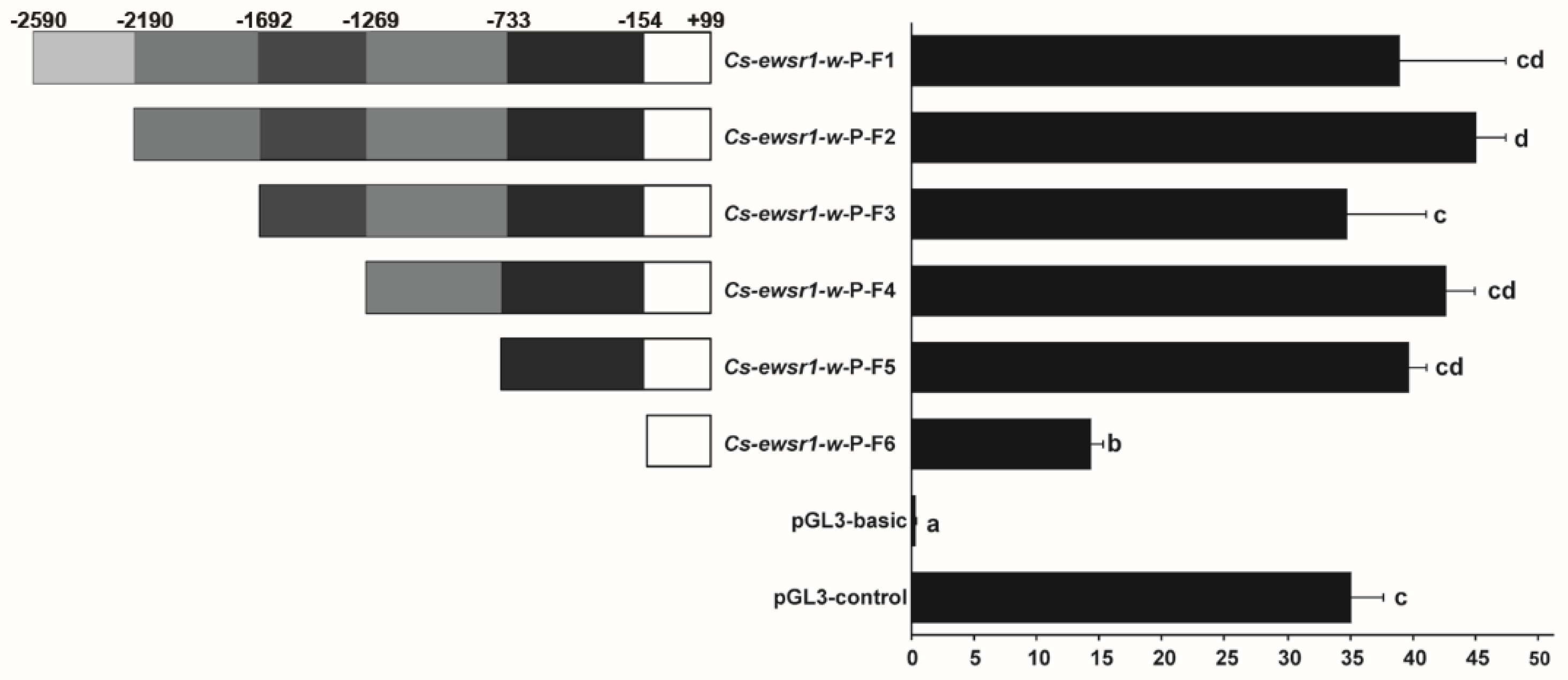
2.6. Promoter Activities Analysis of Cs-ewsr1-w
2.7. The Knockdown Effect of Cs-ewsr1-w siRNA in C. semilaevis Ovarian Cells
3. Results
3.1. Gene Cloning and Characterization of Cs-ewsr1s
3.2. The Expression Patterns of Cs-ewsr1s in Different Tissues and Developmental Stages
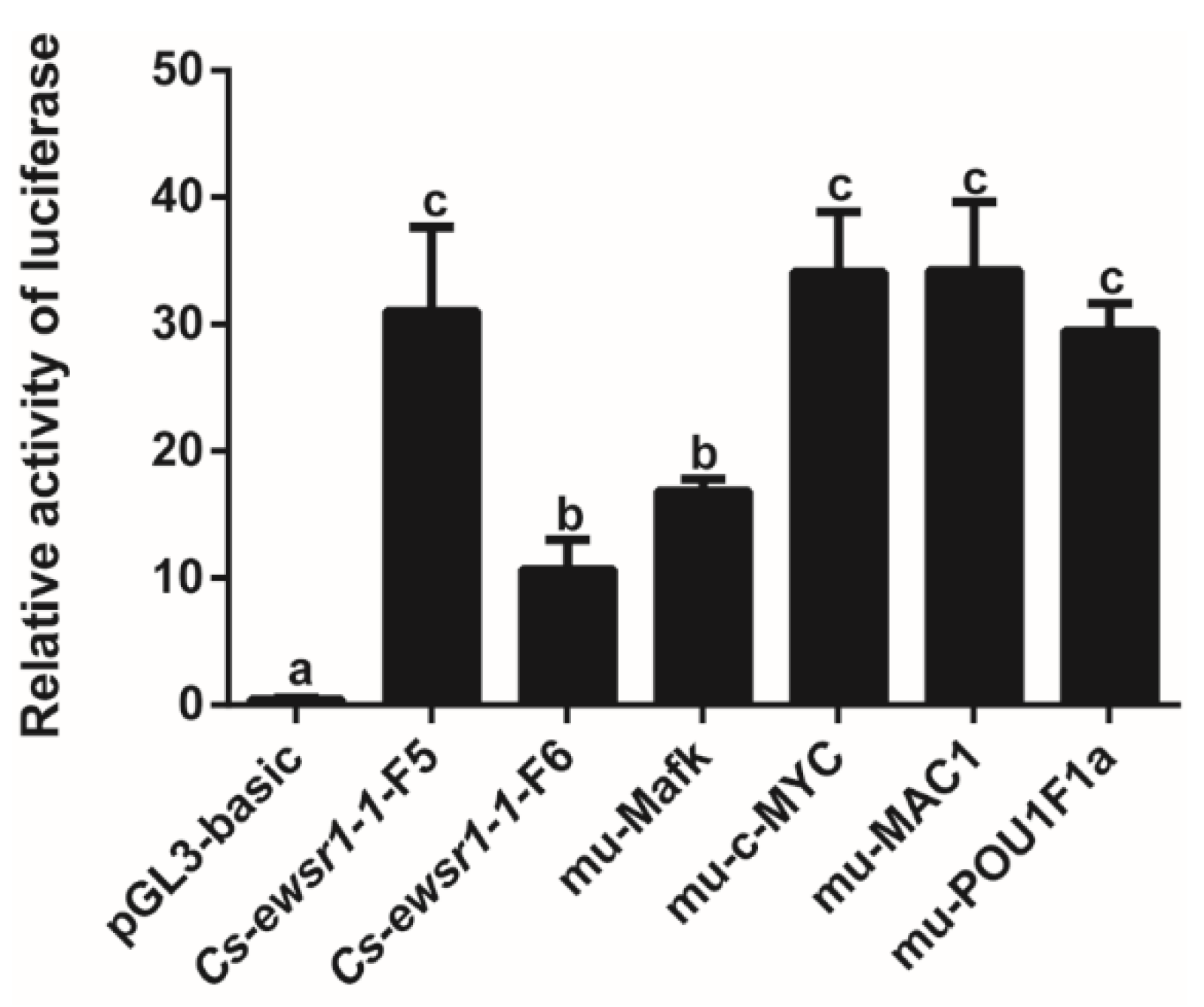
3.3. Promoter Activity of Cs-ewsr1-w Detection and Analysis
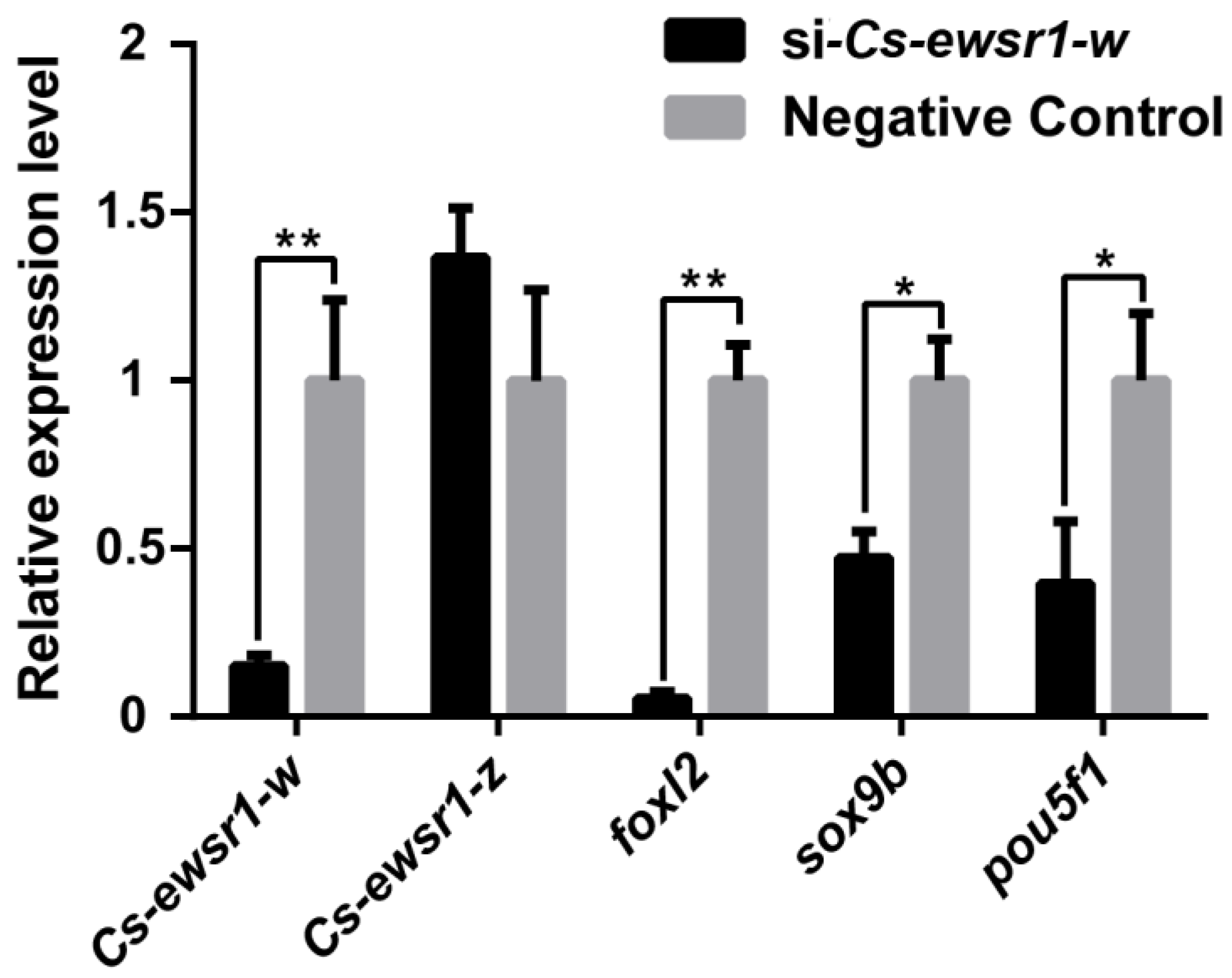
3.4. Expression Patterns of Sex-Related Genes in Cs-ewsr1-w Knockdown Ovarian Cells
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Naqvi, S.; Godfrey, A.K.; Hughes, J.F.; Goodheart, M.L.; Mitchell, R.N.; Page, D.C. Conservation, acquisition, and functional impact of sex-biased gene expression in mammals. Science 2019, 365, eaaw7317. [Google Scholar] [CrossRef]
- Mei, J.; Gui, J. Genetic basis and biotechnological manipulation of sexual dimorphism and sex determination in fish. Sci. China Life Sci. 2015, 58, 124–136. [Google Scholar] [CrossRef]
- Parker, G.A. The evolution of sexual size dimorphism in fish*. J. Fish Biol. 1992, 41, 1–20. [Google Scholar] [CrossRef]
- Foellmer, M.W.; Moya-Laraño, J. Chater 7. Sexual size dimorphism in spiders: patterns and processes. In Sex, Size and Gender Roles: Evolutionary Studies of Sexual Size Dimorphism; Daphne, J.F., Wolf, U.B., Tamás, S., Eds.; Oxford University Press: New York, NY, USA, 2007; pp. 71–81. ISBN 9780191709036. [Google Scholar]
- Lindenfors, P.; Gittleman, J.; Jones, K. Chapter 2. Sexual size dimorphism in mammals. In Sex, Size and Gender Roles: Evolutionary Studies of Sexual Size Dimorphism; Daphne, J.F., Wolf, U.B., Tamás, S., Eds.; Oxford University Press: New York, NY, USA, 2007; pp. 16–26. ISBN 9780191709036. [Google Scholar]
- Székely, T.; Lislevand, T.; Figuerola, J. Chapter 3. Sexual size dimorphism in birds. In Sex, Size and Gender Roles: Evolutionary Studies of Sexual Size Dimorphism; Daphne, J.F., Wolf, U.B., Tamás, S., Eds.; Oxford University Press: New York, NY, USA, 2007; pp. 27–37. ISBN 9780191709036. [Google Scholar]
- Sun, Y.; Yu, H.; Zhang, Q.; Qi, J.; Zhong, Q.; Chen, Y.; Li, C. Molecular characterization and expression pattern of two zona pellucida genes in half-smooth tongue sole (Cynoglossus semilaevis). Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2010, 155, 316–321. [Google Scholar] [CrossRef]
- Bonduriansky, R.; Chenoweth, S. Intralocus sexual conflict. Trends Ecol. Evol. 2009, 24, 280–288. [Google Scholar] [CrossRef]
- Parsch, J.; Ellegren, H. The evolutionary causes and consequences of sex-biased gene expression. Nat. Rev. Genet. 2013, 14, 83–87. [Google Scholar] [CrossRef]
- Williams, T.M.; Carroll, S.B. Genetic and molecular insights into the development and evolution of sexual dimorphism. Nat. Rev. Genet. 2009, 10, 797–804. [Google Scholar] [CrossRef]
- Wang, N.; Wang, R.K.; Wang, R.Q.; Chen, S.L. Transcriptomics analysis revealing candidate networks and genes for the body size sexual dimorphism of Chinese tongue sole (Cynoglossus semilaevis). Funct. Integr. Genom. 2018, 18, 327–339. [Google Scholar] [CrossRef]
- Wang, K.L.; Zhang, H.; Hu, Q.M.; Shao, C.W.; Chen, S.L. Expression and purification of half-smooth tongue sole (Cynoglossus semilaevis) CSDAZL protein. Protein Expr. Purif. 2014, 102, 8–12. [Google Scholar] [CrossRef]
- Wang, N.; Gong, Z.H.; Wang, J.; Xu, W.T.; Yang, Q.; Chen, S.L. Characterization of Chinese tongue sole (Cynoglossus semilaevis) 24-dehydrocholesterol reductase: Expression profile, epigenetic modification, and its knock-down effect. Gen. Comp. Endocrinol. 2021, 312, 113870. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Li, X.H.; Shi, R.; Cheng, P.; Wang, N.; Chen, S.L. The female-biased expression, transcriptional regulation and knock-down effect of insulin-like growth factor binding protein 7 in Chinese tongue sole, Cynoglossus semilaevis. Aquaculture 2022, 551, 737956. [Google Scholar] [CrossRef]
- Xu, W.T.; Cui, Z.K.; Wang, N.; Zhang, M.Q.; Wang, J.; Xu, X.T.; Liu, Y.; Chen, S.L. Transcriptomic analysis revealed gene expression profiles during the sex differentiation of Chinese tongue sole (Cynoglossus semilaevis). Comp. Biochem. Physiol. Part D Genom. Proteom. 2021, 40, 100919. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Nguyen, P.T.; Shim, H.S.; Hyeon, S.J.; Im, H.; Choi, M.H.; Chung, S.; Kowall, N.W.; Lee, S.B.; Ryu, H. EWSR1, a multifunctional protein, regulates cellular function and aging via genetic and epigenetic pathways. Biochim. Biophys. Acta-(BBA) Mol. Basis Dis. 2019, 1865, 1938–1945. [Google Scholar] [CrossRef] [PubMed]
- Rossow, K.L.; Janknecht, R. The Ewing’s sarcoma gene product functions as a transcriptional activator. Cancer Res. 2001, 61, 2690–2695. [Google Scholar]
- Erkizan, H.; Uversky, V.; Toretsky, J. Oncogenic Partnerships: EWS-FLI1 Protein Interactions Initiate Key Pathways of Ewing’s Sarcoma. Clin. Cancer Res. 2010, 16, 4077–4083. [Google Scholar] [CrossRef] [PubMed]
- Morohoshi, F.; Ootsuka, Y.; Arai, K.; Ichikawa, H.; Mitani, S.; Munakata, N.; Ohki, M. Genomic structure of the human RBP56/hTAF(II)68 and FUS/TLS genes. Gene 1998, 221, 191–198. [Google Scholar] [CrossRef]
- Bertolotti, A.; Melot, T.; Acker, J.; Vigneron, M.; Delattre, O.; Tora, L. EWS, but not EWS-FLI-1, is associated with both TFIID and RNA polymerase II: Interactions between two members of the tet family, EWS and HTAF(II)68, and subunits of TFIID and RNA polymerase II complexes. Mol. Cell. Biol. 1998, 18, 1489–1497. [Google Scholar] [CrossRef]
- Azuma, M.; Embree, L.J.; Sabaawy, H.; Hickstein, D.D. Ewing sarcoma protein ewsr1 maintains mitotic integrity and proneural cell survival in the zebrafish embryo. PLoS ONE 2007, 2, e979. [Google Scholar] [CrossRef]
- Li, H.; Watford, W.; Li, C.; Parmelee, A.; Bryant, M.A.; Deng, C.; O’Shea, J.; Lee, S.B. Ewing sarcoma gene EWS is essential for meiosis and B lymphocyte development. J. Clin. Investig. 2007, 117, 1314–1323. [Google Scholar] [CrossRef]
- Araya, N.; Hirota, K.; Shimamoto, Y.; Miyagishi, M.; Yoshida, E.; Ishida, J.; Kaneko, S.; Kaneko, M.; Nakajima, T.; Fukamizu, A. Cooperative interaction of EWS with CREB-binding protein selectively activates hepatocyte nuclear factor 4-mediated transcription. J. Biol. Chem. 2003, 278, 5427–5432. [Google Scholar] [CrossRef]
- Lee, J.; Rhee, B.K.; Bae, G.Y.; Han, Y.M.; Kim, J. Stimulation of Oct-4 activity by Ewing’s sarcoma protein. Stem Cells 2005, 23, 738–751. [Google Scholar] [CrossRef] [PubMed]
- Kovar, H. Dr. Jekyll and Mr. Hyde: The Two Faces of the FUS/EWS/TAF15 Protein Family. Sarcoma 2011, 2011, 837474. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Han, K.; Chen, S.; Cai, M.; Wang, Y.; Zhang, Z. Molecular cloning and expression of Octamer-binding transcription factor (Oct4) in the large yellow croaker, Larimichthys crocea. Gene Expr. Patterns 2018, 27, 16–30. [Google Scholar] [CrossRef]
- Lanier, J.; Quina, L.A.; Eng, S.R.; Cox, E.; Turner, E.E. Brn3a target gene recognition in embryonic sensory neurons. Dev. Biol. 2007, 302, 703–716. [Google Scholar] [CrossRef]
- Tian, H.; Petkov, P.M. Mouse EWSR1 is crucial for spermatid post-meiotic transcription and spermiogenesis. Development 2021, 148, dev199414. [Google Scholar] [CrossRef]
- Sun, Y.X.; Zhu, Y.; Cheng, P.; Zhang, M.Q.; Wang, N.; Cui, Z.K.; Wei, M.; Xu, W.T. A Z-Linked E3 Ubiquitin Ligase Cs-rchy1 Is Involved in Gametogenesis in Chinese Tongue Sole, Cynoglossus semilaevis. Animals 2021, 11, 3265. [Google Scholar] [CrossRef] [PubMed]
- Sun, A.; Wang, T.Z.; Wang, N.; Liu, X.F.; Sha, Z.X.; Chen, S.L. Establishment and characterization of an ovarian cell line from half-smooth tongue sole Cynoglossus semilaevis. J. Fish Biol. 2015, 86, 46–59. [Google Scholar] [CrossRef]
- Liu, Y.; Chen, S.L.; Gao, F.T.; Meng, L.; Hu, Q.M.; Song, W.; Shao, C.W.; Lv, W.Q. SCAR-transformation of sex-specific SSR marker and its application in half-smooth tongue sole (Cynoglossus semiliaevis). Agric. Biotechnol. 2014, 6, 787–792. [Google Scholar] [CrossRef]
- Chen, S.L.; Zhang, G.J.; Shao, C.W.; Huang, Q.F.; Liu, G.; Zhang, P.; Song, W.T.; An, N.; Chalopin, D.; Volff, J.; et al. Whole-genome sequence of a flatfish provides insights into ZW sex chromosome evolution and adaptation to a benthic lifestyle. Nat. Genet. 2014, 46, 253–260. [Google Scholar] [CrossRef]
- Leturnic, I.; Khedkar, S.; Bork, P. SMART: recent updates, new developments and status in 2020. Nucleic Acids Res. 2021, 49, D458–D460. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis acorss computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Messeguer, X.; Escudero, R.; Farré, D.; Nuñez, O.; Martínez, J.; Albà, M.M. PROMO: Detection of known transcription regulatory elements using species-tailored searches. Bioinformatics 2002, 18, 333–334. [Google Scholar] [CrossRef] [PubMed]
- Castro-Mondragon, J.A.; Riudavets-Puig, R.; Rauluseviciute, I.; Lemma, R.B.; Turchi, L.; Blanc-Mathieu, R.; Lucas, J.; Boddie, P.; Khan, A.; Pérez, N.M.; et al. JASPAR 2022: The 9th release of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 2022, 50, D165–D173. [Google Scholar] [CrossRef]
- Paronetto, M.P. Ewing sarcoma protein: A key player in human cancer. Int. J. Cell Biol. 2013, 2013, 642853. [Google Scholar] [CrossRef]
- Bertolotti, A.; Lutz, Y.; Heard, D.J.; Chambon, P.; Tora, L. hTAF(II)68, a novel RNA/ssDNA-binding protein with homology to the pro-oncoproteins TLS/FUS and EWS is associated with both TFIID and RNA polymerase II. EMBO J. 1996, 15, 5022–5031. [Google Scholar] [CrossRef]
- Uhlenhaut, N.H.; Treier, M. Foxl2 function in ovarian development. Mol. Genet. Metab. 2006, 88, 225–234. [Google Scholar] [CrossRef]
- Okada, H.; Hagihara, S.; Yamashita, K.; Ijiri, S.; Adachi, S. Expression pattern of foxl2 and dmrt1 in gonad of Amur sturgeon Acipenser schrenckii in relation to sex differentiation. Aquaculture 2017, 479, 712–720. [Google Scholar] [CrossRef]
- Alam, M.A.; Kobayashi, Y.; Horiguchi, R.; Hirai, T.; Nakamura, M. Molecular cloning and quantitative expression of sexually dimorphic markers Dmrt1 and Foxl2 during female-to-male sex change in Epinephelus merra. Gen. Comp. Endocrinol. 2008, 157, 75–85. [Google Scholar] [CrossRef]
- Yamaguchi, T.; Yamaguchi, S.; Hirai, T.; Kitano, T. Follicle-stimulating hormone signaling and Foxl2 are involved in transcriptional regulation of aromatase gene during gonadal sex differentiation in Japanese flounder, Paralichthys olivaceus. Biochem. Biophys. Res. Commun. 2007, 359, 935–940. [Google Scholar] [CrossRef]
- Ijiri, S.; Kaneko, H.; Kobayashi, T.; Wang, D.S.; Sakai, F.; Paul-Prasanth, B.; Nakamura, M.; Nagahama, Y. Sexual dimorphic expression of genes in gonads during early differentiation of a teleost fish, the Nile tilapia Oreochromis niloticus. Biol. Reprod. 2008, 78, 333–341. [Google Scholar] [CrossRef]
- Dong, X.L.; Chen, S.L.; Ji, X.S.; Shao, C.W. Molecular cloning, characterization and expression analysis of Sox9a and Foxl2 genes in half-smooth tongue sole (Cynoglossus semilaevis). Acta Oceanol. Sin. 2011, 30, 68–77. [Google Scholar] [CrossRef]
- Chiang, E.F.; Pai, C.I.; Wyatt, M.; Yan, Y.L.; Postlethwait, J.; Chung, B. Two sox9 genes on duplicated zebrafish chromosomes: Expression of similar transcription activators in distinct sites. Dev. Biol. 2001, 231, 149–163. [Google Scholar] [CrossRef] [PubMed]
- Shen, X.; Cui, J.; Yang, G.; Gong, Q.; Gu, Q. Expression detection of DMRTs and two sox9 genes in Takifugu rubripes (Tetraodontidae, Vertebrata). J. Ocean Univ. China 2007, 6, 182–186. [Google Scholar] [CrossRef]
- Nakamura, S.; Aoki, Y.; Saito, D.; Kuroki, Y.; Fujiyama, A.; Naruse, K.; Tanaka, M. Sox9b/sox9a2-EGFP transgenic medaka reveals the morphological reorganization of the gonads and a common precursor of both the female and male supporting cells. Mol. Reprod. Dev. 2008, 75, 472–476. [Google Scholar] [CrossRef]
- Nakamura, S.; Watakabe, I.; Nishimura, T.; Toyoda, A.; Taniguchi, Y.; Tanaka, M. Analysis of Medaka sox9 Orthologue Reveals a Conserved Role in Germ Cell Maintenance. PLoS ONE 2012, 7, e29982. [Google Scholar] [CrossRef]
- Li, X.; Yu, H.; Wang, Y.; Liu, X.; Liu, Y.; Qu, J.; Wang, X. Roles of Two Sox9 Genes during Gonadal Development in Japanese Flounder: Sex Differentiation, Spermatogenesis and Gonadal Function Maintenance. Int. J. Mol. Sci. 2018, 19, 512. [Google Scholar] [CrossRef]
- Sun, Y.; Zhang, M.Q.; Cheng, P.; Gong, Z.H.; Li, X.H.; Wang, N.; Wei, M.; Xu, X.W.; Xu, W.T. pitpbeta_w Encoding Phosphatidylinositol Transfer Protein Is Involved in Female Differentiation of Chinese Tongue Sole, Cynoglossus semilaevis. Front. Genet. 2022, 13, 861763. [Google Scholar] [CrossRef]
- Xiaohuan, H.; Yang, Z.; Linyan, L.; Zhenhua, F.; Linyan, Z.; Zhijian, W.; Ling, W.; Deshou, W.; Jing, W. Characterization of the POU5F1 Homologue in Nile Tilapia: From Expression Pattern to Biological Activity. Stem Cells Dev. 2016, 25, 1386–1395. [Google Scholar] [CrossRef]
- Zhong, C.; Liu, M.; Tao, Y.; Wu, X.; Yang, Y.; Wang, T.; Meng, Z.; Xu, H.; Liu, X. Pou5f1 and Nanog Are Reliable Germ Cell-Specific Genes in Gonad of a Protogynous Hermaphroditic Fish, Orange-Spotted Grouper (Epinephelus coioides). Genes 2021, 13, 79. [Google Scholar] [CrossRef]
- Gao, J.; Wang, X.; Zhang, Q. Evolutionary Conservation of pou5f3 Genomic Organization and Its Dynamic Distribution during Embryogenesis and in Adult Gonads in Japanese Flounder Paralichthys olivaceus. Int. J. Mol. Sci. 2017, 18, 231. [Google Scholar] [CrossRef] [PubMed]
- Shijun, L.; Khan, R.; Raza, S.H.A.; Jieyun, H.; Chugang, M.; Kaster, N.; Gong, C.; Chunping, Z.; Schreurs, N.M.; Linsen, Z. Function and characterization of the promoter region of perilipin 1 (PLIN1): Roles of E2F1, PLAG1, C/EBPβ, and SMAD3 in bovine adipocytes. Genomics 2020, 112, 2400–2409. [Google Scholar] [CrossRef] [PubMed]
- Tan, S.; Richmond, T. Eukaryotic transcription factors. Curr. Opin. Struct. Biol. 1998, 8, 41–48. [Google Scholar] [CrossRef]
- de Vooght, K.M.; van Solinge, W.W. Gene promoter analysis in molecular diagnostics: Do or don’t? Expert Rev. Mol. Diagn. 2009, 9, 403–405. [Google Scholar] [CrossRef]
- Yamazaki, H.; Katsuoka, F.; Motohashi, H.; Engel, J.D.; Yamamoto, M. Embryonic lethality and fetal liver apoptosis in mice lacking all three small Maf proteins. Mol. Cell. Biol. 2012, 32, 808–816. [Google Scholar] [CrossRef] [PubMed]
- Kannan, M.B.; Solovieva, V.; Blank, V. The small MAF transcription factors MAFF, MAFG and MAFK: Current knowledge and perspectives. Biochim. Biophys. Acta (BBA)-Mol. Cell Res. 2012, 1823, 1841–1846. [Google Scholar] [CrossRef] [PubMed]

 ” represents RNA recognition motif, “
” represents RNA recognition motif, “ ” respresents Ran binding protein zinc finger domain, “▲” represents Ewsr1 proteins in C. semilaevis.
” respresents Ran binding protein zinc finger domain, “▲” represents Ewsr1 proteins in C. semilaevis.
 ” represents RNA recognition motif, “
” represents RNA recognition motif, “ ” respresents Ran binding protein zinc finger domain, “▲” represents Ewsr1 proteins in C. semilaevis.
” respresents Ran binding protein zinc finger domain, “▲” represents Ewsr1 proteins in C. semilaevis.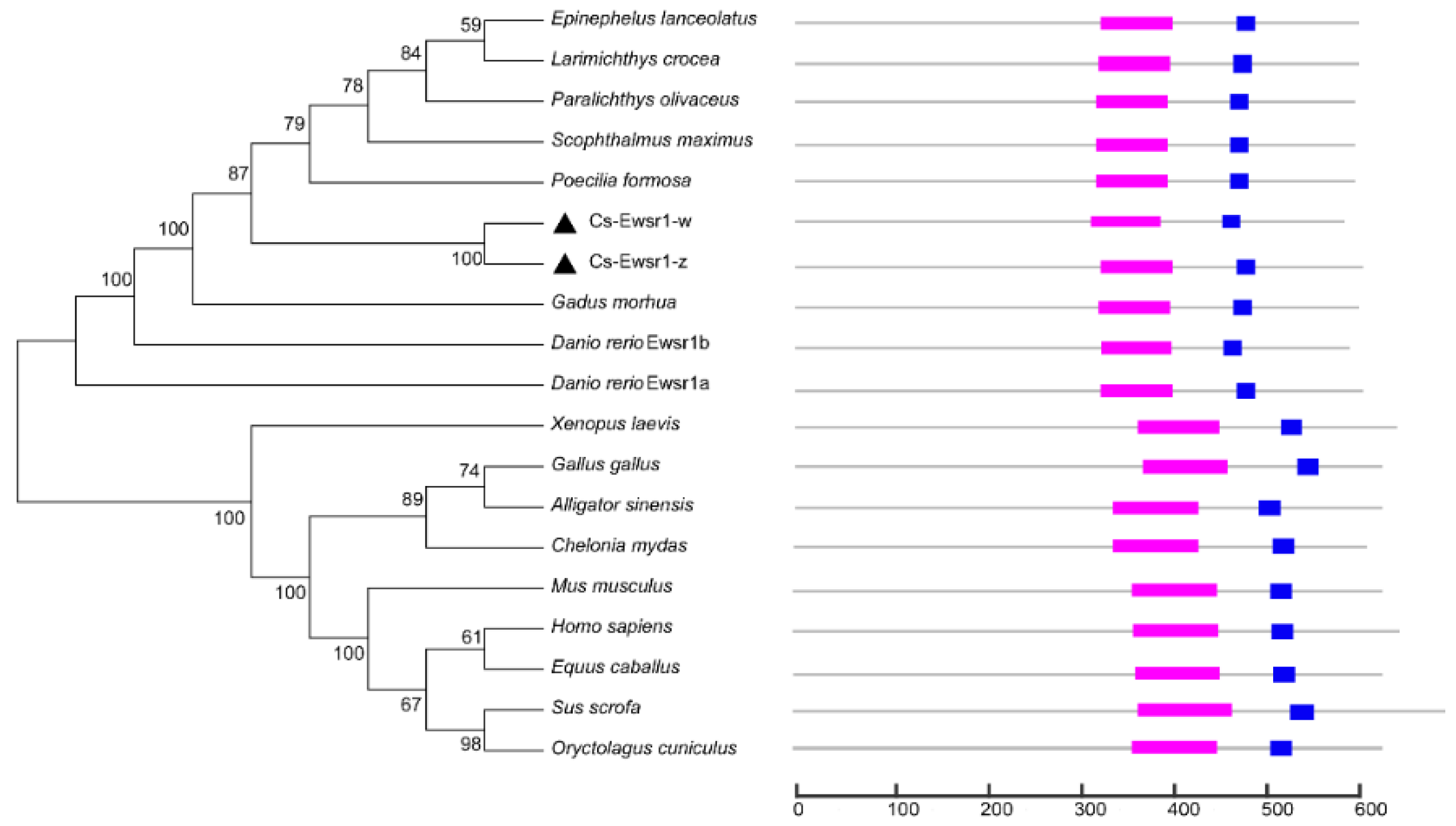

| Symbol | Information | Sequences |
|---|---|---|
| Cs-ewsr1-w-F | CDS cloning | TTCAGTCGTTTAACTCGGAAGT |
| Cs-ewsr1-w-R | CDS cloning | TCACAAGTGGGAAAGTCACCCT |
| Cs-ewsr1-w-5′-1 | 5′-UTR | CTGGAGCAGCAGCAGCTGGAGTACT |
| Cs-ewsr1-w-5′-2 | 5′-UTR | TAACCCTGCTGGGCGTTGGCTTGA |
| Cs-ewsr1-w-3′-1 | 3′-UTR | AATGAGAGGCAGCATGCCAATAAGA |
| Cs-ewsr1-w-3′-2 | 3′-UTR | GGTCGGCGGTCCTCCATTCCCCCCT |
| Cs-ewsr1-w-RT-F | qPCR | GCGGGCCCCCCATGGACC |
| Cs-ewsr1-w-RT-R | qPCR | CAAATTTCACAAGTGGGA |
| Cs-ewsr1-w-P-F1 | promoter | AGATCTGCGATCTAAGTAAGCTGACGCTGGCATGTATGTT |
| Cs-ewsr1-w-P-F2 | promoter | AGATCTGCGATCTAAGTAAGCTGAGGACCACAACGACCCA |
| Cs-ewsr1-w-P-F3 | promoter | AGATCTGCGATCTAAGTAAGCTTGCGAACAAATCACTGCG |
| Cs-ewsr1-w-P-F4 | promoter | AGATCTGCGATCTAAGTAAGCTAGGTGTATCCTAAACAGAAA |
| Cs-ewsr1-w-P-F5 | promoter | AGATCTGCGATCTAAGTAAGCTATCGGATCGGAAAGGAAA |
| Cs-ewsr1-w-P-F6 | promoter | AGATCTGCGATCTAAGTAAGCTCGGTCTTCCCATCACTAA |
| Cs-ewsr1-w-P-R | promoter | CAACAGTACCGGAATGCCAAGCTTTCCGAGTTAAACGACTGAAGA |
| mu-Mafk-F | TF binding site mutation | CAAACCATGTTGCCTCGTACGCTAAGTAAGCCCTACT |
| mu-Mafk-R | TF binding site mutation | CAAACCATGTTGCCTTCAGTATTAAGTAAGCCCTACT |
| mu-c-Myc-F | TF binding site mutation | ATTAGTTTCATTTGTAGACGAACAGAGAGCTCGGGT |
| mu-c-Myc-R | TF binding site mutation | ACCCGAGCTCTCTGTTCGTCTACAAATGAAACTAAT |
| mu-MAC1-F | TF binding site mutation | AGAGCTCGGGTCTCTACGATGATCATTCATCTTCACA |
| mu-MAC1-R | TF binding site mutation | TGTGAAGATGAATGATCATCGTAGAGACCCGAGCTCT |
| mu-POU1F1a-F | TF binding site mutation | AGTAAGCCCTACTCTCGGCTGTGTCAGAGAACCGC |
| mu-POU1F1a-R | TF binding site mutation | GCGGTTCTCTGACACAGCCGAGAGTAGGGCTTACT |
| Cs-ewsr1-w-siRNA | siRNA | CGUUUAACUCGGAAGUUGUGA |
| sox9b-F | qPCR | AAGAACCACACAGATCAAGACAGA |
| sox9b-R | qPCR | TAGTCATACTGTGCTCTGGTGATG |
| foxl2-F | qPCR | GAGGAAGGGCAACTACTGGA |
| foxl2-R | qPCR | CAGCGACCAGGAGTTGTTCA |
| pou5f1-F | qPCR | CCATCTGCCGCTTTGAGG |
| pou5f1-R | qPCR | CCTGGGTGTTGGGTTTGG |
| Cs-ewsr1-z-F | CDS cloning | ATGGCGTCGACACAGGATTACAGCT |
| Cs-ewsr1-z-R | CDS cloning | TTAGTAAGGTCTGTCTCGGCGCTCC |
| Cs-ewsr1-z-5′-1 | 5′-UTR | CAGCAGCTGGAGTACTGTCATATCC |
| Cs-ewsr1-z-5′-2 | 5′-UTR | CCCCTGCTGGGCGCTGGTCTGG |
| Cs-ewsr1-z-3′-1 | 3′-UTR | GATGAGAGGTGGCATGCCAATGAGA |
| Cs-ewsr1-z-3′-2 | 3′-UTR | AGGCGGAGGTCCTCCATTTCCCCCT |
| Cs-ewsr1-z-RT-F | qPCR | GGATATGACAGTACTCCAGCT |
| Cs-ewsr1-z-RT-R | qPCR | TCCTGCAGGCTGGCTATAGCTAC |
| sex-F | sex identification | CCTAAATGATGGATGTAGATTCTGTC |
| sex-R | sex identification | GATCCAGAGAAAATAAACCCAGG |
| Species | Accession No. |
|---|---|
| Larimichthys crocea | XP_019130992.1 |
| Paralichthys olivaceus | XP_019956591.1 |
| Poecilia formosa | XP_007559175.1 |
| Gadus morhua | XP_030215256.1 |
| Scophthalmus maximus | XP_035495384.1 |
| Danio rerio-Ewsr1a | XP_021334784.1 |
| Danio rerio-Ewsr1b | NP_997795.1 |
| Epinephelus lanceolatus | XP_033475149.1 |
| Alligator sinensis | XP_025048938.1 |
| Homo sapiens | XP_011528297.1 |
| Gallus gallus | XP_015150339.2 |
| Mus musculus | NP_001269990.1 |
| Chelonia mydas | XP_037735001.1 |
| Sus scrofa | XP_020927659.1 |
| Equus caballus | XP_023502669.1 |
| Xenopus laevis | XP_018095208.1 |
| Oryctolagus cuniculus | XP_017206143.1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cheng, P.; Chen, Z.; Xu, W.; Wang, N.; Yang, Q.; Shi, R.; Li, X.; Cui, Z.; Cheng, J.; Chen, S. Potential Involvement of ewsr1-w Gene in Ovarian Development of Chinese Tongue Sole, Cynoglossus semilaevis. Animals 2022, 12, 2503. https://doi.org/10.3390/ani12192503
Cheng P, Chen Z, Xu W, Wang N, Yang Q, Shi R, Li X, Cui Z, Cheng J, Chen S. Potential Involvement of ewsr1-w Gene in Ovarian Development of Chinese Tongue Sole, Cynoglossus semilaevis. Animals. 2022; 12(19):2503. https://doi.org/10.3390/ani12192503
Chicago/Turabian StyleCheng, Peng, Zhangfan Chen, Wenteng Xu, Na Wang, Qian Yang, Rui Shi, Xihong Li, Zhongkai Cui, Jiayu Cheng, and Songlin Chen. 2022. "Potential Involvement of ewsr1-w Gene in Ovarian Development of Chinese Tongue Sole, Cynoglossus semilaevis" Animals 12, no. 19: 2503. https://doi.org/10.3390/ani12192503
APA StyleCheng, P., Chen, Z., Xu, W., Wang, N., Yang, Q., Shi, R., Li, X., Cui, Z., Cheng, J., & Chen, S. (2022). Potential Involvement of ewsr1-w Gene in Ovarian Development of Chinese Tongue Sole, Cynoglossus semilaevis. Animals, 12(19), 2503. https://doi.org/10.3390/ani12192503






