Transcriptome Analyses Provide Insights into the Auditory Function in Trachemys scripta elegans
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
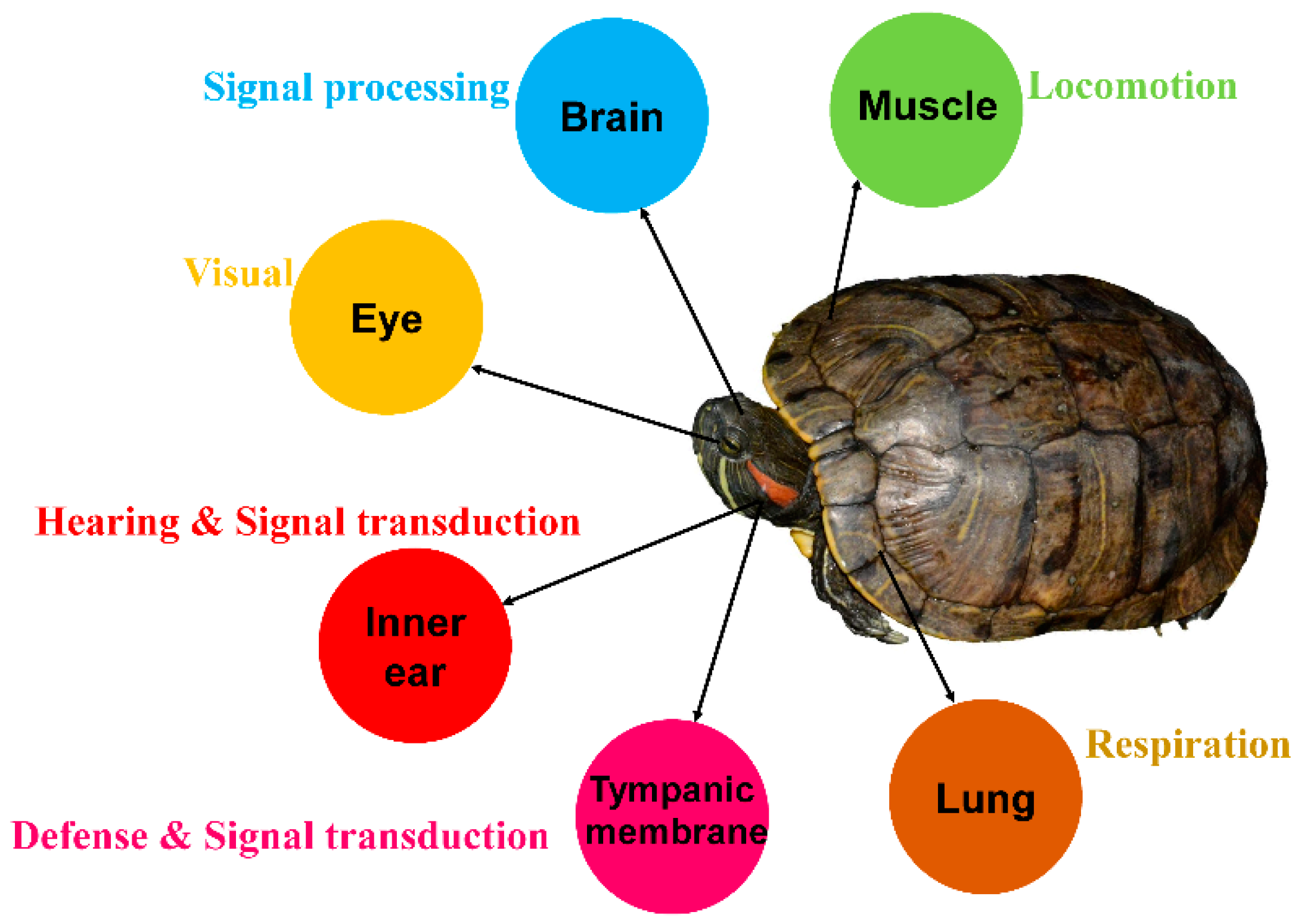
2.1. Collecting Samples
2.2. RNA Extraction and Illumina Sequencing
2.3. Transcriptomic Analyses
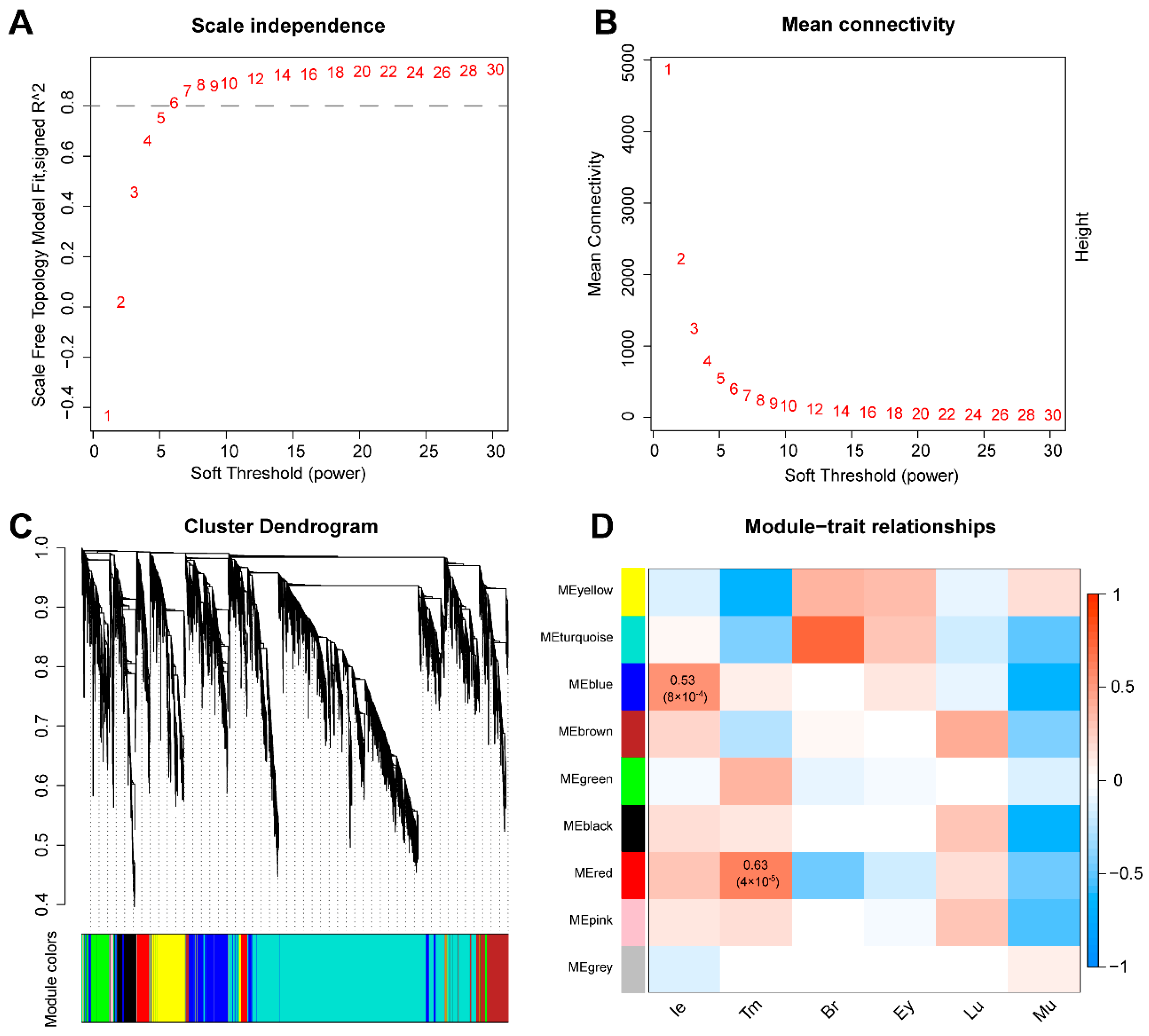
2.4. Weighted Gene Co-Expression Network Analysis (WGCNA)
2.5. Screening of Differently Expressed Genes (DEGs)
2.6. Enrichment Analysis of Gene Functions
3. Results
3.1. Transcriptome Sequencing Analysis
3.2. Co-Expression Network Construction
3.3. Functional Enrichment Analysis of Significant Modules
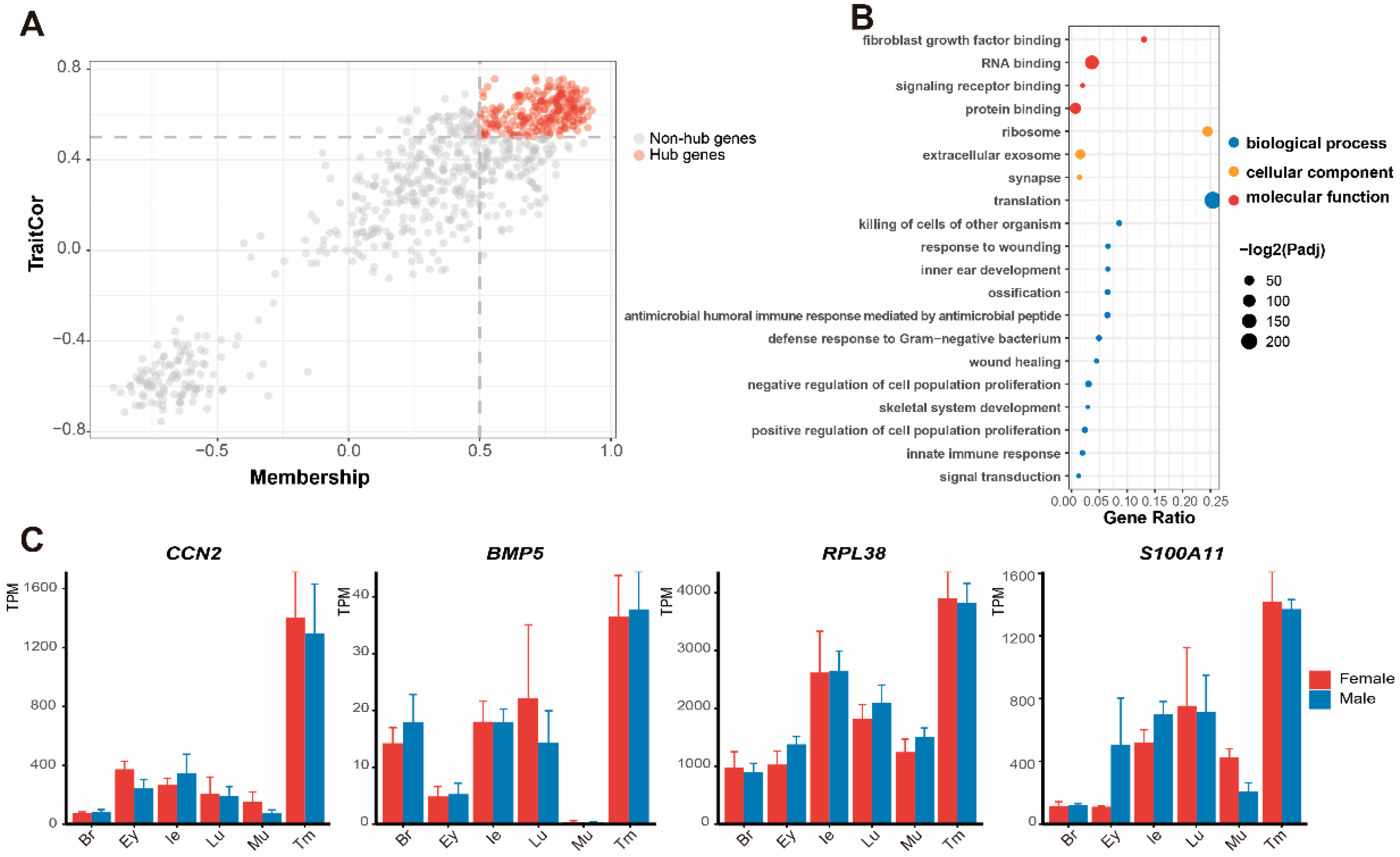
3.4. Expression of DEGs and Screening of Genes in Pathways
4. Discussion
4.1. Molecular Functions in the Inner Ear
4.2. Molecular Function of the Tympanic Membrane
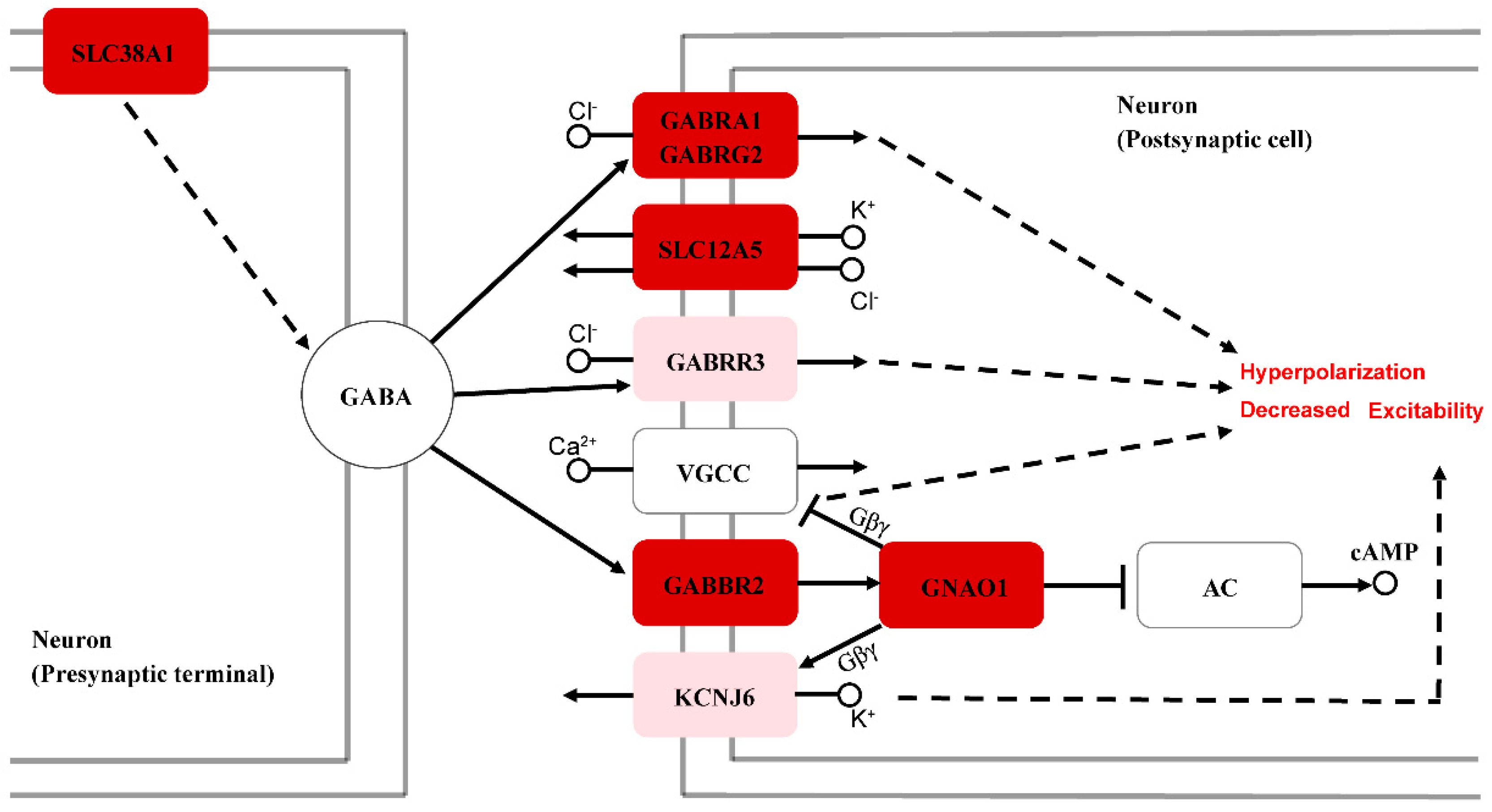
4.3. Molecular Mechanisms Underlying the Differences in Hearing Sensitivity between Sexes
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lee, N.; Christensen-Dalsgaard, J.; White, L.A.; Schrode, K.M.; Bee, M.A. Lung mediated auditory contrast enhancement improves the Signal-to-noise ratio for communication in frogs. Curr. Biol. 2021, 31, 1488–1498. [Google Scholar] [CrossRef] [PubMed]
- Yu, K.; Wood, W.E.; Theunissen, F.E. High-capacity auditory memory for vocal communication in a social songbird. Sci. Adv. 2020, 6, eabe0440. [Google Scholar] [CrossRef] [PubMed]
- Egnor, S.R.; Seagraves, K.M. The contribution of ultrasonic vocalizations to mouse courtship. Curr. Opin. Neurobiol. 2016, 38, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Manley, G.A. Some aspects of the evolution of hearing in vertebrates. Nature 1971, 230, 506–509. [Google Scholar] [CrossRef] [PubMed]
- Flaherty, M.; Libert, K.; Monson, B.B. Extended high-frequency hearing and head orientation cues benefit children during speech-in-speech recognition. Hear. Res. 2021, 406, 108230. [Google Scholar] [CrossRef]
- Baotic, A.; Sicks, F.; Stoeger, A.S. Nocturnal “humming” vocalizations: Adding a piece to the puzzle of giraffe vocal communication. BMC Res. Notes 2015, 8, 425. [Google Scholar] [CrossRef]
- Nachtigall, P.E.; Supin, A.Y.; Smith, A.B.; Pacini, A.F. Expectancy and conditioned hearing levels in the bottlenose dolphin (Tursiops truncatus). J. Exp. Biol. 2016, 219, 844–850. [Google Scholar] [CrossRef]
- Heffner, R.S.; Koay, G.; Heffner, H.E. Hearing in American leaf-nosed bats. IV: The Common vampire bat, Desmodus rotundus. Hear. Res. 2013, 296, 42–50. [Google Scholar] [CrossRef]
- Simmons, A.M.; Hom, K.N.; Simmons, J.A. Big brown bats (Eptesicus fuscus) maintain hearing sensitivity after exposure to intense band-limited noise. J. Acoust. Soc. Am. 2017, 141, 1481. [Google Scholar] [CrossRef]
- Duque, F.G.; Carruth, L.L. Vocal communication in hummingbirds. Brain Behav. Evol. 2022, 97, 241–252. [Google Scholar] [CrossRef]
- Feng, A.S.; Narins, P.M. Ultrasonic communication in concave-eared torrent frogs (Amolops tormotus). J. Comp. Physiol. A Neuroethol. Sens. Neural Behav. Physiol. 2008, 194, 159–167. [Google Scholar] [CrossRef] [PubMed]
- Chapuis, L.; Kerr, C.C.; Collin, S.P.; Hart, N.S.; Sanders, K.L. Underwater hearing in sea snakes (Hydrophiinae): First evidence of auditory evoked potential thresholds. J. Exp. Biol. 2019, 222, jeb198184. [Google Scholar] [CrossRef] [PubMed]
- Berman, D.S.; Regal, P.J. The loss of the ophidian middleear. Evolution 1967, 21, 641–643. [Google Scholar] [CrossRef] [PubMed]
- Wever, E.G.; Vernon, J.A. The sensitivity of the turtle’s ear as shown by its electrical potentials. Proc. Natl. Acad. Sci. USA 1956, 42, 213–220. [Google Scholar] [CrossRef] [PubMed]
- Martin, K.J.; Alessi, S.C.; Gaspard, J.C.; Tucker, A.D.; Bauer, G.B.; Mann, D.A. Underwater hearing in the loggerhead turtle (Caretta caretta): A comparison of behavioral and auditory evoked potential audiograms. J. Exp. Biol. 2012, 215, 3001–3009. [Google Scholar] [CrossRef]
- Piniak, W.E.; Mann, D.A.; Harms, C.A.; Jones, T.T.; Eckert, S.A. Hearing in the Juvenile Green Sea Turtle (Chelonia mydas): A Comparison of Underwater and Aerial Hearing Using Auditory Evoked Potentials. PLoS ONE 2016, 11, e0159711. [Google Scholar] [CrossRef]
- Ridgway, S.H.; Wever, E.G.; McCormick, J.G.; Palin, J.; Anderson, J.H. Hearing in the giant sea turtle, Chelonia mydas. Proc. Natl. Acad. Sci. USA 1969, 64, 884–890. [Google Scholar] [CrossRef]
- Fay, R.R.; Popper, A.N. Evolution of hearing in vertebrates: The inner ears and processing. Hear. Res. 2000, 149, 1–10. [Google Scholar] [CrossRef]
- Anthwal, N.; Thompson, H. The development of the mammalian outer and middle ear. J. Anat. 2016, 228, 217–232. [Google Scholar] [CrossRef]
- Van Dijk, P.; Mason, M.J.; Schoffelen, R.L.; Narins, P.M.; Meenderink, S.W. Mechanics of the frog ear. Hear. Res. 2011, 273, 46–58. [Google Scholar] [CrossRef]
- Montefeltro, F.C.; Andrade, D.V.; Larsson, H.C. The evolution of the meatal chamber in crocodyliforms. J. Anat. 2016, 228, 838–863. [Google Scholar] [CrossRef] [PubMed]
- Takechi, M.; Kitazawa, T.; Hirasawa, T.; Hirai, T.; Iseki, S.; Kurihara, H.; Kuratani, S. Developmental mechanisms of the tympanic membrane in mammals and non-mammalian amniotes. Congenit. Anom. 2016, 56, 12–17. [Google Scholar] [CrossRef] [PubMed]
- Christensen-Dalsgaard, J.; Brandt, C.; Willis, K.L.; Christensen, C.B.; Ketten, D.; Edds-Walton, P.; Fay, R.R.; Madsen, P.T.; Carr, C.E. Specialization for underwater hearing by the tympanic middle ear of the turtle, Trachemys scripta elegans. Proc. R. Soc. B 2012, 279, 2816–2824. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Corey, D.P. An inner ear gene expression database. J. Assoc. Res. Otolaryngol. 2002, 3, 140–148. [Google Scholar] [CrossRef]
- Cho, Y.; Gong, T.W.; Stöver, T.; Lomax, M.I.; Altschuler, R.A. Gene expression profiles of the rat cochlea, cochlear nucleus, and inferior colliculus. J. Assoc. Res. Otolaryngol. 2002, 3, 54–67. [Google Scholar] [CrossRef]
- Chen, Z.; Corey, D.P. Understanding inner ear development with gene expression profiling. J. Neurobiol. 2002, 53, 276–285. [Google Scholar] [CrossRef]
- Schimmang, T.; Maconochie, M. Gene expression profiling of the inner ear. J. Anat. 2016, 228, 255–269. [Google Scholar] [CrossRef]
- Dong, D.; Lei, M.; Liu, Y.; Zhang, S. Comparative inner ear transcriptome analysis between the Rickett’s big-footed bats (Myotis ricketti) and the greater short-nosed fruit bats (Cynopterus sphinx). BMC Genom. 2013, 14, 916. [Google Scholar] [CrossRef]
- Chen, Z.; Liu, Y.; Liang, R.; Cui, C.; Zhu, Y.; Zhang, F.; Zhang, J.; Chen, X. Comparative transcriptome analysis provides insights into the molecular mechanisms of high-frequency hearing differences between the sexes of Odorrana tormota. BMC Genom. 2022, 23, 296. [Google Scholar] [CrossRef]
- Wang, T.; Li, H.; Cui, J.; Zhai, X.; Shi, H.; Wang, J. Auditory brainstem responses in the red-eared slider Trachemys scripta elegans (Testudoformes: Emydidae) reveal sexually dimorphic hearing sensitivity. J. Comp. Physiol. A Neuroethol. Sens. Neural Behav. Physiol. 2019, 205, 847–854. [Google Scholar] [CrossRef] [Green Version]
- Zhu, W.; Liu, L.; Wang, X.; Gao, X.; Jiang, J.; Wang, B. Transcriptomics reveals the molecular processes of light-induced rapid darkening of the non-obligate cave dweller Oreolalax rhodostigmatus (Megophryidae, Anura) and their genetic basis of pigmentation strategy. BMC Genom. 2018, 19, 422. [Google Scholar] [CrossRef] [PubMed]
- Brian Simison, W.; Parham, J.F.; Papenfuss, T.J.; Lam, A.W.; Henderson, J.B. An Annotated Chromosome-Level Reference Genome of the Red-Eared Slider Turtle (Trachemys scripta elegans). Genome Biol. Evol. 2020, 12, 456–462. [Google Scholar] [CrossRef] [PubMed]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Bu, D.; Luo, H.; Huo, P.; Wang, Z.; Zhang, S.; He, Z.; Wu, Y.; Zhao, L.; Liu, J.; Guo, J.; et al. KOBAS-i: Intelligent prioritization and exploratory visualization of biological functions for gene enrichment analysis. Nucleic Acids Res. 2021, 49, W317–W325. [Google Scholar] [CrossRef] [PubMed]
- Brown, R.; Groves, A.K. Hear, Hear for Notch: Control of Cell Fates in the Inner Ear by Notch Signaling. Biomolecules 2020, 10, 370. [Google Scholar] [CrossRef]
- Spokony, R.F.; Aoki, Y.; Saint-Germain, N.; Magner-Fink, E.; Saint-Jeannet, J.P. The transcription factor Sox9 is required for cranial neural crest development in Xenopus. Development 2002, 129, 421–432. [Google Scholar] [CrossRef]
- Cheung, M.; Briscoe, J. Neural crest development is regulated by the transcription factor Sox9. Development 2003, 130, 5681–5693. [Google Scholar] [CrossRef]
- Saint-Germain, N.; Lee, Y.H.; Zhang, Y.; Sargent, T.D.; Saint-Jeannet, J.P. Specification of the otic placode depends on Sox9 function in Xenopus. Development 2004, 131, 1755–1763. [Google Scholar] [CrossRef]
- Bagheri-Fam, S.; Barrionuevo, F.; Dohrmann, U.; Günther, T.; Schüle, R.; Kemler, R.; Mallo, M.; Kanzler, B.; Scherer, G. Long-range upstream and downstream enhancers control distinct subsets of the complex spatiotemporal Sox9 expression pattern. Dev. Biol. 2006, 291, 382–397. [Google Scholar] [CrossRef] [Green Version]
- Pirvola, U.; Ylikoski, J.; Trokovic, R.; Hébert, J.M.; McConnell, S.K.; Partanen, J. FGFR1 is required for the development of the auditory sensory epithelium. Neuron 2002, 35, 671–680. [Google Scholar] [CrossRef]
- Liu, W.; Rask-Andersen, H. Na/K-ATPase Gene Expression in the Human Cochlea: A Study Using mRNA in situ Hybridization and Super-Resolution Structured Illumination Microscopy. Front. Mol. Neurosci. 2022, 15, 857216. [Google Scholar] [CrossRef] [PubMed]
- Pivovarov, A.S.; Calahorro, F.; Walker, R.J. Na(+)/K(+)-pump and neurotransmitter membrane receptors. Invertebr. Neurosci. 2018, 19, 1. [Google Scholar] [CrossRef] [PubMed]
- Kamaid, A.; Neves, J.; Giráldez, F. Id gene regulation and function in the prosensory domains of the chicken inner ear: A link between Bmp signaling and Atoh1. J. Neurosci. 2010, 30, 11426–11434. [Google Scholar] [CrossRef] [PubMed]
- Jones, J.M.; Montcouquiol, M.; Dabdoub, A.; Woods, C.; Kelley, M.W. Inhibitors of differentiation and DNA binding (Ids) regulate Math1 and hair cell formation during the development of the organ of Corti. J. Neurosci. 2006, 26, 550–558. [Google Scholar] [CrossRef]
- Ichimiya, I.; Kurono, Y.; Mogi, G. Immunological potential of the tympanic membrane. Observation under normal and inflammatory conditions. Am. J. Otolaryngol. 1997, 18, 165–172. [Google Scholar] [CrossRef]
- Kingsley, D.M. The TGF-beta superfamily: New members, new receptors, and new genetic tests of function in different organisms. Genes Dev. 1994, 8, 133–146. [Google Scholar] [CrossRef]
- King, J.A.; Marker, P.C.; Seung, K.J.; Kingsley, D.M. BMP5 and the molecular, skeletal, and soft-tissue alterations in short ear mice. Dev. Biol. 1994, 166, 112–122. [Google Scholar] [CrossRef]
- Noben-Trauth, K.; Latoche, J.R. Ectopic mineralization in the middle ear and chronic otitis media with effusion caused by RPL38 deficiency in the Tail-short (Ts) mouse. J. Biol. Chem. 2011, 286, 3079–3093. [Google Scholar] [CrossRef]
- Ramazani, Y.; Knops, N.; Elmonem, M.A.; Nguyen, T.Q.; Arcolino, F.O.; van den Heuvel, L.; Levtchenko, E.; Kuypers, D.; Goldschmeding, R. Connective tissue growth factor (CTGF) from basics to clinics. Matrix Biol. 2018, 68–69, 44–66. [Google Scholar] [CrossRef]
- Zhang, L.; Zhu, T.; Miao, H.; Liang, B. The Calcium Binding Protein S100A11 and Its Roles in Diseases. Front. Cell Dev. Biol. 2021, 9, 693262. [Google Scholar] [CrossRef] [PubMed]
- He, H.; Li, J.; Weng, S.; Li, M.; Yu, Y. S100A11: Diverse function and pathology corresponding to different target proteins. Cell Biochem. Biophys. 2009, 55, 117–126. [Google Scholar] [CrossRef] [PubMed]
- Ben-Ari, Y.; Gaiarsa, J.L.; Tyzio, R.; Khazipov, R. GABA: A pioneer transmitter that excites immature neurons and generates primitive oscillations. Physiol. Rev. 2007, 87, 1215–1284. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Sun, H.; Li, X.; Li, Y.; Zhao, S.; Zhang, D.; Yao, Z.; Li, J. A local GABAergic system is functionally expressed in human fallopian tube. Biochem. Biophys. Res. Commun. 2010, 398, 237–241. [Google Scholar] [CrossRef]
- Lu, W.; Bromley-Coolidge, S.; Li, J. Regulation of GABAergic synapse development by postsynaptic membrane proteins. Brain Res. Bull. 2017, 129, 30–42. [Google Scholar] [CrossRef]
- Farrant, M.; Nusser, Z. Variations on an inhibitory theme: Phasic and tonic activation of GABA(A) receptors. Nat. Rev. Neurosci. 2005, 6, 215–229. [Google Scholar] [CrossRef]
- Rivera, C.; Voipio, J.; Kaila, K. Two developmental switches in GABAergic signalling: The K+-Cl− cotransporter KCC2 and carbonic anhydrase CAVII. J. Physiol. 2005, 562, 27–36. [Google Scholar] [CrossRef]
- Kaila, K.; Price, T.J.; Payne, J.A.; Puskarjov, M.; Voipio, J. Cation-chloride cotransporters in neuronal development, plasticity and disease. Nat. Rev. Neurosci. 2014, 15, 637–654. [Google Scholar] [CrossRef]
- Bormann, J. Electrophysiology of GABAA and GABAB receptor subtypes. Trends Neurosci. 1988, 11, 112–116. [Google Scholar] [CrossRef]
- Padgett, C.L.; Slesinger, P.A. GABAB Receptor Coupling to G-proteins and Ion Channels. Adv. Pharmacol. 2010, 58, 123–147. [Google Scholar]
- Zurawski, Z.; Page, B.; Chicka, M.C.; Brindley, R.L.; Wells, C.A.; Preininger, A.M.; Hyde, K.; Gilbert, J.A.; Cruz-Rodriguez, O.; Currie, K.P.M.; et al. Gbetagamma directly modulates vesicle fusion by competing with synaptotagmin for binding to neuronal SNARE proteins embedded in membranes. J. Biol. Chem. 2017, 292, 12165–12177. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huff, R.M.; Axton, J.M.; Neer, E.J. Physical and immunological characterization of a guanine nucleotide-binding protein purified from bovine cerebral cortex. J. Biol. Chem. 1985, 260, 10864–10871. [Google Scholar] [CrossRef]
- Feng, H.; Khalil, S.; Neubig, R.R.; Sidiropoulos, C. A mechanistic review on GNAO1-associated movement disorder. Neurobiol. Dis. 2018, 116, 131–141. [Google Scholar] [CrossRef] [PubMed]
- Logothetis, D.E.; Kurachi, Y.; Galper, J.; Neer, E.J.; Clapham, D.E. The beta gamma subunits of GTP-binding proteins activate the muscarinic K+ channel in heart. Nature 1987, 325, 321–326. [Google Scholar] [CrossRef]
- Whorton, M.R.; MacKinnon, R. X-ray structure of the mammalian GIRK2-betagamma G-protein complex. Nature 2013, 498, 190–197. [Google Scholar] [CrossRef]
- Hibino, H.; Inanobe, A.; Furutani, K.; Murakami, S.; Findlay, I.; Kurachi, Y. Inwardly rectifying potassium channels: Their structure, function, and physiological roles. Physiol. Rev. 2010, 90, 291–366. [Google Scholar] [CrossRef]
- Lüscher, C.; Slesinger, P.A. Emerging roles for G protein-gated inwardly rectifying potassium (GIRK) channels in health and disease. Nat. Rev. Neurosci. 2010, 11, 301–315. [Google Scholar] [CrossRef]
- Dascal, N. Signalling via the G protein-activated K+ channels. Cell. Signal. 1997, 9, 551–573. [Google Scholar] [CrossRef]
- Cortez, M.A.; Shen, L.; Wu, Y.; Aleem, I.S.; Trepanier, C.H.; Sadeghnia, H.R.; Ashraf, A.; Kanawaty, A.; Liu, C.C.; Stewart, L.; et al. Infantile spasms and Down syndrome: A new animal model. Pediatric Res. 2009, 65, 499–503. [Google Scholar] [CrossRef]
- Bormann, J.; Feigenspan, A. GABAC receptors. Trends Neurosci. 1995, 18, 515–519. [Google Scholar] [CrossRef]
- Qureshi, T.; Sørensen, C.; Berghuis, P.; Jensen, V.; Dobszay, M.B.; Farkas, T.; Dalen, K.T.; Guo, C.; Hassel, B.; Utheim, T.P.; et al. The Glutamine Transporter Slc38a1 Regulates GABAergic Neurotransmission and Synaptic Plasticity. Cereb. Cortex 2019, 29, 5166–5179. [Google Scholar] [CrossRef] [PubMed]
- Qureshi, T.; Bjørkmo, M.; Nordengen, K.; Gundersen, V.; Utheim, T.P.; Watne, L.O.; Storm-Mathisen, J.; Hassel, B.; Chaudhry, F.A. Slc38a1 Conveys Astroglia-Derived Glutamine into GABAergic Interneurons for Neurotransmitter GABA Synthesis. Cells 2020, 9, 1686. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lu, N.; Chen, B.; Qing, J.; Lei, J.; Wang, T.; Shi, H.; Wang, J. Transcriptome Analyses Provide Insights into the Auditory Function in Trachemys scripta elegans. Animals 2022, 12, 2410. https://doi.org/10.3390/ani12182410
Lu N, Chen B, Qing J, Lei J, Wang T, Shi H, Wang J. Transcriptome Analyses Provide Insights into the Auditory Function in Trachemys scripta elegans. Animals. 2022; 12(18):2410. https://doi.org/10.3390/ani12182410
Chicago/Turabian StyleLu, Ningning, Bo Chen, Jiao Qing, Jinhong Lei, Tongliang Wang, Haitao Shi, and Jichao Wang. 2022. "Transcriptome Analyses Provide Insights into the Auditory Function in Trachemys scripta elegans" Animals 12, no. 18: 2410. https://doi.org/10.3390/ani12182410




