Investigation of Sperm and Seminal Plasma Candidate MicroRNAs of Bulls with Differing Fertility and In Silico Prediction of miRNA-mRNA Interaction Network of Reproductive Function
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Semen Sample Processing
2.2.1. Individual Progressive Motility (%)
2.2.2. Abnormal Sperm (%)
2.3. RNA Isolation
2.3.1. Sperm
2.3.2. Seminal Plasma
2.4. Complementary DNA Synthesis of Sperm and Seminal Plasma miRNAs
2.5. Sperm and Seminal Plasma Mature Mirna Profiling Using Real-Time PCR
2.6. Bovine Mature miRNA PCR Array Analysis
2.7. Bioinformatics Analysis
2.7.1. Conserved Nucleotide Sequences
2.7.2. Identification of Target and Predicted Genes of Differentially Expressed miRNAs
2.7.3. Construction of Protein-Protein Interaction Network and Screening of Hub Genes
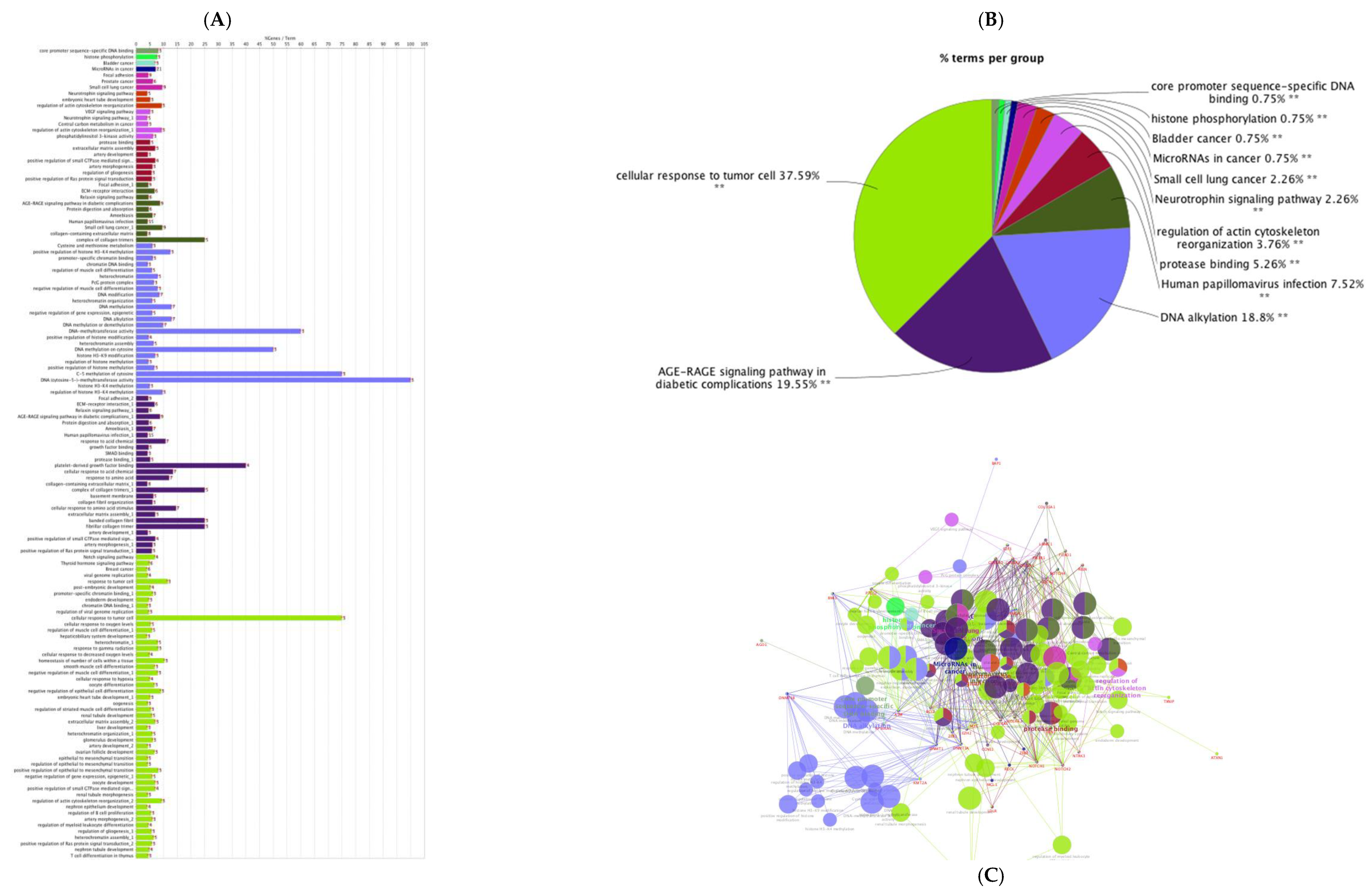
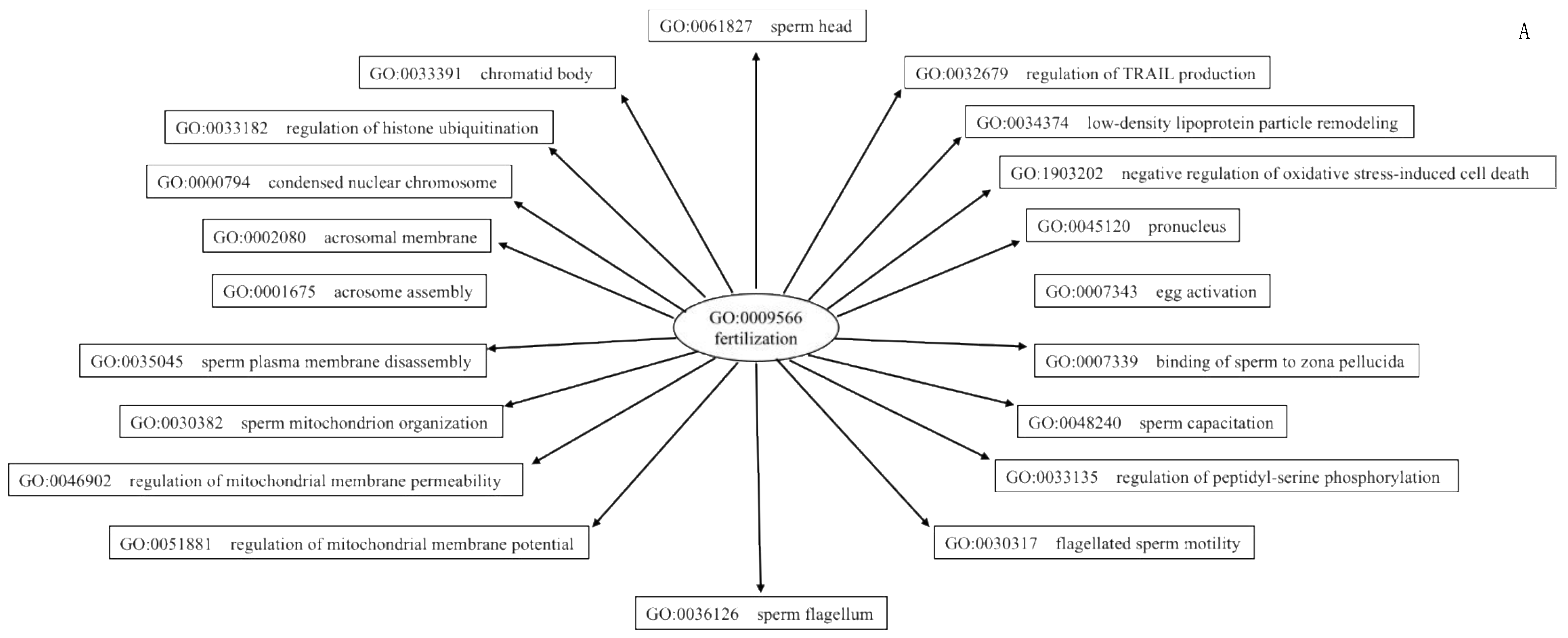
2.7.4. Gene Ontology and Functional Annotation Analysis
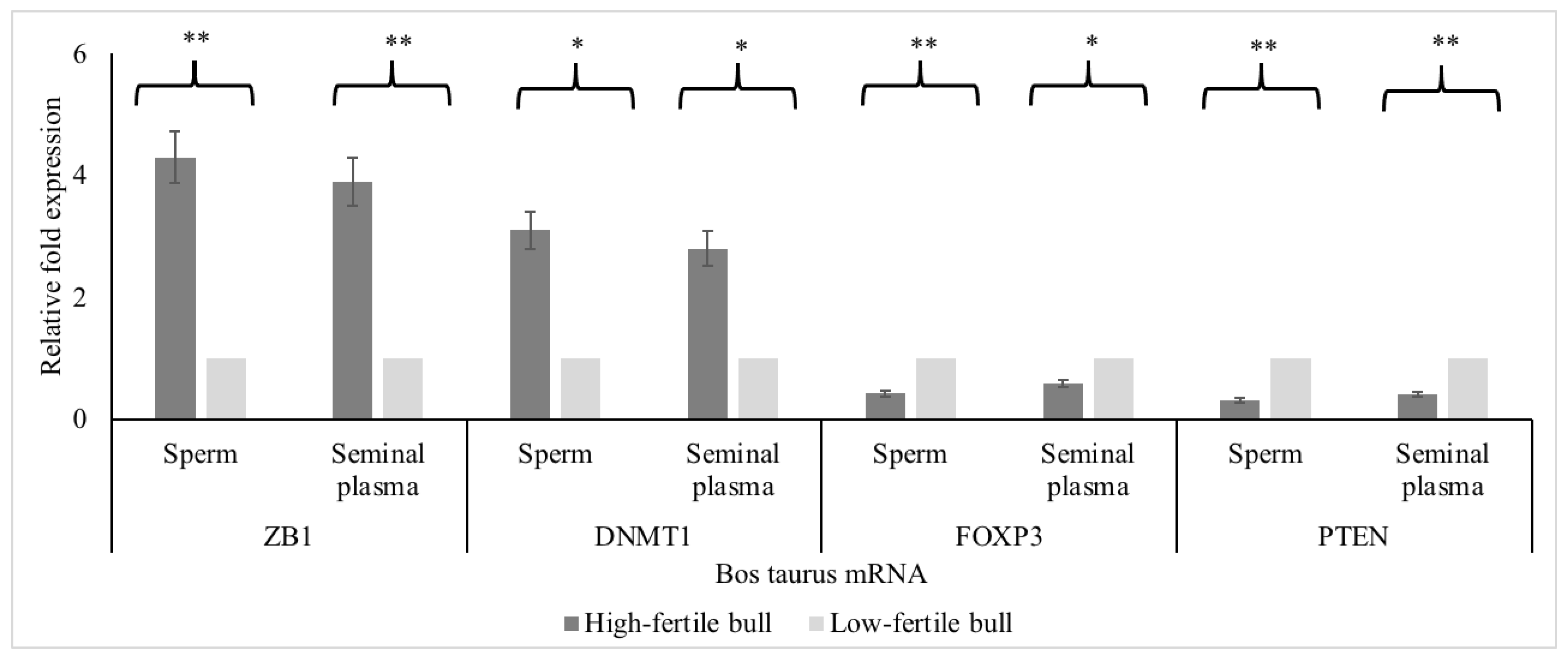
2.7.5. Real-Time Polymerase Chain Reaction for Determining mRNA Expression of Hub Genes
2.7.6. Protein Immunoblots
2.7.7. Statistical Analyses to Determine Differences in mRNA Expression
3. Results
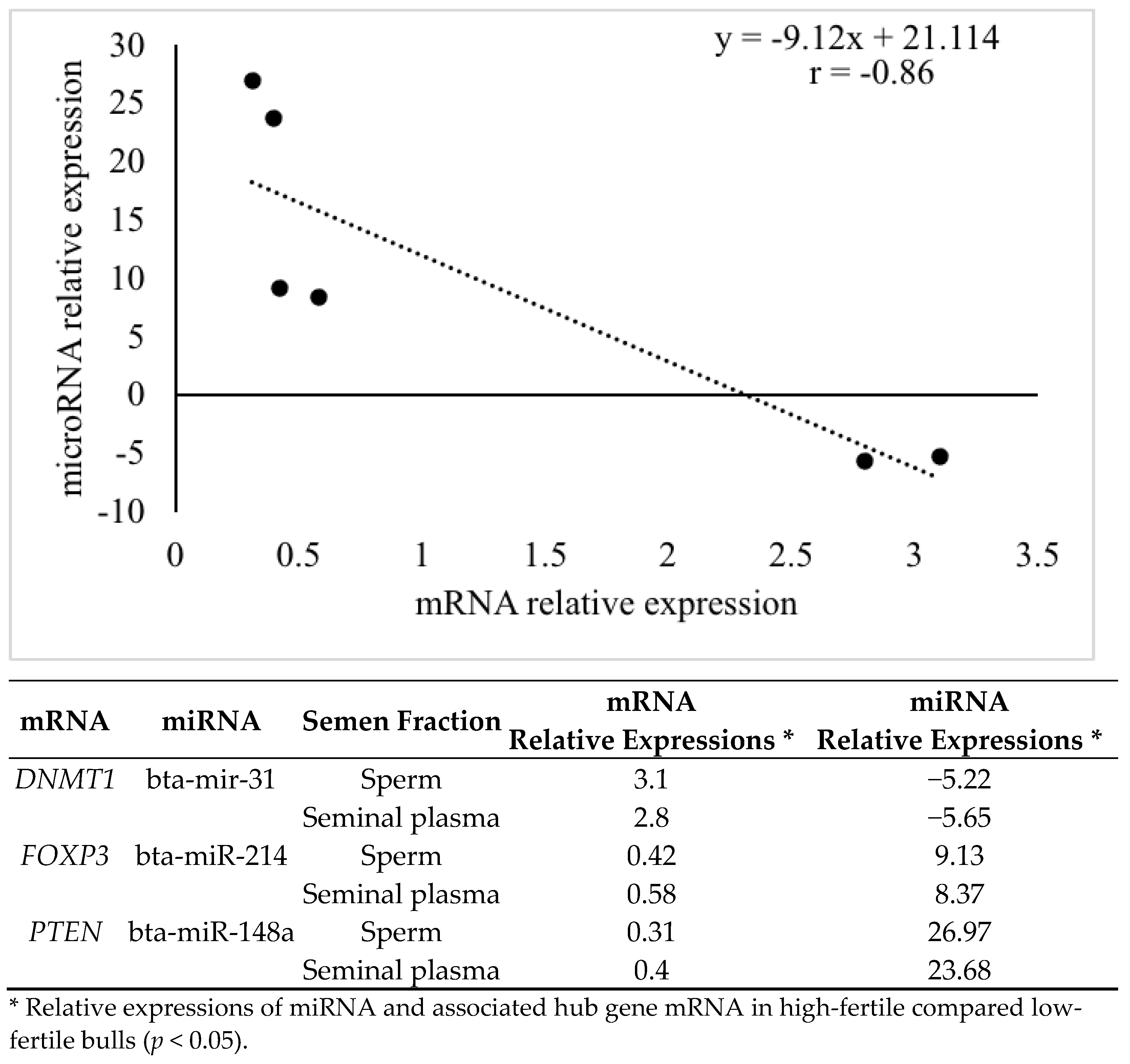
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fang, X.; Qin, L.; Yu, H.; Jiang, P.; Xia, L.; Gao, Z.; Yang, R.; Zhao, Y.; Yu, X.; Zhao, Z. Comprehensive Analysis of miRNAs and Target mRNAs between Immature and Mature Testis Tissue in Chinese Red Steppes Cattle. Animals 2021, 11, 3024. [Google Scholar] [CrossRef] [PubMed]
- Kasimanickam, V.R.; Kasimanickam, R.K. Differential expression of microRNAs in sexually immature and mature canine testes. Theriogenology 2015, 83, 394–398. [Google Scholar] [CrossRef] [PubMed]
- Vendrell-Flotats, M.; García-Martínez, T.; Martínez-Rodero, I.; López-Béjar, M.; LaMarre, J.; Yeste, M.; Mogas, T. In vitro maturation in the presence of Leukemia Inhibitory Factor modulates gene and miRNA expression in bovine oocytes and embryos. Sci. Rep. 2020, 10, 17777. [Google Scholar] [CrossRef] [PubMed]
- Costa, M.C.; Leitão, A.L.; Enguita, F.J. MicroRNA Profiling in Plasma or Serum Using Quantitative RT-PCR. RNA Mapp. 2014, 1182, 121–129. [Google Scholar] [CrossRef]
- Kasimanickam, V.; Kastelic, J. Circulating cell-free mature microRNAs and their target gene prediction in bovine metritis. Sci. Rep. 2016, 6, 29509. [Google Scholar] [CrossRef]
- Roberts, T.C.; Coenen-Stass, A.M.L.; Betts, C.A.; Wood, M.J.A. Detection and quantification of extracellular microRNAs in murine biofluids. Biol. Proced. Online 2014, 16, 5. [Google Scholar] [CrossRef]
- Weber, J.A.; Baxter, D.H.; Zhang, S.; Huang, D.Y.; Huang, K.H.; Lee, M.J.; Galas, D.J.; Wang, K. The MicroRNA Spectrum in 12 Body Fluids. Clin. Chem. 2010, 56, 1733–1741. [Google Scholar] [CrossRef] [PubMed]
- Turchinovich, A.; Weiz, L.; Burwinkel, B. Extracellular miRNAs: The mystery of their origin and function. Trends Biochem. Sci. 2012, 37, 460–465. [Google Scholar] [CrossRef]
- Kasimanickam, V.; Kasimanickam, R. An Efficient Approach for RNA Extraction from Boar Sperm and Seminal Plasma. Bio-Protocol 2019, 9, e3284. [Google Scholar] [CrossRef]
- Kasimanickam, V.; Buhr, M.; Kasimanickam, R. Patterns of expression of sperm and seminal plasma microRNAs in boar semen. Theriogenology 2019, 125, 87–92. [Google Scholar] [CrossRef]
- Rodríguez-Martínez, H.; Kvist, U.; Ernerudh, J.; Sanz, L.; Calvete, J.J. Seminal Plasma Proteins: What role do they play? Am. J. Reprod. Immunol. 2011, 66 (Suppl. 1), 11–22. [Google Scholar] [CrossRef] [PubMed]
- Caballero, I.; Parrilla, I.; Almiñana, C.; del Olmo, D.; Roca, J.; Martínez, E.; Vázquez, J. Seminal plasma proteins as modulators of the sperm function and their application in sperm biotechnologies. Reprod. Domest. Anim. 2012, 47 (Suppl. S3), 12–21. [Google Scholar] [CrossRef]
- Chabory, E.; Damon, C.; Lenoir, A.; Henry-Berger, J.; Vernet, P.; Cadet, R.; Saez, F.; Drevet, J.R. Mammalian glutathione peroxidases control acquisition and maintenance of spermatozoa integrity 1. J. Anim. Sci. 2010, 88, 1321–1331. [Google Scholar] [CrossRef] [PubMed]
- Juyena, N.S.; Stelletta, C. Seminal Plasma: An Essential Attribute to Spermatozoa. J. Androl. 2012, 33, 536–551. [Google Scholar] [CrossRef] [PubMed]
- Mann, T. The Biochemistry of Semen and of the Male Reproductive Tract, 2nd ed.; Methuen: London, UK, 1964. [Google Scholar]
- Taylor, A.; Robson, A.; Houghton, B.C.; Jepson, C.A.; Ford, W.C.L.; Frayne, J. Epididymal specific, selenium-independent GPX5 protects cells from oxidative stress-induced lipid peroxidation and DNA mutation. Hum. Reprod. 2013, 28, 2332–2342. [Google Scholar] [CrossRef] [PubMed]
- Fagerlind, M.; Stålhammar, H.; Olsson, B.; Klinga-Levan, K. Expression of miRNAs in Bull Spermatozoa Correlates with Fertility Rates. Reprod. Domest. Anim. 2015, 50, 587–594. [Google Scholar] [CrossRef]
- Selth, L.A.; Roberts, M.; Chow, C.W.K.; Marshall, V.R.; Doi, S.; Vincent, A.D.; Butler, L.; Lavin, M.; Tilley, W.D.; Gardiner, R.A. Human seminal fluid as a source of prostate cancer-specific microRNA biomarkers. Endocr.-Relat. Cancer 2014, 21, L17–L21. [Google Scholar] [CrossRef]
- Du, Y.; Wang, X.; Wang, B.; Chen, W.; He, R.; Zhang, L.; Xing, X.; Su, J.; Wang, Y.; Zhang, Y. Deep sequencing analysis of microRNAs in bovine sperm. Mol. Reprod. Dev. 2014, 81, 1042–1052. [Google Scholar] [CrossRef]
- Wu, W.; Hu, Z.; Qin, Y.; Dong, J.; Dai, J.; Lu, C.; Zhang, W.; Shen, H.; Xia, Y.; Wang, X. Seminal plasma microRNAs: Potential biomarkers for spermatogenesis status. Mol. Hum. Reprod. 2012, 18, 489–497. [Google Scholar] [CrossRef]
- Turri, F.; Capra, E.; Lazzari, B.; Cremonesi, P.; Stella, A.; Pizzi, F. A Combined Flow Cytometric Semen Analysis and miRNA Profiling as a Tool to Discriminate Between High- and Low-Fertility Bulls. Front. Vet. Sci. 2021, 8, 703101. [Google Scholar] [CrossRef]
- Nixon, B.; Stanger, S.J.; Mihalas, B.P.; Reilly, J.N.; Anderson, A.L.; Tyagi, S.; Holt, J.E.; McLaughlin, E.A. The microRNA signature of mouse spermatozoa is substantially modified during epididymal maturation. Biol. Reprod. 2015, 93, 91. [Google Scholar] [CrossRef] [PubMed]
- Jodar, M. Sperm and seminal plasma RNAs: What roles do they play beyond fertilization? Reproduction 2019, 158, R113–R123. [Google Scholar] [CrossRef] [PubMed]
- Sharma, U.; Sun, F.; Conine, C.C.; Reichholf, B.; Kukreja, S.; Herzog, V.A.; Ameres, S.L.; Rando, O.J. Small RNAs are trafficked from the epididymis to developing mammalian sperm. Dev. Cell 2018, 46, 481–494.e6. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.-M.; Pang, R.T.K.; Chiu, P.C.N.; Wong, B.P.C.; Lao, K.; Lee, K.-F.; Yeung, W.S.B. Sperm-borne microRNA-34c is required for the first cleavage division in mouse. Proc. Natl. Acad. Sci. USA 2012, 109, 490–494. [Google Scholar] [CrossRef] [PubMed]
- Yuan, S.; Schuster, A.; Tang, C.; Yu, T.; Ortogero, N.; Bao, J.; Zheng, H.; Yan, W. Sperm-borne miRNAs and endo-siRNAs are important for fertilization and preimplantation embryonic development. Development 2016, 143, 635–647. [Google Scholar] [CrossRef]
- Kozomara, A.; Birgaoanu, M.; Griffiths-Jones, S. miRBase: From microRNA sequences to function. Nucleic Acids Res. 2019, 47, D155–D162. [Google Scholar] [CrossRef] [PubMed]
- Kozomara, A.; Griffiths-Jones, S. miRBase: Annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014, 42, D68–D73. [Google Scholar] [CrossRef]
- Chang, L.; Zhou, G.; Soufan, O.; Xia, J. miRNet 2.0: Network-based visual analytics for miRNA functional analysis and systems biology. Nucleic Acids Res. 2020, 48, W244–W251. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Nastou, K.C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.T.; Legeay, M.; Fang, T.; Bork, P.; et al. The STRING database in 2021: Customizable protein–protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021, 49, D605–D612. [Google Scholar] [CrossRef]
- Gustavsen, J.A.; Pai, S.; Isserlin, R.; Demchak, B.; Pico, A.R. RCy3: Network biology using Cytoscape from within R. F1000Research 2019, 8, 1774. [Google Scholar] [CrossRef]
- Chin, C.-H.; Chen, S.-H.; Wu, H.-H.; Ho, C.-W.; Ko, M.-T.; Lin, C.-Y. cytoHubba: Identifying hub objects and sub-networks from complex interactome. BMC Syst. Biol. 2014, 8 (Suppl. 4), S11. [Google Scholar] [CrossRef] [PubMed]
- Bindea, G.; Mlecnik, B.; Hackl, H.; Charoentong, P.; Tosolini, M.; Kirilovsky, A.; Fridman, W.-H.; Pagès, F.; Trajanoski, Z.; Galon, J. ClueGO: A Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 2009, 25, 1091–1093. [Google Scholar] [CrossRef] [PubMed]
- Kasimanickam, V.R.; Kasimanickam, R.K.; Kastelic, J.P.; Stevenson, J.S. Associations of adiponectin and fertility estimates in Holstein bulls. Theriogenology 2013, 79, 766–777.e3. [Google Scholar] [CrossRef] [PubMed]
- Kasimanickam, R.; Kasimanickam, V.; Arangasamy, A.; Kastelic, J. Associations of hypoosmotic swelling test, relative sperm volume shift, aquaporin7 mRNA abundance and bull fertility estimates. Theriogenology 2017, 89, 162–168. [Google Scholar] [CrossRef] [PubMed]
- Kasimanickam, V.R.; Kasimanickam, R.K. Sertoli, Leydig, and Spermatogonial Cells’ Specific Gene and Protein Expressions as Dog Testes Evolve from Immature into Mature States. Animals 2022, 12, 271. [Google Scholar] [CrossRef]
- Wang, S.; Ma, G.; Zhu, H.; Lv, C.; Chu, H.; Tong, N.; Wu, D.; Qiang, F.; Gong, W.; Zhao, Q.; et al. miR-107 regulates tumor progression by targeting NF1 in gastric cancer. Sci. Rep. 2016, 6, 36531. [Google Scholar] [CrossRef]
- Sellem, E.; Marthey, S.; Rau, A.; Jouneau, L.; Bonnet, A.; Perrier, J.-P.; Fritz, S.; Le Danvic, C.; Boussaha, M.; Kiefer, H.; et al. A comprehensive overview of bull sperm-borne small non-coding RNAs and their diversity across breeds. Epigenet. Chromatin 2020, 13, 19. [Google Scholar] [CrossRef]
- Liu, T.; Cheng, W.; Gao, Y.; Wang, H.; Liu, Z. Microarray analysis of microRNA expression patterns in the semen of infertile men with semen abnormalities. Mol. Med. Rep. 2012, 6, 535–542. [Google Scholar] [CrossRef]
- Khor, E.-S.; Noor, S.M.; Wong, P.-F. MiR-107 inhibits the sprouting of intersegmental vessels of zebrafish embryos. Protoplasma 2022, 259, 691–702. [Google Scholar] [CrossRef]
- Abu-Halima, M.; Hammadeh, M.; Schmitt, J.; Leidinger, P.; Keller, A.; Meese, E.; Backes, C. Altered microRNA expression profiles of human spermatozoa in patients with different spermatogenic impairments. Fertil. Steril. 2013, 99, 1249–1255.e16. [Google Scholar] [CrossRef]
- Chen, Z.; Lai, T.-C.; Jan, Y.-H.; Lin, F.-M.; Wang, W.-C.; Xiao, H.; Wang, Y.-T.; Sun, W.; Cui, X.; Li, Y.-S.; et al. Hypoxia-responsive miRNAs target argonaute 1 to promote angiogenesis. J. Clin. Investig. 2013, 123, 1057–1067. [Google Scholar] [CrossRef] [PubMed]
- Meinhardt, G.; Haider, S.; Kunihs, V.; Saleh, L.; Pollheimer, J.; Fiala, C.; Hetey, S.; Feher, Z.; Szilagyi, A.; Than, N.G.; et al. Pivotal role of the transcriptional co-activator YAP in trophoblast stemness of the developing human placenta. Proc. Natl. Acad. Sci. USA 2020, 117, 13562–13570. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Shin, K.-T.; Cui, X.-S. Analysis of Cyclin E1 Functions in Porcine Preimplantation Embryonic Development by Fluorescence Microscopy. Microsc. Microanal. 2017, 23, 69–76. [Google Scholar] [CrossRef]
- Boissière, A.; Gala, A.; Ferrières-Hoa, A.; Mullet, T.; Baillet, S.; Petiton, A.; Torre, A.; Hamamah, S. Cell-free and intracellular nucleic acids: New non-invasive biomarkers to explore male infertility. Basic Clin. Androl. 2017, 27, 7. [Google Scholar] [CrossRef] [PubMed]
- Leung, C.; Zhu, M.; Zernicka-Goetz, M. Polarity in Cell-Fate Acquisition in the Early Mouse Embryo. Curr. Top. Dev. Biol. 2016, 120, 203–234. [Google Scholar] [CrossRef] [PubMed]
- Ngondo, R.P.; Cirera-Salinas, D.; Yu, J.; Wischnewski, H.; Bodak, M.; Vandormael-Pournin, S.; Geiselmann, A.; Wettstein, R.; Luitz, J.; Cohen-Tannoudji, M.; et al. Argonaute 2 Is Required for Extra-embryonic Endoderm Differentiation of Mouse Embryonic Stem Cells. Stem Cell Rep. 2018, 10, 461–476. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Xiang, C.; Zheng, X. miR-132 serves as a diagnostic biomarker in gestational diabetes mellitus and its regulatory effect on trophoblast cell viability. Diagn. Pathol. 2019, 14, 119. [Google Scholar] [CrossRef]
- Kagedan, D.; Lecker, I.; Batruch, I.; Smith, C.; Kaploun, I.; Lo, K.; Grober, E.; Diamandis, E.P.; Jarvi, K.A. Characterization of the seminal plasma proteome in men with prostatitis by mass spectrometry. Clin. Proteom. 2012, 9, 2. [Google Scholar] [CrossRef]
- Ding, R.; Guo, F.; Zhang, Y.; Liu, X.-M.; Xiang, Y.-Q.; Zhang, C.; Liu, Z.-W.; Sheng, J.-Z.; Huang, H.-F.; Zhang, J.; et al. Integrated Transcriptome Sequencing Analysis Reveals Role of miR-138-5p/ TBL1X in Placenta from Gestational Diabetes Mellitus. Cell. Physiol. Biochem. 2018, 51, 630–646. [Google Scholar] [CrossRef]
- Yin, A.; Chen, Q.; Zhong, M.; Jia, B. MicroRNA-138 improves LPS-induced trophoblast dysfunction through targeting RELA and NF-κB signaling. Cell Cycle 2021, 20, 508–521. [Google Scholar] [CrossRef]
- Zhang, J.Y.; Dong, H.S.; Oqani, R.K.; Lin, T.; Kang, J.W.; Jin, D.I. Distinct roles of ROCK1 and ROCK2 during development of porcine preimplantation embryos. Reproduction 2014, 148, 99–107. [Google Scholar] [CrossRef] [PubMed]
- Morton, S.U.; Scherz, P.J.; Cordes, K.R.; Ivey, K.N.; Stainier, D.Y.R.; Srivastava, D. microRNA-138 modulates cardiac patterning during embryonic development. Proc. Natl. Acad. Sci. USA 2008, 105, 17830–17835. [Google Scholar] [CrossRef] [PubMed]
- Barceló, M.; Castells, M.; Bassas, L.; Vigués, F.; Larriba, S. Semen miRNAs Contained in Exosomes as Non-Invasive Biomarkers for Prostate Cancer Diagnosis. Sci. Rep. 2019, 9, 13772. [Google Scholar] [CrossRef] [PubMed]
- Tesfaye, D.; Worku, D.; Rings, F.; Phatsara, C.; Tholen, E.; Schellander, K.; Hoelker, M. Identification and expression profiling of microRNAs during bovine oocyte maturation using heterologous approach. Mol. Reprod. Dev. 2009, 76, 665–677. [Google Scholar] [CrossRef] [PubMed]
- Schrader, M.; Müller-Tidow, C.; Ravnik, S.; Müller, M.; Schulze, W.; Diederichs, S.; Serve, H.; Miller, K. Cyclin A1 and gametogenesis in fertile and infertile patients: A potential new molecular diagnostic marker. Hum. Reprod. 2002, 17, 2338–2343. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Murphy, M.; Stinnakre, M.-G.; Senamaud-Beaufort, C.; Winston, N.J.; Sweeney, C.; Kubelka, M.; Carrington, M.; Bréchot, C.; Sobczak-Thépot, J. Delayed early embryonic lethality following disruption of the murine cyclin A2 gene. Nat. Genet. 1997, 15, 83–86. [Google Scholar] [CrossRef]
- Kasimanickam, R.; Kasimanickam, V. mRNA Expressions of Candidate Genes in Gestational Day 16 Conceptus and Corresponding Endometrium in Repeat Breeder Dairy Cows with Suboptimal Uterine Environment Following Transfer of Different Quality Day 7 Embryos. Animals 2020, 11, 1092. [Google Scholar] [CrossRef]
- Ibrahim, L.A.; Rizo, J.A.; Fontes, P.L.P.; Lamb, G.C.; Bromfield, J.J. Seminal plasma modulates expression of endometrial inflammatory meditators in the bovine†. Biol. Reprod. 2019, 100, 660–671. [Google Scholar] [CrossRef]
- Li, X.; Ma, X.; Tian, F.; Wu, F.; Zhang, J.; Zeng, W.; Lin, Y.; Zhang, Y. Downregulation of CCNA2 disturbs trophoblast migration, proliferation, and apoptosis during the pathogenesis of recurrent miscarriage. Am. J. Reprod. Immunol. 2019, 82, e13144. [Google Scholar] [CrossRef]
- Stanton, B.R.; Perkins, A.S.; Tessarollo, L.; Sassoon, D.A.; Parada, L.F. Loss of N-myc function results in embryonic lethality and failure of the epithelial component of the embryo to develop. Genes Dev. 1992, 6, 2235–2247. [Google Scholar] [CrossRef]
- Abu-Halima, M.; Galata, V.; Backes, C.; Keller, A.; Hammadeh, M.; Meese, E. MicroRNA signature in spermatozoa and seminal plasma of proven fertile men and in testicular tissue of men with obstructive azoospermia. Andrologia 2020, 52, e13503. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.-C.; Kuo, M.-W.; Yu, J.; Kuo, H.-H.; Lin, R.-J.; Lo, W.-L.; Yu, A. c-Myb Is an Evolutionary Conserved miR-150 Target and miR-150/c-Myb Interaction Is Important for Embryonic Development. Mol. Biol. Evol. 2008, 25, 2189–2198. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Lagah, S.V.; Nagoorvali, D.; Kumar, B.B.; Singh, M.K.; Singla, S.K.; Manik, R.S.; Palta, P.; Chauhan, M.S. Supplementation of Glial Cell Line-Derived Neurotrophic Factor, Fibroblast Growth Factor 2, and Epidermal Growth Factor Promotes Self-Renewal of Putative Buffalo (Bubalus bubalis) Spermatogonial Stem Cells by Upregulating the Expression of miR-20b, miR-21, and miR-106a. Cell. Reprogram. 2019, 21, 11–17. [Google Scholar] [CrossRef] [PubMed]
- Gad, A.; Sánchez, J.M.; Browne, J.A.; Nemcova, L.; Laurincik, J.; Prochazka, R.; Lonergan, P. Plasma extracellular vesicle miRNAs as potential biomarkers of superstimulatory response in cattle. Sci. Rep. 2020, 10, 19130. [Google Scholar] [CrossRef]
- Lee, J.; Lee, S.; Son, J.; Lim, H.; Kim, E.; Kim, D.; Ha, S.; Hur, T.; Lee, S.; Choi, I. Analysis of circulating-microRNA expression in lactating Holstein cows under summer heat stress. PLoS ONE 2020, 15, e0231125. [Google Scholar] [CrossRef]
- Carreras-Badosa, G.; Bonmatí, A.; Ortega, F.J.; Mercader, J.M.; Guindo-Martínez, M.; Torrents, D.; Prats-Puig, A.; Martinez-Calcerrada, J.-M.; De Zegher, F.; Ibanez, L.; et al. Dysregulation of Placental miRNA in Maternal Obesity Is Associated with Pre- and Postnatal Growth. J. Clin. Endocrinol. Metab. 2017, 102, 2584–2594. [Google Scholar] [CrossRef]
- Chan, O.C.; Chow, P.H. Total ablation of paternal accessory sex glands curtails developmental potential in preimplantation embryos in the golden hamster. Anat. Embryol. 2001, 204, 117–122. [Google Scholar] [CrossRef]
- Martinez, R.M.; Liang, L.; Racowsky, C.; Dioni, L.; Mansur, A.; Adir, M.; Bollati, V.; Baccarelli, A.A.; Hauser, R.; Machtinger, R. Extracellular microRNAs profile in human follicular fluid and IVF outcomes. Sci. Rep. 2018, 8, 17036. [Google Scholar] [CrossRef]
- Nadkarni, S.; Smith, J.; Sferruzzi-Perri, A.N.; Ledwozyw, A.; Kishore, M.; Haas, R.; Mauro, C.; Williams, D.J.; Farsky, S.H.P.; Marelli-Berg, F.M.; et al. Neutrophils induce proangiogenic T cells with a regulatory phenotype in pregnancy. Proc. Natl. Acad. Sci. USA 2016, 113, E8415–E8424. [Google Scholar] [CrossRef]
- Martinez, C.A.; Cambra, J.M.; Parrilla, I.; Roca, J.; Ferreira-Dias, G.; Pallares, F.J.; Lucas, X.; Vazquez, J.M.; Martinez, E.A.; Gil, M.A.; et al. Seminal plasma modifies the transcriptional pattern of the endometrium and advances embryo development in pigs. Front. Vet. Sci. 2019, 6, 465. [Google Scholar] [CrossRef]
- Robertson, S.A.; Care, A.S.; Moldenhauer, L.M. Regulatory T cells in embryo implantation and the immune response to pregnancy. J. Clin. Investig. 2018, 128, 4224–4235. [Google Scholar] [CrossRef]
- Robertson, S.A.; Guerin, L.R.; Moldenhauer, L.M.; Hayball, J.D. Activating T regulatory cells for tolerance in early pregnancy—The contribution of seminal fluid. J. Reprod. Immunol. 2009, 83, 109–116. [Google Scholar] [CrossRef] [PubMed]
- O’Flaherty, C.; de Lamirande, E.; Gagnon, C. Phosphorylation of the Arginine-X-X-(Serine/Threonine) motif in human sperm proteins during capacitation: Modulation and protein kinase A dependency. Mol. Hum. Reprod. 2004, 10, 355–363. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.Z.; Tschopp, O.; Hemmings-Mieszczak, M.; Feng, J.; Brodbeck, D.; Perentes, E.; Hemmings, B.A. Protein kinase B alpha/Akt1 regulates placental development and fetal growth. J. Biol. Chem. 2003, 278, 32124–32131. [Google Scholar] [CrossRef] [PubMed]
- Albert, C.; Luque, G.M.; Courchamp, F. The twenty most charismatic species. PLoS ONE 2014, 9, e99433. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Yin, Z.J.; Feng, Y.F.; Zhang, X.D.; Wu, T.; Ding, Y.Y.; Ye, P.F.; Fu, K.; Zhang, M.Q. Identification and differential expression of microRNAs in the ovaries of pigs (Sus scrofa) with high and low litter sizes. Anim. Genet. 2016, 47, 543–551. [Google Scholar] [CrossRef]
- Wang, W.; Peng, M.; Yuan, H.; Liu, C.; Zhang, Y.; Fang, Y.; Su, Y.; Zhang, X.; Zhang, H.; Tang, Y.; et al. Studying the mechanism of sperm DNA damage caused by folate deficiency. J. Cell. Mol. Med. 2022, 26, 776–788. [Google Scholar] [CrossRef]
- Hu, L.; Wu, C.; Guo, C.; Li, H.; Xiong, C. Identification of microRNAs predominately derived from testis and epididymis in human seminal plasma. Clin. Biochem. 2014, 47, 967–972. [Google Scholar] [CrossRef]
- Kresowik, J.D.; Devor, E.J.; Van Voorhis, B.J.; Leslie, K.K. MicroRNA-31 is Significantly Elevated in Both Human Endometrium and Serum During the Window of Implantation: A Potential Biomarker for Optimum Receptivity1. Biol. Reprod. 2014, 91, 17. [Google Scholar] [CrossRef] [PubMed]
- Rouas, R.; Fayyad-Kazan, H.; El Zein, N.; Lewalle, P.; Rothé, F.; Simion, A.; Akl, H.; Mourtada, M.; El Rifai, M.; Burny, A.; et al. Human natural Treg microRNA signature: Role of microRNA-31 and microRNA-21 in FOXP3 expression. Eur. J. Immunol. 2009, 39, 1608–1618. [Google Scholar] [CrossRef]
- Abu-Halima, M.; Abu Khaizaran, Z.; Ayesh, B.M.; Fischer, U.; Abu Khaizaran, S.; Al-Battah, F.; Hammadeh, M.; Keller, A.; Meese, E. MicroRNAs in combined spent culture media and sperm are associated with embryo quality and pregnancy outcome. Fertil. Steril. 2020, 113, 970–980.e2. [Google Scholar] [CrossRef] [PubMed]
- Xia, H.-F.; Jin, X.-H.; Song, P.-P.; Cui, Y.; Liu, C.-M.; Ma, X. Temporal and Spatial Regulation of miR-320 in the Uterus during Embryo Implantation in the Rat. Int. J. Mol. Sci. 2010, 11, 719–730. [Google Scholar] [CrossRef] [PubMed]
- Gross, N.; Kropp, J.; Khatib, H. Sexual Dimorphism of miRNAs Secreted by Bovine In vitro-produced Embryos. Front. Genet. 2017, 8, 39. [Google Scholar] [CrossRef]
- Zhou, R.; Wang, R.; Qin, Y.; Ji, J.; Xu, M.; Wu, W.; Chen, M.; Wu, D.; Song, L.; Shen, H.; et al. Mitochondria-related miR-151a-5p reduces cellular ATP production by targeting CYTB in asthenozoospermia. Sci. Rep. 2015, 5, 17743. [Google Scholar] [CrossRef]
- Zou, Y.; Jiang, Z.; Yu, X.; Zhang, Y.; Sun, M.; Wang, W.; Ge, Z.; De, W.; Sun, L. MiR-101 regulates apoptosis of trophoblast HTR-8/SVneo cells by targeting endoplasmic reticulum (ER) protein 44 during preeclampsia. J. Hum. Hypertens. 2014, 28, 610–616. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Ruan, Y.C.; Guo, J.; Chen, H.; Tsang, L.L.; Zhang, X.; Jiang, C.; Chan, H.C. Regulation of miR-101/miR-199a-3p by the epithelial sodium channel during embryo implantation: Involvement of CREB phosphorylation. Reproduction 2014, 148, 559–568. [Google Scholar] [CrossRef] [PubMed]
- Ruan, Y.C.; Guo, J.H.; Liu, X.; Zhang, R.; Tsang, L.L.; Da Dong, J.; Chen, H.; Yu, M.K.; Jiang, X.; Zhang, X.H.; et al. Activation of the epithelial Na+ channel triggers prostaglandin E2 release and production required for embryo implantation. Nat. Med. 2012, 18, 1112–1117. [Google Scholar] [CrossRef]
- Farina, F.M.; Hall, I.F.; Serio, S.; Zani, S.; Climent, M.; Salvarani, N.; Carullo, P.; Civilini, E.; Condorelli, G.; Elia, L.; et al. miR-128-3p Is a Novel Regulator of Vascular Smooth Muscle Cell Phenotypic Switch and Vascular Diseases. Circ. Res. 2020, 126, e120–e135. [Google Scholar] [CrossRef]
- Li, J.; Aung, L.H.H.; Long, B.; Qin, D.; An, S.; Li, P. miR-23a binds to p53 and enhances its association with miR-128 promoter. Sci. Rep. 2015, 5, 16422. [Google Scholar] [CrossRef]
- Ponsuksili, S.; Tesfaye, D.; Schellander, K.; Hoelker, M.; Hadlich, F.; Schwerin, M.; Wimmers, K. Differential Expression of miRNAs and Their Target mRNAs in Endometria Prior to Maternal Recognition of Pregnancy Associates with Endometrial Receptivity for In Vivo- and In Vitro-Produced Bovine Embryos1. Biol. Reprod. 2014, 91, 135. [Google Scholar] [CrossRef]
- Marques, C.J.; Pinho, M.J.; Carvalho, F.; Bieche, I.; Barros, A.; Sousa, M. DNA methylation imprinting marks and DNA methyltransferase expression in human spermatogenic cell stages. Epigenetics 2011, 6, 1354–1361. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Ko, Y.; Park, H.; Zhang, H.; Jeong, Y.; Kim, Y.; Noh, M.; Park, S.; Kim, Y.-M.; Kwon, Y.-G. MicroRNA-148a/b-3p regulates angiogenesis by targeting neuropilin-1 in endothelial cells. Exp. Mol. Med. 2019, 51, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Frazier, S.; McBride, M.W.; Mulvana, H.; Graham, D. From animal models to patients: The role of placental microRNAs, miR-210, miR-126, and miR-148a/152 in preeclampsia. Clin. Sci. 2020, 134, 1001–1025. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Ni, T.; Dang, Y.; Ding, L.; Jiang, J.; Li, J.; Xia, M.; Yu, N.; Ma, J.; Yan, J.; et al. MiR-148a-3p may contribute to flawed decidualization in recurrent implantation failure by modulating HOXC8. J. Assist. Reprod. Genet. 2020, 37, 2535–2544. [Google Scholar] [CrossRef]
- Miao, H.; Miao, C.; Han, J.; Li, N. Downregulation of miR-200a Protects Mouse Leydig Cells Against Triptolide by Triggering Autophagy. Drug Des. Dev. Ther. 2020, 14, 4845–4854. [Google Scholar] [CrossRef]
- Berardi, E.; Pues, M.; Thorrez, L.; Sampaolesi, M. miRNAs in ESC differentiation. Am. J. Physiol. Circ. Physiol. 2012, 303, H931–H939. [Google Scholar] [CrossRef]
- Shen, L.-J.; He, J.-L.; Yang, D.-H.; Ding, Y.-B.; Chen, X.-M.; Geng, Y.-Q.; Liu, S.-J.; Liu, X.-Q.; Wang, Y.-X. Mmu-microRNA-200a Overexpression Leads to Implantation Defect by Targeting Phosphatase and Tensin Homolog in Mouse Uterus. Reprod. Sci. 2013, 20, 1518–1528. [Google Scholar] [CrossRef]
- Zheng, Q.; Zhang, D.; Yang, Y.U.; Cui, X.; Sun, J.; Liang, C.; Qin, H.; Yang, X.; Liu, S.; Yan, Q. MicroRNA-200c impairs uterine receptivity formation by targeting FUT4 and α1,3-fucosylation. Cell Death Differ. 2017, 24, 2161–2172. [Google Scholar] [CrossRef]
- Saha, S.; Choudhury, J.; Ain, R. MicroRNA-141-3p and miR-200a-3p regulate insulin-like growth factor 2 during mouse placental development. Mol. Cell. Endocrinol. 2015, 414, 186–193. [Google Scholar] [CrossRef]
- Yu, B.; Chen, X.; Li, J.; Gu, Q.; Zhu, Z.; Li, C.; Su, L.; Liu, B. microRNA-29c inhibits cell proliferation by targeting NASP in human gastric cancer. BMC Cancer 2017, 17, 109. [Google Scholar] [CrossRef]
- Long, M.; Wan, X.; LA, X.; Gong, X.; Cai, X. miR-29c is downregulated in the ectopic endometrium and exerts its effects on endometrial cell proliferation, apoptosis and invasion by targeting c-Jun. Int. J. Mol. Med. 2015, 35, 1119–1125. [Google Scholar] [CrossRef] [PubMed]
- Griffiths, M.; Van Sinderen, M.; Rainczuk, K.; Dimitriadis, E. miR-29c overexpression and COL4A1 downregulation in infertile human endometrium reduces endometrial epithelial cell adhesive capacity in vitro implying roles in receptivity. Sci. Rep. 2019, 9, 8644. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.; Zou, Q.-Y.; Li, H.; Wang, R.-F.; Liu, A.-X.; Magness, R.R.; Zheng, J. Preeclampsia Downregulates MicroRNAs in Fetal Endothelial Cells: Roles of miR-29a/c-3p in Endothelial Function. J. Clin. Endocrinol. Metab. 2017, 102, 3470–3479. [Google Scholar] [CrossRef] [PubMed]
- Bao, J.; Li, D.; Wang, L.; Wu, J.; Hu, Y.; Wang, Z.; Chen, Y.; Cao, X.; Jiang, C.; Yan, W.; et al. MicroRNA-449 and MicroRNA-34b/c Function Redundantly in Murine Testes by Targeting E2F Transcription Factor-Retinoblastoma Protein (E2F-pRb) Pathway. J. Biol. Chem. 2012, 287, 21686–21698. [Google Scholar] [CrossRef]
- Yuan, S.; Tang, C.; Zhang, Y.; Wu, J.; Bao, J.; Zheng, H.; Xu, C.; Yan, W. mir-34b/c and mir-449a/b/c are required for spermatogenesis, but not for the first cleavage division in mice. Biol. Open 2015, 4, 212–223. [Google Scholar] [CrossRef]
- Eikmans, M.; Anholts, J.D.H.; Blijleven, L.; Meuleman, T.; Van Beelen, E.; Van Der Hoorn, M.-L.P.; Claas, F.H.J. Optimization of microRNA Acquirement from Seminal Plasma and Identification of Diminished Seminal microRNA-34b as Indicator of Low Semen Concentration. Int. J. Mol. Sci. 2020, 21, 4089. [Google Scholar] [CrossRef]
- Capalbo, A.; Ubaldi, F.M.; Cimadomo, D.; Noli, L.; Khalaf, Y.; Farcomeni, A.; Ilic, D.; Rienzi, L. MicroRNAs in spent blastocyst culture medium are derived from trophectoderm cells and can be explored for human embryo reproductive competence assessment. Fertil. Steril. 2016, 105, 225–235.e3. [Google Scholar] [CrossRef]
- Wu, J.; Bao, J.; Kim, M.; Yuan, S.; Tang, C.; Zheng, H.; Mastick, G.S.; Xu, C.; Yan, W. Two miRNA clusters, miR-34b/c and miR-449, are essential for normal brain development, motile ciliogenesis, and spermatogenesis. Proc. Natl. Acad. Sci. USA 2014, 111, E2851–E2857. [Google Scholar] [CrossRef]
- Rolland, A.; Lavigne, R.; Dauly, C.; Calvel, P.; Kervarrec, C.; Freour, T.; Evrard, B.; Rioux-Leclercq, N.; Auger, J.; Pineau, C. Identification of genital tract markers in the human seminal plasma using an integrative genomics approach. Hum. Reprod. 2013, 28, 199–209. [Google Scholar] [CrossRef]
- Tompkins, A.; Chatterjee, D.; Maddox, M.; Wang, J.; Arciero, E.; Camussi, G.; Quesenberry, P.J.; Renzulli, J.F. The emergence of extracellular vesicles in urology: Fertility, cancer, biomarkers and targeted pharmacotherapy. J. Extracell. Vesicles 2015, 4, 23815. [Google Scholar] [CrossRef]
- Robertson, S.A.; Sharkey, D.J. Seminal fluid and fertility in women. Fertil. Steril. 2016, 106, 511–519. [Google Scholar] [CrossRef] [PubMed]
- Bai, R.; Latifi, Z.; Kusama, K.; Nakamura, K.; Shimada, M.; Imakawa, K. Induction of immune-related gene expression by seminal exosomes in the porcine endometrium. Biochem. Biophys. Res. Commun. 2018, 495, 1094–1101. [Google Scholar] [CrossRef] [PubMed]
- Schjenken, J.; Robertson, S.; Schjenken, J.; Robertson, S. Seminal Fluid and Immune Adaptation for Pregnancy—Comparative Biology in Mammalian Species. Reprod. Domest. Anim. 2014, 49 (Suppl. S3), 27–36. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Yang, C.; Chen, X.; Yao, B.; Yang, C.; Zhu, C.; Li, L.; Wang, J.; Li, X.; Shao, Y.; et al. Altered Profile of Seminal Plasma MicroRNAs in the Molecular Diagnosis of Male Infertility. Clin. Chem. 2011, 57, 1722–1731. [Google Scholar] [CrossRef] [PubMed]
- Abu-Halima, M.; Ludwig, N.; Hart, M.; Leidinger, P.; Backes, C.; Keller, A.; Hammadeh, M.; Meese, E. Altered micro-ribonucleic acid expression profiles of extracellular microvesicles in the seminal plasma of patients with oligoasthenozoospermia. Fertil. Steril. 2016, 106, 1061–1069.e3. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Wu, H.; Shen, D.; Wang, S.; Zhang, L.; Wang, X.; Gao, B.; Wu, T.; Li, B.; Li, K.; et al. Comparative profiling of small RNAs of pig seminal plasma and ejaculated and epididymal sperm. Reproduction 2017, 153, 785–796. [Google Scholar] [CrossRef] [PubMed]
- Dupont, C.; Kappeler, L.; Saget, S.; Grandjean, V.; Lévy, R. Role of miRNA in the Transmission of Metabolic Diseases Associated with Paternal Diet-Induced Obesity. Front. Genet. 2019, 10, 337. [Google Scholar] [CrossRef] [PubMed]
- Faulkner, L.C.; Hopwood, M.L.; Wiltbank, J.N. Seminal Vesiculectomy in Bulls. Reproduction 1968, 16, 179–182. [Google Scholar] [CrossRef][Green Version]
- Ortiz, W.; Rizo, J.; Carvalheira, L.; Ahmed, B.; Cortes, E.E.; Harstine, B.; Bromfield, J.; Hansen, P. Effects of intrauterine infusion of seminal plasma at artificial insemination on fertility of lactating Holstein cows. J. Dairy Sci. 2019, 102, 6587–6594. [Google Scholar] [CrossRef]
- Odhiambo, J.; Poole, D.; Hughes, L.; DeJarnette, J.; Inskeep, E.; Dailey, R. Pregnancy outcome in dairy and beef cattle after artificial insemination and treatment with seminal plasma or transforming growth factor beta-1. Theriogenology 2009, 72, 566–571. [Google Scholar] [CrossRef]
- Plante, G.; Prud’Homme, B.; Fan, J.; Lafleur, M.; Manjunath, P. Evolution and function of mammalian binder of sperm proteins. Cell Tissue Res. 2016, 363, 105–127. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.Y.; Lee, K.; Tang, P.L.; Chow, P.H. Ablation of paternal accessory sex glands is detrimental to embryo development during implantation. Anat. Embryol. 2001, 203, 255–263. [Google Scholar] [CrossRef] [PubMed]
- Gardela, J.; Ruiz-Conca, M.; Wright, D.; López-Béjar, M.; Martínez, C.A.; Rodríguez-Martínez, H.; Álvarez-Rodríguez, M. Semen modulates cell proliferation and differentiation-related transcripts in the pig peri-ovulatory endometrium. Biology 2022, 11, 616. [Google Scholar] [CrossRef]
- Wong, C.; Lee, K.; Lo, K.; Chan, O.; Goggins, W.; Chow, P. Ablation of paternal accessory sex glands imparts physical and behavioural abnormalities to the progeny: An in vivo study in the golden hamster. Theriogenology 2007, 68, 654–662. [Google Scholar] [CrossRef] [PubMed]
- Woidacki, K.; Meyer, N.; Schumacher, A.; Goldschmidt, A.; Maurer, M.; Zenclussen, A.C. Transfer of regulatory T cells into abortion-prone mice promotes the expansion of uterine mast cells and normalizes early pregnancy angiogenesis. Sci. Rep. 2015, 5, 13938. [Google Scholar] [CrossRef]
- Urner, F.; Leppens-Luisier, G.; Sakkas, D. Protein Tyrosine Phosphorylation in Sperm During Gamete Interaction in the Mouse: The Influence of Glucose1. Biol. Reprod. 2001, 64, 1350–1357. [Google Scholar] [CrossRef]
- Stenzinger, A.; Märker, D.; Koch, P.-S.; Hoffmann, J.; Baal, N.; Steger, K.; Wimmer, M. Protein Tyrosine Phosphatase Interacting Protein 51 (PTPIP51) mRNA Expression and Localization and Its In Vitro Interacting Partner Protein Tyrosine Phosphatase 1B (PTP1B) in Human Placenta of the First, Second, and Third Trimester. J. Histochem. Cytochem. 2009, 57, 143–153. [Google Scholar] [CrossRef]
- Chen, M.-C.; Lin, J.-A.; Lin, H.-T.; Chen, S.-Y.; Yen, G.-C. Potential effect of advanced glycation end products (AGEs) on spermatogenesis and sperm quality in rodents. Food Funct. 2019, 10, 3324–3333. [Google Scholar] [CrossRef]
- Alexander, K.L.; Mejia, C.; Jordan, C.; Nelson, M.B.; Howell, B.; Jones, C.M.; Reynolds, P.; Arroyo, J.A. Differential Receptor for Advanced Glycation End Products Expression in Preeclamptic, Intrauterine Growth Restricted, and Gestational Diabetic Placentas. Am. J. Reprod. Immunol. 2016, 75, 172–180. [Google Scholar] [CrossRef]
- Lee, F.S.; Rajagopal, R.; Chao, M.V. Distinctive features of Trk neurotrophin receptor transactivation by G protein-coupled receptors. Cytokine Growth Factor Rev. 2002, 13, 11–17. [Google Scholar] [CrossRef]
- Crown, A.; Clifton, D.K.; Steiner, R.A. Neuropeptide Signaling in the Integration of Metabolism and Reproduction. Neuroendocrinology 2007, 86, 175–182. [Google Scholar] [CrossRef] [PubMed]
- Parrilla, I.; Martinez, E.A.; Gil, M.A.; Cuello, C.; Roca, J.; Rodriguez-Martinez, H.; Martinez, C.A. Boar seminal plasma: Current insights on its potential role for assisted reproductive technologies in swine. Anim. Reprod. 2020, 17, e20200022. [Google Scholar] [CrossRef] [PubMed]
- Allen, C.C.; Alves, B.R.C.; Li, X.; Tedeschi, L.O.; Zhou, H.; Paschal, J.C.; Riggs, P.; Braga-Neto, U.M.; Keisler, D.; Williams, G.L.; et al. Gene expression in the arcuate nucleus of heifers is affected by controlled intake of high- and low-concentrate diets1. J. Anim. Sci. 2012, 90, 2222–2232. [Google Scholar] [CrossRef] [PubMed]
- Paluch, E.; Heisenberg, C.-P. Biology and Physics of Cell Shape Changes in Development. Curr. Biol. 2009, 19, R790–R799. [Google Scholar] [CrossRef]
- Bromfield, J.J.; Schjenken, J.E.; Chin, P.Y.; Care, A.S.; Jasper, M.J.; Robertson, S.A. Maternal tract factors contribute to paternal seminal fluid impact on metabolic phenotype in offspring. Proc. Natl. Acad. Sci. USA 2014, 111, 2200–2205. [Google Scholar] [CrossRef]
- Ata, B.; Abou-Setta, A.M.; Seyhan, A.; Buckett, W. Application of seminal plasma to female genital tract prior to embryo transfer in assisted reproductive technology cycles (IVF, ICSI and frozen embryo transfer). Cochrane Database Syst. Rev. 2018, 2018, CD011809. [Google Scholar] [CrossRef]
- Sellem, E.; Marthey, S.; Rau, A.; Jouneau, L.; Bonnet, A.; Le Danvic, C.; Guyonnet, B.; Kiefer, H.; Jammes, H.; Schibler, L. Dynamics of cattle sperm sncRNAs during maturation, from testis to ejaculated sperm. Epigenet. Chromatin 2021, 14, 24. [Google Scholar] [CrossRef]
- Montalbetti, N.; Li, Q.; Timpanaro, G.A.; González-Perrett, S.; Dai, X.-Q.; Chen, X.-Z.; Cantiello, H.F. Cytoskeletal regulation of calcium-permeable cation channels in the human syncytiotrophoblast: Role of gelsolin. J. Physiol. 2005, 566, 309–325. [Google Scholar] [CrossRef]
- Xia, S.; Lim, Y.B.; Zhang, Z.; Wang, Y.; Zhang, S.; Lim, C.T.; Yim, E.K.; Kanchanawong, P. Nanoscale Architecture of the Cortical Actin Cytoskeleton in Embryonic Stem Cells. Cell Rep. 2019, 28, 1251–1267.e7. [Google Scholar] [CrossRef]
- Soda, T.; Miyagawa, Y.; Fukuhara, S.; Tanaka, H. Physiological role of actin regulation in male fertility: Insight into actin capping proteins in spermatogenic cells. Reprod. Med. Biol. 2020, 19, 120–127. [Google Scholar] [CrossRef]
- Capco, D.G.; Tutnick, J.M.; Bement, W.M. The role of protein kinase C in reorganization of the cortical cytoskeleton during the transition from oocyte to fertilization-competent egg. J. Exp. Zool. 1992, 264, 395–405. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.-E.; Ozawa, M.; Zhang, K.; Johnson, S.E.; Ealy, A.D. The requirement for protein kinase C delta (PRKCD) during preimplantation bovine embryo development. Reprod. Fertil. Dev. 2016, 28, 482–490. [Google Scholar] [CrossRef] [PubMed]
- Bidarimath, M.; Edwards, A.K.; Wessels, J.M.; Khalaj, K.; Kridli, R.T.; Tayade, C. Distinct microRNA expression in endometrial lymphocytes, endometrium, and trophoblast during spontaneous porcine fetal loss. J. Reprod. Immunol. 2015, 107, 64–79. [Google Scholar] [CrossRef] [PubMed]
- Cardoso, R.; Alves, B.R.C.; Prezotto, L.D.; Thorson, J.F.; Tedeschi, L.O.; Keisler, D.H.; Park, C.S.; Amstalden, M.; Williams, G.L. Use of a stair-step compensatory gain nutritional regimen to program the onset of puberty in beef heifers1. J. Anim. Sci. 2014, 92, 2942–2949. [Google Scholar] [CrossRef] [PubMed]
- Gasser, C.L.; Behlke, E.J.; Grum, D.E.; Day, M.L. Effect of timing of feeding a high-concentrate diet on growth and attainment of puberty in early-weaned heifers1. J. Anim. Sci. 2006, 84, 3118–3122. [Google Scholar] [CrossRef] [PubMed]
- Pan, J.-A.; Lin, H.; Yu, J.-Y.; Zhang, H.-L.; Zhang, J.-F.; Wang, C.-Q.; Gu, J. MiR-21-3p Inhibits Adipose Browning by Targeting FGFR1 and Aggravates Atrial Fibrosis in Diabetes. Oxidative Med. Cell. Longev. 2021, 2021, 9987219. [Google Scholar] [CrossRef] [PubMed]
- Thomas, S.; Kratzsch, D.; Schaab, M.; Scholz, M.; Grunewald, S.; Thiery, J.; Paasch, U.; Kratzsch, J. Seminal plasma adipokine levels are correlated with functional characteristics of spermatozoa. Fertil. Steril. 2013, 99, 1256–1263.e3. [Google Scholar] [CrossRef]
- Kasimanickam, R.K.; Kasimanickam, V.R.; Olsen, J.R.; Jeffress, E.J.; Moore, D.A.; Kastelic, J.P. Associations among serum pro- and anti-inflammatory cytokines, metabolic mediators, body condition, and uterine disease in postpartum dairy cows. Reprod. Biol. Endocrinol. 2013, 11, 103. [Google Scholar] [CrossRef]
- Chimento, A.; De Luca, A.; Nocito, M.C.; Avena, P.; La Padula, D.; Zavaglia, L.; Pezzi, V. Role of GPER-Mediated Signaling in Testicular Functions and Tumorigenesis. Cells 2020, 9, 2115. [Google Scholar] [CrossRef]
- de Oliveira, V.; Schaefer, J.; Calder, M.; Lydon, J.P.; Demayo, F.J.; Bhattacharya, M.; Radovick, S.; Babwah, A.V. Uterine Gαq/11 signaling, in a progesterone-dependent manner, critically regulates the acquisition of uterine receptivity in the female mouse. FASEB J. 2019, 33, 9374–9387. [Google Scholar] [CrossRef]
- Yoo, J.-Y.; Ahn, J.I.; Kim, T.H.; Yu, S.; Ahn, J.Y.; Lim, J.M.; Jeong, J.-W. G-protein coupled receptor 64 is required for decidualization of endometrial stromal cells. Sci. Rep. 2017, 7, 5021. [Google Scholar] [CrossRef] [PubMed]
- Layden, B.T.; Newman, M.; Chen, F.; Fisher, A.; Lowe, W.L., Jr. G Protein Coupled Receptors in Embryonic Stem Cells: A Role for Gs-Alpha Signaling. PLoS ONE 2010, 5, e9105. [Google Scholar] [CrossRef] [PubMed]
- Torregrosa-Carrión, R.; Piñeiro-Sabarís, R.; Siguero-Álvarez, M.; Grego-Bessa, J.; Luna-Zurita, L.; Fernandes, V.S.; MacGrogan, D.; Stainier, D.Y.R.; de la Pompa, J.L. Adhesion G protein–coupled receptor Gpr126/Adgrg6 is essential for placental development. Sci. Adv. 2021, 7, eabj5445. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Gao, H.; Li, W.; Liu, C. Essential Role of Histone Replacement and Modifications in Male Fertility. Front. Genet. 2019, 10, 962. [Google Scholar] [CrossRef] [PubMed]
- Couldrey, C.; Carlton, M.B.L.; Nolan, P.; Colledge, W.H.; Evans, M.J. A retroviral Gene Trap Insertion into the Histone 3.3A Gene Causes Partial Neonatal Lethality, Stunted Growth, Neuromuscular Deficits and Male Sub-fertility in Transgenic Mice. Hum. Mol. Genet. 1999, 8, 2489–2495. [Google Scholar] [CrossRef]
- Tang, M.C.W.; Jacobs, S.A.; Mattiske, D.M.; Soh, Y.M.; Graham, A.N.; Tran, A.; Lim, S.L.; Hudson, D.F.; Kalitsis, P.; O’Bryan, M.K.; et al. Contribution of the Two Genes Encoding Histone Variant H3.3 to Viability and Fertility in Mice. PLoS Genet. 2015, 11, e1004964. [Google Scholar] [CrossRef] [PubMed]
- Yuen, B.T.K.; Bush, K.; Barrilleaux, B.L.; Cotterman, R.; Knoepfler, P.S. Histone H3.3 regulates dynamic chromatin states during spermatogenesis. Development 2014, 141, 3483–3494. [Google Scholar] [CrossRef]
- Deppe, M.; Morales, P.; Sánchez, R. Effect of Protease Inhibitors on the Acrosome Reaction and Sperm-Zona Pellucida Binding in Bovine Sperm. Reprod. Domest. Anim. 2008, 43, 713–719. [Google Scholar] [CrossRef]
- Shang, X.; Shen, C.; Liu, J.; Tang, L.; Zhang, H.; Wang, Y.; Wu, W.; Chi, J.; Zhuang, H.; Fei, J.; et al. Serine protease PRSS55 is crucial for male mouse fertility via affecting sperm migration and sperm–egg binding. Cell. Mol. Life Sci. 2018, 75, 4371–4384. [Google Scholar] [CrossRef]
- Chen, C.P.; Posy, S.; Ben-Shaul, A.; Shapiro, L.; Honig, B.H. Specificity of cell–cell adhesion by classical cadherins: Critical role for low-affinity dimerization through β-strand swapping. Proc. Natl. Acad. Sci. USA 2005, 102, 8531–8536. [Google Scholar] [CrossRef]
- Vazquez-Levin, M.H.; Marín-Briggiler, C.I.; Caballero, J.N.; Veiga, M.F. Epithelial and neural cadherin expression in the mammalian reproductive tract and gametes and their participation in fertilization-related events. Dev. Biol. 2015, 401, 2–16. [Google Scholar] [CrossRef] [PubMed]
- Tom, P.F.; Fleming, T.P.; Sheth, B.; Fesenko, I. Cell adhesion in the preimplantation mammalian embryo and its role in trophectoderm differentiation and blastocyst morphogenesis. Front. Biosci. 2001, 6, D1000–D1007. [Google Scholar] [CrossRef]
- Muñoz-Espín, D.; Cañamero, M.; Maraver, A.; Gómez-López, G.; Contreras, J.; Murillo-Cuesta, S.; Rodríguez-Baeza, A.; Varela-Nieto, I.; Ruberte, J.; Collado, M.; et al. Programmed Cell Senescence during Mammalian Embryonic Development. Cell 2013, 155, 1104–1118. [Google Scholar] [CrossRef] [PubMed]
- Barbu, M.; Thompson, D.; Suciu, N.; Voinea, S.; Cretoiu, D.; Predescu, D. The Roles of MicroRNAs in Male Infertility. Int. J. Mol. Sci. 2021, 22, 2910. [Google Scholar] [CrossRef]
- Burrola-Barraza, M.; Hernández-Seáñez, R.; Barcelo-Fimbres, M.; Rodríguez-Almeida, F.; González-Rodríguez, E.; García-Quiñónez, S.; Grado-Ahuir, J.; Moreno-Brito, V. Dicer gene expression during early bovine embryo development. Mol. Reprod. Dev. 2011, 78, 622. [Google Scholar] [CrossRef]
- Sun, M.; Chen, H.; Liu, J.; Tong, C.; Meng, T. MicroRNA-34a inhibits human trophoblast cell invasion by targeting MYC. BMC Cell Biol. 2015, 16, 21. [Google Scholar] [CrossRef]
- Belleannee, C.; Légaré, C.; Calvo, E.; Thimon, V.; Sullivan, R. microRNA signature is altered in both human epididymis and seminal microvesicles following vasectomy. Hum. Reprod. 2013, 28, 1455–1467. [Google Scholar] [CrossRef]
- Reilly, J.N.; McLaughlin, E.; Stanger, S.J.; Anderson, A.L.; Hutcheon, K.; Church, K.; Mihalas, B.; Tyagi, S.; Holt, J.E.; Eamens, A.; et al. Characterisation of mouse epididymosomes reveals a complex profile of microRNAs and a potential mechanism for modification of the sperm epigenome. Sci. Rep. 2016, 6, 31794. [Google Scholar] [CrossRef]
- Sharma, U.; Conine, C.C.; Shea, J.M.; Boskovic, A.; Derr, A.G.; Bing, X.Y.; Belleannee, C.; Kucukural, A.; Serra, R.W.; Sun, F.; et al. Biogenesis and function of tRNA fragments during sperm maturation and fertilization in mammals. Science 2016, 351, 391–396. [Google Scholar] [CrossRef]
- Kasimanickam, R.; Thatcher, C.; Nebel, R.; Cassell, B. Relationships among lipid peroxidation, glutathione peroxidase, superoxide dismutase, sperm parameters, and competitive index in dairy bulls. Theriogenology 2007, 67, 1004–1012. [Google Scholar] [CrossRef]
- Kasimanickam, R.; Nebel, R.; Peeler, I.; Silvia, W.; Wolf, K.; McAllister, A.; Cassell, B. Breed differences in competitive indices of Holstein and Jersey bulls and their association with sperm DNA fragmentation index and plasma membrane integrity. Theriogenology 2006, 66, 1307–1315. [Google Scholar] [CrossRef] [PubMed]
- Keles, E.; Malama, E.; Bozukova, S.; Siuda, M.; Wyck, S.; Witschi, U.; Bauersachs, S.; Bollwein, H. The micro-RNA content of unsorted cryopreserved bovine sperm and its relation to the fertility of sperm after sex-sorting. BMC Genom. 2021, 22, 30. [Google Scholar] [CrossRef] [PubMed]
- Orang, A.V.; Safaralizadeh, R.; Kazemzadeh-Bavili, M. Mechanisms of miRNA-Mediated Gene Regulation from Common Downregulation to mRNA-Specific Upregulation. Int. J. Genom. 2014, 2014, 970607. [Google Scholar] [CrossRef]
- Wang, F.; Wong, S.C.C.; Chan, L.W.C.; Cho, W.C.S.; Yip, S.P.; Yung, B.Y.M. Multiple Regression Analysis of mRNA-miRNA Associations in Colorectal Cancer Pathway. BioMed Res. Int. 2014, 2014, 676724. [Google Scholar] [CrossRef]
- Höfner, L.; Luther, A.-M.; Waberski, D. The role of seminal plasma in the liquid storage of spermatozoa. Anim. Reprod. Sci. 2020, 220, 106290. [Google Scholar] [CrossRef]
- Zoca, S.M.; Northrop-Albrecht, E.J.; Walker, J.A.; Cushman, R.A.; Perry, G.A. Proteomics dataset of epididymal fluid, seminal plasma, and proteins loosely attached to epididymal and ejaculated sperm from Angus bulls. Data Brief 2022, 42, 108150. [Google Scholar] [CrossRef]


| Layout | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | bta-let-7f | bta-miR-101 | bta-miR-103 | bta-miR-125a | bta-miR-125b | bta-miR-126-3p | bta-miR-128 | bta-miR-145 | bta-miR-148a | bta-miR-151-3p | bta-miR-151-5p | bta-miR-16b |
| B | bta-miR-181a | bta-miR-18a | bta-miR-18b | bta-miR-199a-5p | bta-miR-205 | bta-miR-20a | bta-miR-21-5p | bta-miR-221 | bta-miR-222 | bta-miR-26a | bta-miR-26b | bta-miR-27a-3p |
| C | bta-miR-27b | bta-miR-29a | bta-miR-300-5p | bta-miR-30d | bta-miR-31 | bta-miR-320a | bta-miR-34b | bta-miR-484 | bta-miR-499 | bta-miR-99a-5p | bta-miR-7a-5p | bta-let-7d |
| D | bta-let-7g | bta-let-7i | bta-miR-17-5p | bta-miR-107 | bta-miR-10a | bta-miR-10b | bta-miR-122 | bta-miR-124b | bta-miR-127 | bta-miR-132 | bta-miR-138 | bta-miR-139 |
| E | bta-miR-140 | bta-miR-142-3p | bta-miR-142-5p | bta-miR-148b | bta-miR-150 | bta-miR-15b | bta-miR-17-3p | bta-miR-17-5p | bta-miR-181b | bta-miR-181c | bta-miR-186 | bta-miR-191 |
| F | bta-miR-192 | bta-miR-193a-3p | bta-miR-193a-5p | bta-miR-199a-3p | bta-miR-199b | bta-miR-200a | bta-miR-200b | bta-miR-200c | bta-miR-20b | bta-miR-210 | bta-miR-21-3p | bta-miR-214 |
| G | bta-miR-215 | bta-miR-218 | bta-miR-22-5p | bta-miR-23a | bta-miR-23b-3p | bta-miR-24-3p | bta-miR-25 | bta-miR-29b | bta-miR-29c | bta-miR-30a-5p | bta-miR-30c | bta-miR-30e-5p |
| H | cel-miR39-3p | cel-miR39-3p | SNORD42B | SNORD69 | SNORD61 | SNORD68 | SNORD96A | RNU6-6P | miRTC | miRTC | PPC | PPC |
| Gene | Forward Primer | Reverse Primer | Product Length | Accession Number |
|---|---|---|---|---|
| ZEB1 | AAAGCAGCAGGGCGAGTTAT | TATGGGGTTGGCACTTGGTG | 181 | NM_001206590.1 |
| DNMT1 | TATCGGCTGTTCGGCAACAT | GGCAGCCTCCTCCTTGATTT | 153 | NM_182651.2 |
| FOXP3 | CAGCGGACACTCAACGAGAT | AACTCATCCACGGTCCACAC | 164 | XM_024987818.1 |
| PTEN | GCAGCTTCTGCCATCTCTCT | ATGCTTTGAATCCAAAAACCTTACT | 235 | NM_001319898.1 |
| GADPH | GTGAAGGTCGGAGTGAACGG | ATTGATGGCGACGATGTCCA | 93 | NM_001034034.2 |
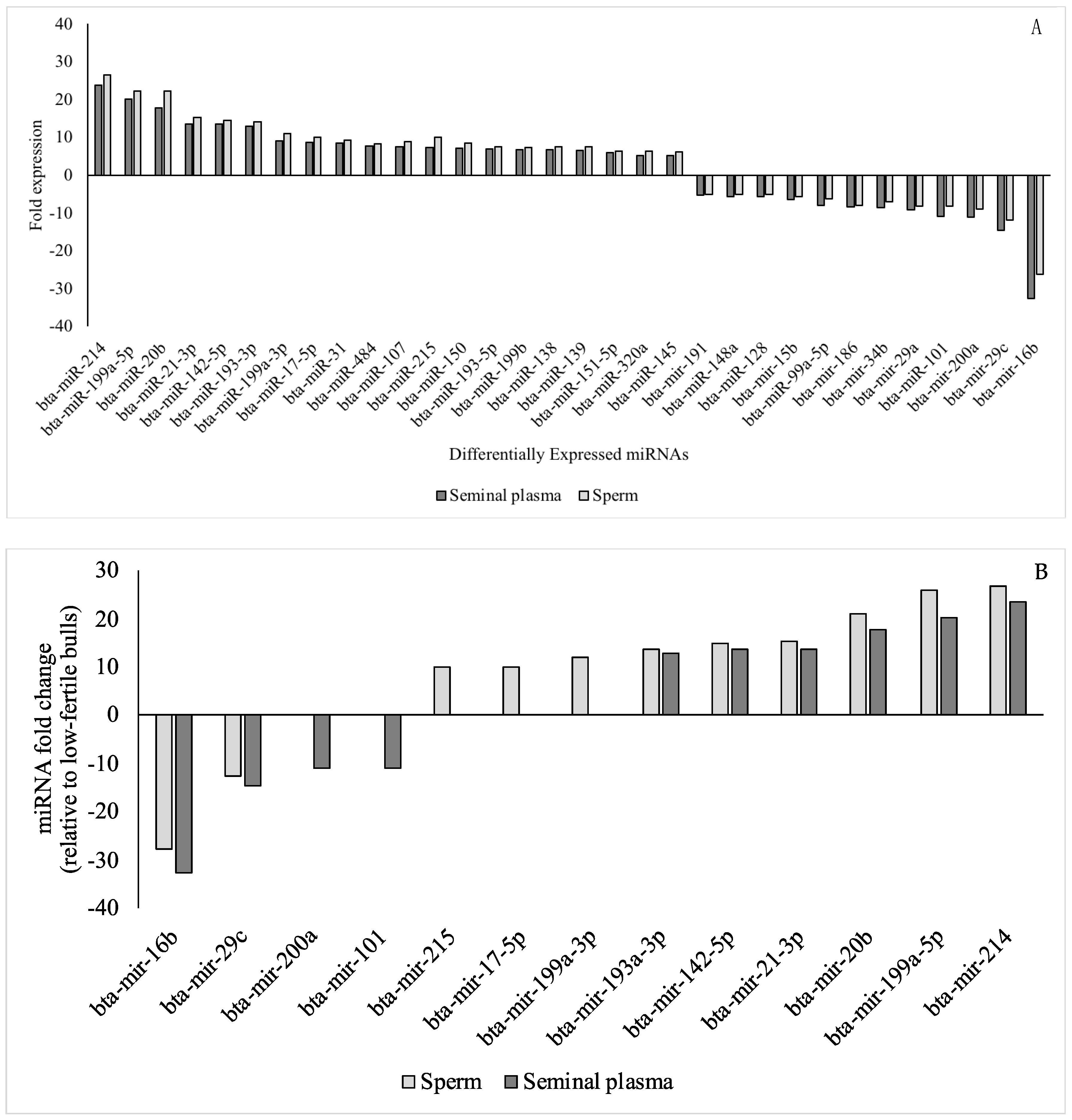
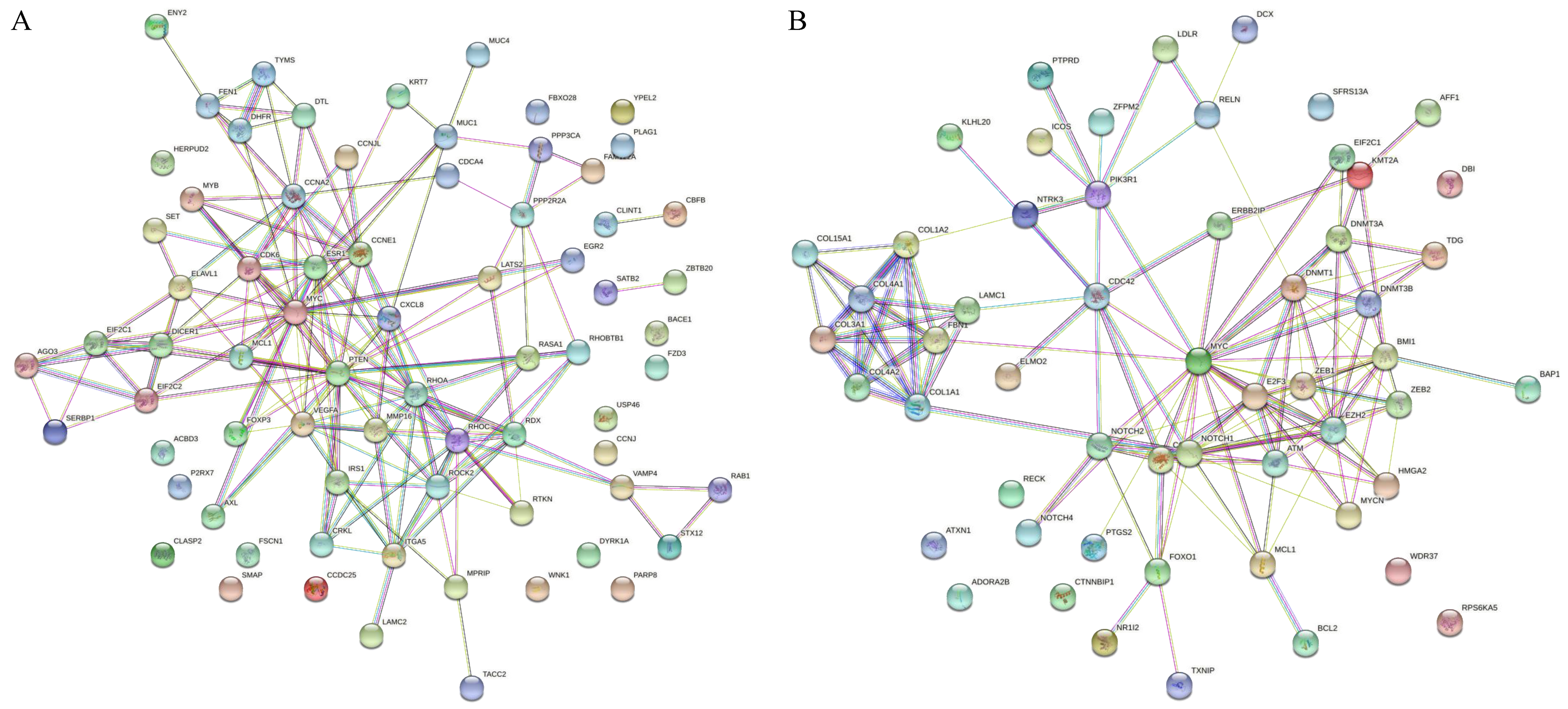
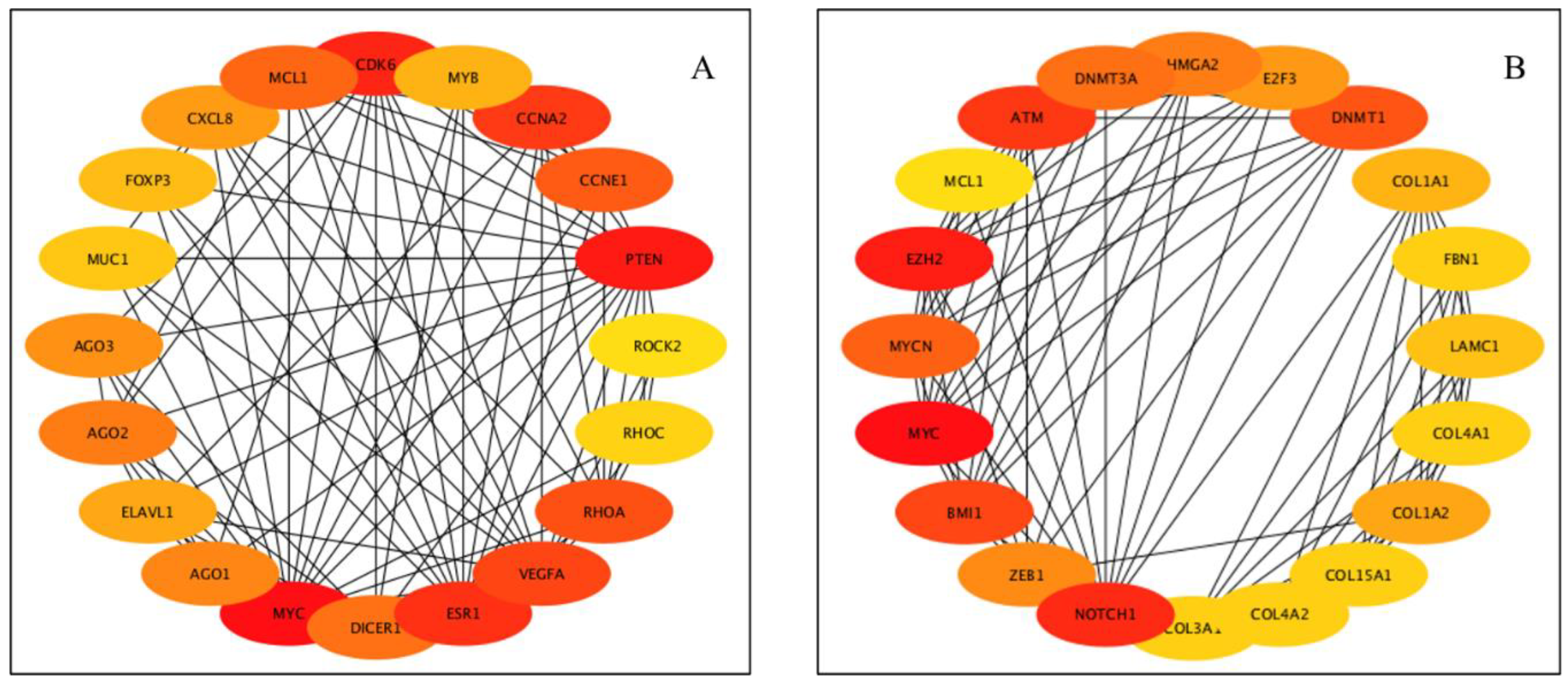
| Upregulated miRNAs | Hub Genes | Reproductive Functions | Species | References |
|---|---|---|---|---|
| miR-107 | AGO1, AGO2, AGO3, CCNE1, CDK6 | Sperm function | Bovine & human | [37,38,39] |
| Angiogenesis | Human | [40] | ||
| Embryo development | Porcine & invertebrates | [41,42,43] | ||
| Placental development | Human | [44] | ||
| miR-132 | RHOC | Sperm maturation, sperm parameters | Human & murine | [45,46,47,48] |
| Trophoblast development | Human | [25] | ||
| Embryo development | Murine | [49] | ||
| miR-138 | ROCK2 | Placental development | Human | [46,50] |
| Embryo development | Swine | [51] | ||
| Embryo-placenta interaction | Murine | [52] | ||
| Organogenesis | Human | [53] | ||
| miR-145 | CCNA2, MYC, MUC1, CXCL8 | Spermatogenesis and function | Human | [54,55] |
| Embryo development | Bovine & murine | [24,56] | ||
| Trophoblast development | Human | [57] | ||
| Embryo implantation | Bovine & Murine | [58,59,60] | ||
| miR-150 | CCNE1, MYB | Spermatogenesis | Human | [61] |
| Embryo development | Human | [62] | ||
| Trophoblast development | Human | [63] | ||
| miR-20b | VEGFA, ESR1 | Spermatogenesis | Buffalo | [64] |
| Pregnancy establishment | Bovine | [65,66] | ||
| Trophoblast development | Human | [67] | ||
| Immune response, uterine receptivity | Swine, murine & hamster | [68,69,70,71,72,73,74,75] | ||
| miR-214 | PTEN | Spermatogenesis, sperm DNA | Canine & murine | [76] |
| Embryo development | Human | [69] | ||
| Uterine capacity and liter size | Swine | [77] | ||
| miR-31 | RHOA, ELAVL1, FOXP3, DICER1 | Sperm DNA | Human | [78] |
| Fertility, embryo development | Human, mouse, rat & invertebrates | [79] | ||
| Embryo implantation | Human | [80,81] | ||
| Organogenesis | Human, mouse, rat & invertebrates | [79] | ||
| miR-320a | MCL | Embryo development | Human, bovine & rat | [17,82,83,84] |
| miR-101 | MYCN, MCL1, ATM, EZH2 | Sperm parameters | Human | [85] |
| Placental development | Human | [86] | ||
| Embryo implantation | Murine | [87,88] | ||
| miR-128 | BMI1, E2F3 | Spermatogenesis, sperm maturation | Murine | [86] |
| Angiogenesis | Human & rat | [89,90] | ||
| Endometrium programming | Bovine | [91] | ||
| Trophoblast development | Human | [17] | ||
| miR-148a | DNMT1 | Spermatogenesis | Human | [92] |
| Angiogenesis | Human | [93] | ||
| Placenta development | Human & rat | [94,95] | ||
| Embryo development | Swine | [78] (Weng, Peng) | ||
| miR-200a | ZB1 | Spermatogenesis | Canine, Murine | [76,96] |
| Embryo development | Murine | [97] | ||
| Embryo implantation/Uterine receptivity | Murine | [98,99] | ||
| Trophoblast development | Murine | [100] | ||
| miR-29c | COL1A1, COL1A2, COL4A2, FBN1, LAMC1, COL4A1, COL3A1, COL15A1, DNMT3A | Sperm DNA | Human | [101] |
| Endometrium programming | Human | [102,103] | ||
| Embryo implantation | Human | [103,104] | ||
| miR-34b | NOCH1, MYC, HMGA2 | Spermatogenesis | Murine | [105,106,107] |
| Embryo implantation | Human | [108] | ||
| Fetal development | Murine | [109] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kasimanickam, V.; Kumar, N.; Kasimanickam, R. Investigation of Sperm and Seminal Plasma Candidate MicroRNAs of Bulls with Differing Fertility and In Silico Prediction of miRNA-mRNA Interaction Network of Reproductive Function. Animals 2022, 12, 2360. https://doi.org/10.3390/ani12182360
Kasimanickam V, Kumar N, Kasimanickam R. Investigation of Sperm and Seminal Plasma Candidate MicroRNAs of Bulls with Differing Fertility and In Silico Prediction of miRNA-mRNA Interaction Network of Reproductive Function. Animals. 2022; 12(18):2360. https://doi.org/10.3390/ani12182360
Chicago/Turabian StyleKasimanickam, Vanmathy, Nishant Kumar, and Ramanathan Kasimanickam. 2022. "Investigation of Sperm and Seminal Plasma Candidate MicroRNAs of Bulls with Differing Fertility and In Silico Prediction of miRNA-mRNA Interaction Network of Reproductive Function" Animals 12, no. 18: 2360. https://doi.org/10.3390/ani12182360
APA StyleKasimanickam, V., Kumar, N., & Kasimanickam, R. (2022). Investigation of Sperm and Seminal Plasma Candidate MicroRNAs of Bulls with Differing Fertility and In Silico Prediction of miRNA-mRNA Interaction Network of Reproductive Function. Animals, 12(18), 2360. https://doi.org/10.3390/ani12182360





