Digestive Tract Morphology and Gut Microbiota Jointly Determine an Efficient Digestive Strategy in Subterranean Rodents: Plateau Zokor
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. Food Consumption and Digestibility
2.3. Digestive Tract Morphology
2.4. DNA Extraction and MiSeq Sequencing of 16S rRNA Gene Amplicons
2.5. Bioinformatics Analysis
2.6. Statistical Analysis
2.7. Predicted Metagenomes
3. Result
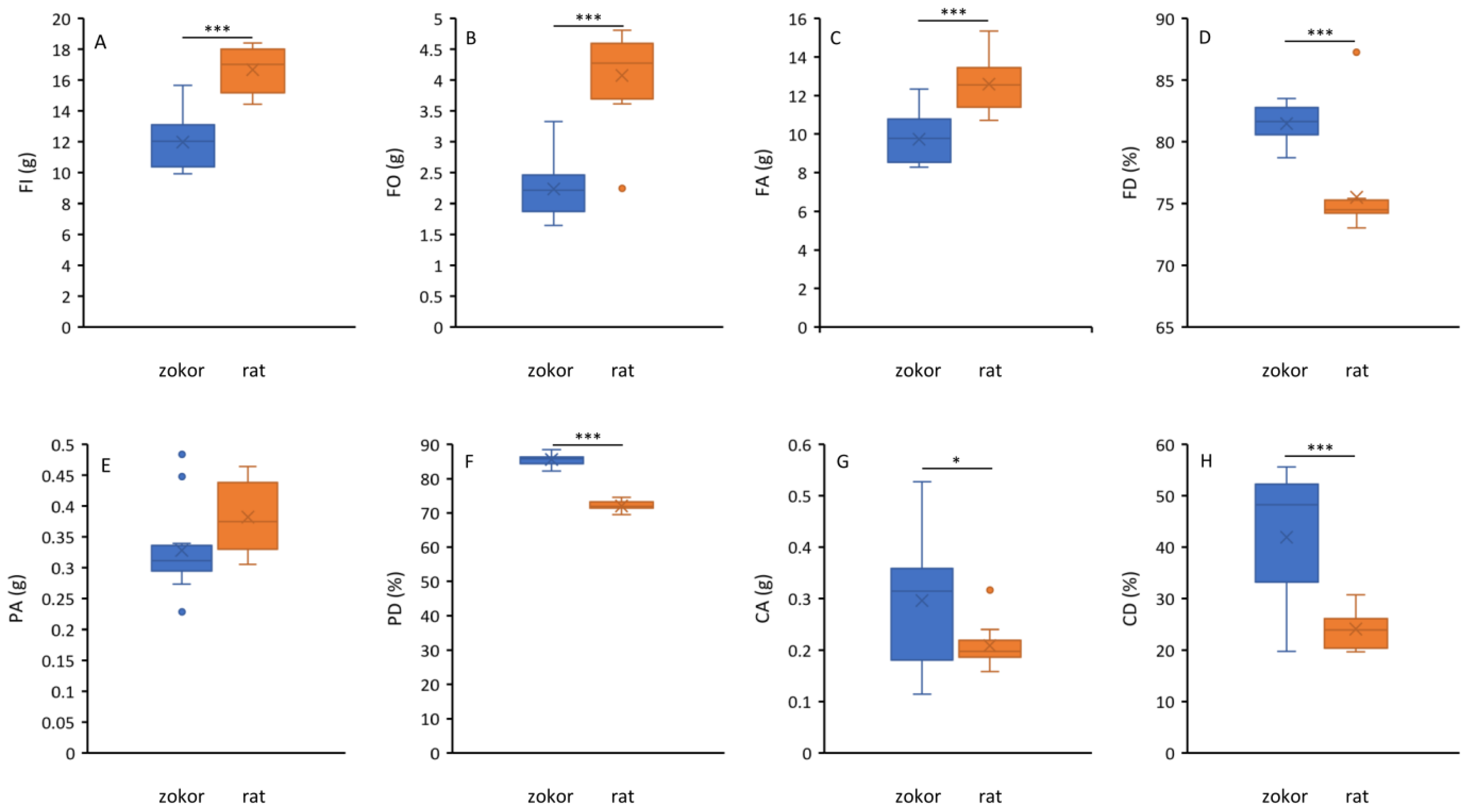
3.1. Food Consumption and Digestibility
3.2. Digestive Tract Morphology
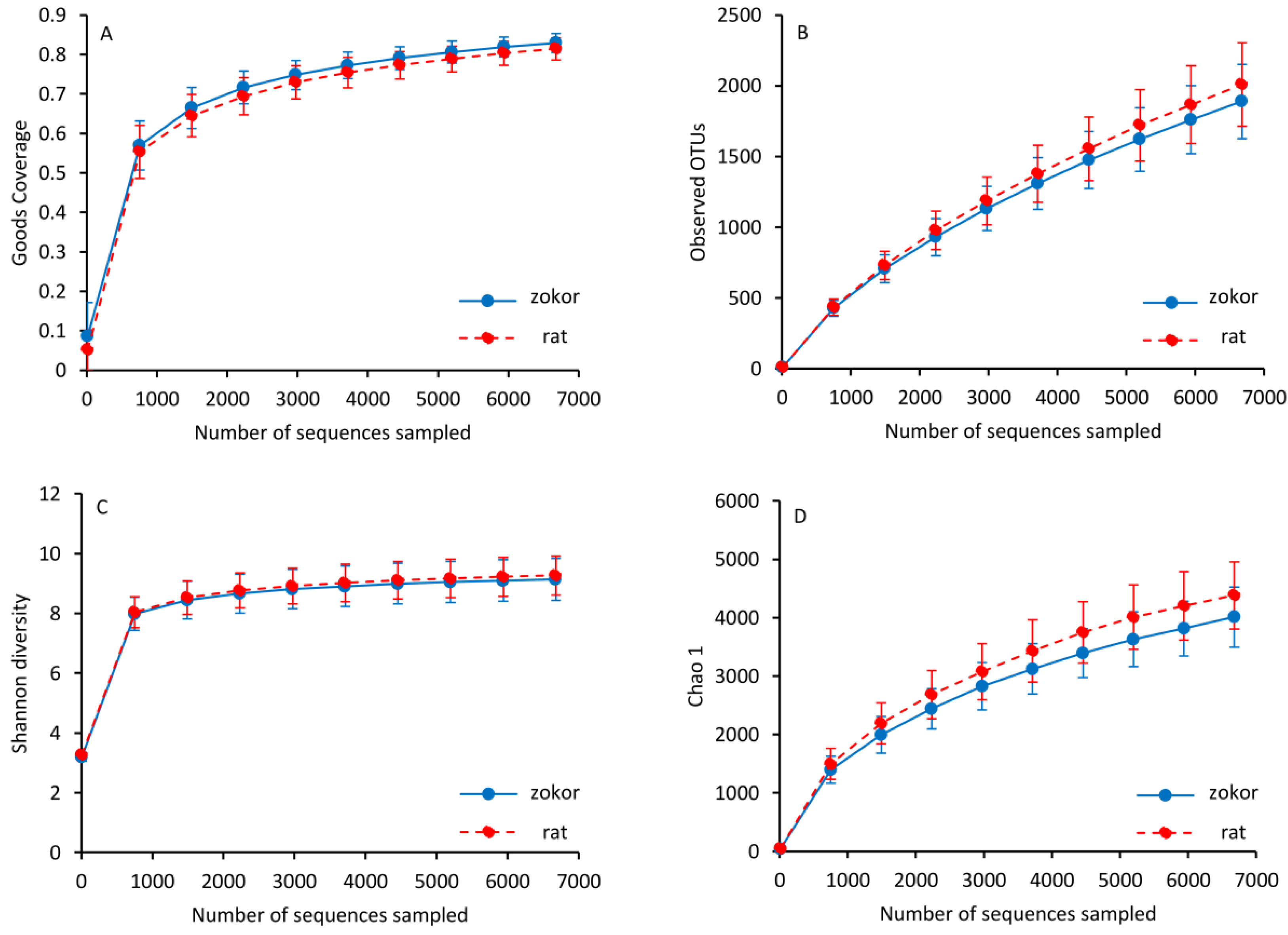
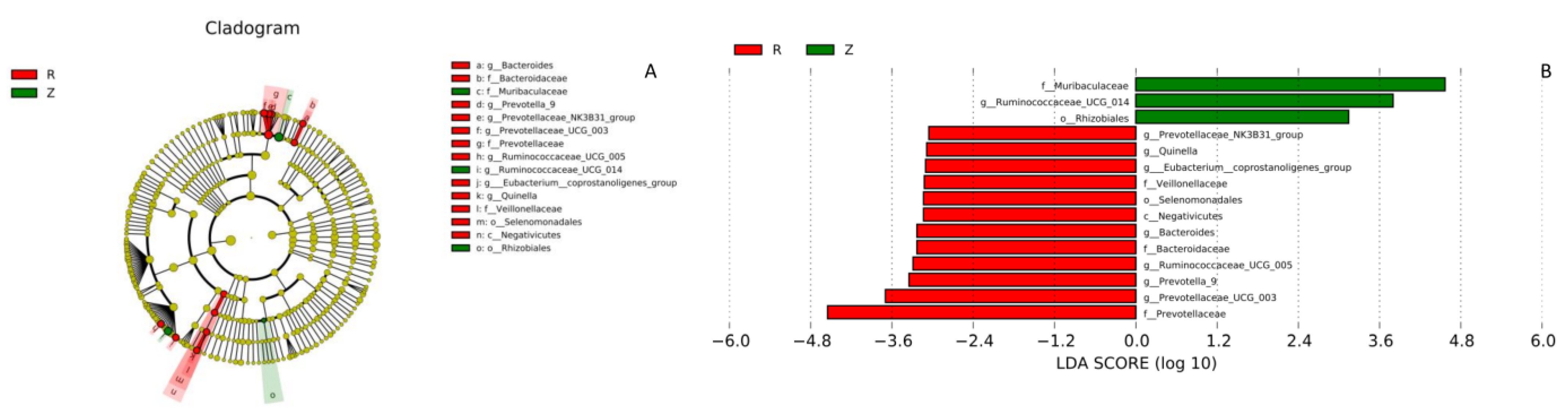
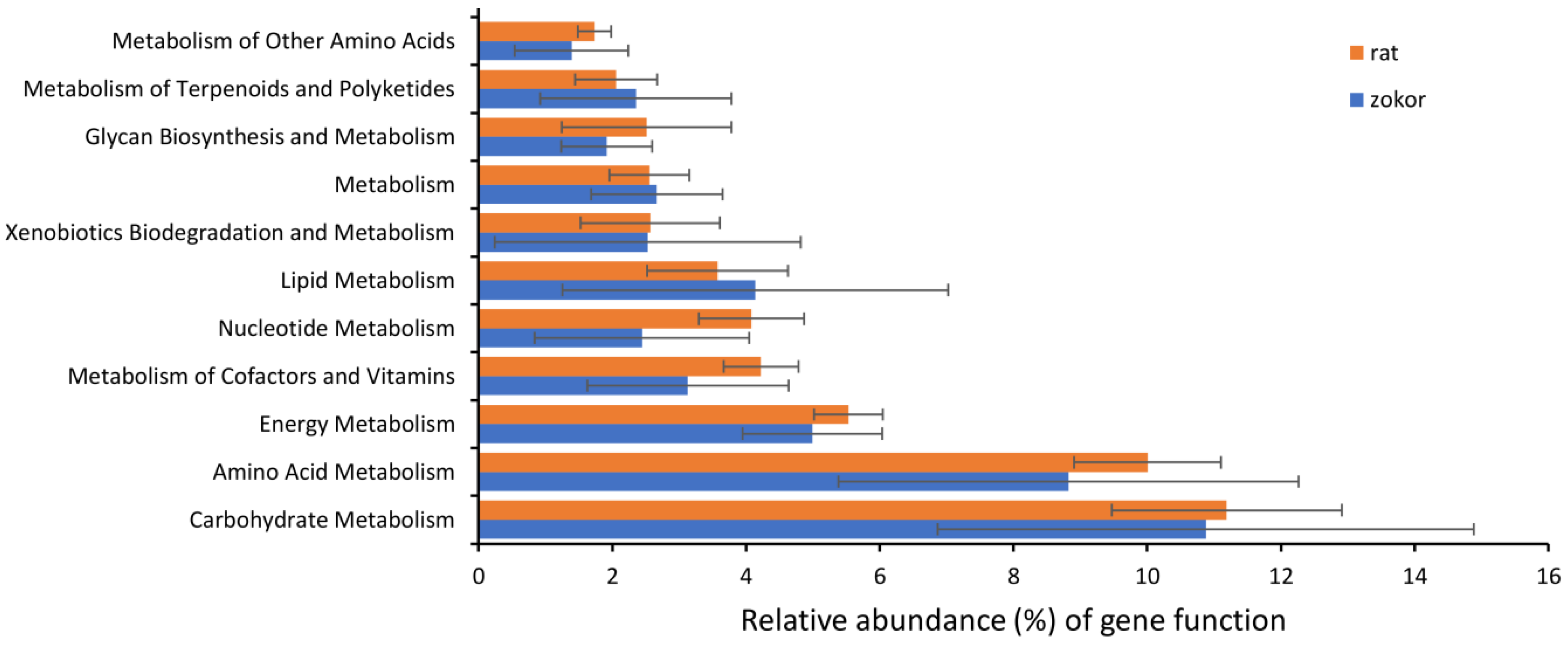
3.3. Gut Microbiota
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Batzli, G.; Broussard, A.; Oliver, R. The integrated processing response in herbivorous small mammals. In The Digestive System in Mammals: Food, Form, and Function; Cambridge University Press: Cambridge, UK, 1994; pp. 324–336. [Google Scholar]
- Hume, I. Optimal digestive strategies in mammalian herbivores. Physiol. Zool. 1989, 62, 1145–1163. [Google Scholar] [CrossRef]
- Rogers, P.J.; Blundell, J.E. Mechanisms of diet selection: The translation of needs into behaviour. Proc. Nutr. Soc. 1991, 50, 65–70. [Google Scholar] [CrossRef] [PubMed]
- Gross, J.E.; Wang, Z.; Wunder, B.A. Effects of food quality and energy needs: Changes in gut morphology and capacity of Microtus ochrogaster. J. Mammal. 1985, 66, 661–667. [Google Scholar] [CrossRef]
- Karasov, W.H. Digestive plasticity in avian energetics and feeding ecology. In Avian Energetics and Nutritional Ecology; Springer: Berlin/Heidelberg, Germany, 1996; pp. 61–84. [Google Scholar]
- McWilliams, S.R.; Karasov, W.H. Phenotypic flexibility in digestive system structure and function in migratory birds and its ecological significance. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2001, 128, 577–591. [Google Scholar] [CrossRef]
- Schieck, J.O.; Millar, J.S. Alimentary tract measurements as indicators of diets of small mammals. Mammalia 1985, 49, 93–104. [Google Scholar] [CrossRef]
- Perrin, M.; Curtis, B. Comparative morphology of the digestive system of 19 species of Southern African myomorph rodents in relation to diet and evolution. Afr. Zool. 1980, 15, 22–33. [Google Scholar] [CrossRef]
- Tremaroli, V.; Bäckhed, F. Functional interactions between the gut microbiota and host metabolism. Nature 2012, 489, 242–249. [Google Scholar] [CrossRef]
- Turnbaugh, P.J.; Gordon, J.I. The core gut microbiome, energy balance and obesity. J. Physiol. 2009, 587, 4153–4158. [Google Scholar] [CrossRef]
- Hooper, L.V.; Gordon, J.I. Commensal host-bacterial relationships in the gut. Science 2001, 292, 1115–1118. [Google Scholar] [CrossRef]
- Li, H.; Qu, J.; Li, T.; Wirth, S.; Zhang, Y.; Zhao, X.; Li, X. Diet simplification selects for high gut microbial diversity and strong fermenting ability in high-altitude pikas. Appl. Microbiol. Biotechnol. 2018, 102, 6739–6751. [Google Scholar] [CrossRef]
- Schaubeck, M.; Clavel, T.; Calasan, J.; Lagkouvardos, I.; Haange, S.B.; Jehmlich, N.; Basic, M.; Dupont, A.; Hornef, M.; Von Bergen, M.; et al. Dysbiotic gut microbiota causes transmissible Crohn’s disease-like ileitis independent of failure in antimicrobial defence. Gut 2016, 65, 225–237. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suzuki, T.A.; Worobey, M. Geographical variation of human gut microbial composition. Biol. Lett. 2014, 10, 20131037. [Google Scholar] [CrossRef] [PubMed]
- Carmody, R.N.; Gerber, G.K.; Luevano, J.M., Jr.; Gatti, D.M.; Somes, L.; Svenson, K.L.; Turnbaugh, P.J. Diet dominates host genotype in shaping the murine gut microbiota. Cell Host Microbe 2015, 17, 72–84. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maurice, C.F.; Knowles, S.C.; Ladau, J.; Pollard, K.S.; Fenton, A.; Pedersen, A.B.; Turnbaugh, P.J. Marked seasonal variation in the wild mouse gut microbiota. ISME J. 2015, 9, 2423–2434. [Google Scholar] [CrossRef] [Green Version]
- Turnbaugh, P.J.; Bäckhed, F.; Fulton, L.; Gordon, J.I. Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host Microbe 2008, 3, 213–223. [Google Scholar] [CrossRef] [Green Version]
- Pérez-Cobas, A.E.; Maiques, E.; Angelova, A.; Carrasco, P.; Moya, A.; Latorre, A. Diet shapes the gut microbiota of the omnivorous cockroach Blattella germanica. FEMS Microbiol. Ecol. 2015, 91, fiv022. [Google Scholar] [CrossRef] [Green Version]
- Ley, R.E.; Hamady, M.; Lozupone, C.; Turnbaugh, P.J.; Ramey, R.R.; Bircher, J.S.; Schlegel, M.L.; Tucker, T.A.; Schrenzel, M.D.; Knight, R.; et al. Evolution of mammals and their gut microbes. Science 2008, 320, 1647–1651. [Google Scholar] [CrossRef] [Green Version]
- Vendrame, S.; Guglielmetti, S.; Riso, P.; Arioli, S.; Klimis-Zacas, D.; Porrini, M. Six-week consumption of a wild blueberry powder drink increases bifidobacteria in the human gut. J. Agric. Food Chem. 2011, 59, 12815–12820. [Google Scholar] [CrossRef]
- Graf, D.; Di Cagno, R.; Fåk, F.; Flint, H.J.; Nyman, M.; Saarela, M.; Watzl, B. Contribution of diet to the composition of the human gut microbiota. Microb. Ecol. Health Dis. 2015, 26, 26164. [Google Scholar] [CrossRef]
- Zhu, L.; Wu, Q.; Dai, J.; Zhang, S.; Wei, F. Evidence of cellulose metabolism by the giant panda gut microbiome. Proc. Natl. Acad. Sci. USA 2011, 108, 17714–17719. [Google Scholar] [CrossRef] [Green Version]
- Zhang, T.; Nevo, E.; Tang, L.; Su, J.; Lin, G. Plateau zokors on the Qinghai-Tibetan Plateau follow Bergmann’s rule latitudinally, but not altitudinally. Mamm. Biol. 2012, 77, 108–112. [Google Scholar] [CrossRef]
- Lin, G.-H.; Xie, J.-X.; Cui, X.-F.; Nevo, E.; Su, J.-P.; Zhang, T.-Z. Effects of supplemental dietary tannic acid on digestion in plateau zokors (Eospalax baileyi). In Annales Zoologici Fennici; BioOne: Washington, WA, USA, 2012; Volume 49, pp. 371–377. [Google Scholar]
- Luna, F.; Antinuchi, C.D. Effect of tunnel inclination on digging energetics in the tuco-tuco, Ctenomys talarum (Rodentia: Ctenomyidae). Naturwissenschaften 2007, 94, 100–106. [Google Scholar] [CrossRef]
- Luna, F.; Antinuchi, C.D. Energetics and thermoregulation during digging in the rodent tuco-tuco (Ctenomys talarum). Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2007, 146, 559–564. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhang, Z.; Liu, J. Burrowing rodents as ecosystem engineers: The ecology and management of plateau zokors Myospalax fontanierii in alpine meadow ecosystems on the Tibetan Plateau. Mammal. Rev. 2003, 33, 284–294. [Google Scholar] [CrossRef] [Green Version]
- Seymour, R.; Withers, P.; Weathers, W. Energetics of burrowing, running, and free-living in the Namib Desert golden mole (Eremitalpa namibensis). J. Zool. 1998, 244, 107–117. [Google Scholar] [CrossRef]
- Xie, J.-X.; Lin, G.-H.; Liu, C.-X.; Yang, C.-H.; Deng, X.-G.; Cui, X.-F.; Li, B.; Zhang, T.-Z.; Su, J.-P. Diet selection in overwinter caches of plateau zokor (Eospalax baileyi). Acta Theriol. 2014, 59, 337–345. [Google Scholar] [CrossRef]
- Xie, J.-X.; Lin, G.-H.; Zhang, T.-Z.; Su, J.P. Foraging strategy of plateau zokors (Eospalax baileyi Thomas) when collecting food for winter caches. Pol. J. Ecol. 2014, 62, 173–182. [Google Scholar]
- Caporaso, J.G.; Lauber, C.L.; Costello, E.K.; Berg-Lyons, D.; Gonzalez, A.; Stombaugh, J.; Knights, D.; Gajer, P.; Ravel, J.; Fierer, N.; et al. Moving pictures of the human microbiome. Genome Biol. 2011, 12, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Caporaso, J.G.; Lauber, C.L.; Walters, W.A.; Berg-Lyons, D.; Huntley, J.; Fierer, N.; Owens, S.M.; Betley, J.; Fraser, L.; Bauer, M.; et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012, 6, 1621–1624. [Google Scholar] [CrossRef] [Green Version]
- Toju, H.; Tanabe, A.S.; Yamamoto, S.; Sato, H. High-coverage ITS primers for the DNA-based identification of ascomycetes and basidiomycetes in environmental samples. PLoS ONE 2012, 7, e40863. [Google Scholar]
- Li, H.; Li, T.; Yao, M.; Li, J.; Zhang, S.; Wirth, S.; Cao, W.; Lin, Q.; Li, X. Pika gut may select for rare but diverse environmental bacteria. Front. Microbiol. 2016, 7, 1269. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Li, T.; Beasley, D.E.; Heděnec, P.; Xiao, Z.; Zhang, S.; Li, J.; Lin, Q.; Li, X. Diet diversity is associated with beta but not alpha diversity of pika gut microbiota. Front. Microbiol. 2016, 7, 1169. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bengtsson-Palme, J.; Hartmann, M.; Eriksson, K.M.; Pal, C.; Thorell, K.; Larsson, D.G.J.; Nilsson, R.H. METAXA2: Improved identification and taxonomic classification of small and large subunit rRNA in metagenomic data. Mol. Ecol. Resour. 2015, 15, 1403–1414. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C.; Haas, B.J.; Clemente, J.C.; Quince, C.; Knight, R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 2011, 27, 2194–2200. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, W.; Godzik, A. Cd-hit: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 2006, 22, 1658–1659. [Google Scholar] [CrossRef] [Green Version]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef] [Green Version]
- Wickham, H.; Chang, W.; Henry, L.; Pedersen, T.L.; Takahashi, K.; Wilke, C.; Woo, K.; Yutani, H.; Dunnington, D. ggplot2: Create elegant data visualisations using the grammar of graphics. R Package Version 2016, 2. [Google Scholar]
- Qin, W.; Song, P.; Lin, G.; Huang, Y.; Wang, L.; Zhou, X.; Li, S.; Zhang, T. Gut microbiota plasticity influences the adaptability of wild and domestic animals in co-inhabited areas. Front. Microbiol. 2020, 11, 125. [Google Scholar] [CrossRef]
- Oksanen, J.; Blanchet, F.G.; Kindt, R.; Legendre, P.; Minchin, P.; O’hara, R.; Simpson, G.; Solymos, P.; Stevens, M.; Wagner, H.; et al. Community ecology package. R Package Version 2013, 2, 321–326. [Google Scholar]
- Perry, M. Flexible heatmaps for functional genomics and sequence feature S. R Package Version 1.0 2016, 1. [Google Scholar]
- Davis, T. Optparse: Command line option parser. R package version 2014, 39, 58–61. [Google Scholar]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, 1–18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Langille, M.G.; Zaneveld, J.; Caporaso, J.G.; McDonald, D.; Knights, D.; Reyes, J.A.; Clemente, J.C.; Burkepile, D.E.; Thurber, R.L.V.; Knight, R.; et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 2013, 31, 814–821. [Google Scholar] [CrossRef]
- Wunder, B.A. Morphophysiological indicators of the energy state of small mammals. In Mammalian Energetics; Cornell University Press: Ithaca, NY, USA, 2019; pp. 83–104. [Google Scholar]
- Liu, Q.-S.; Wang, D.-H. Effects of diet quality on phenotypic flexibility of organ size and digestive function in Mongolian gerbils (Meriones unguiculatus). J. Comp. Physiol. B 2007, 177, 509–518. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.-H.; Pei, Y.-X.; Yang, J.-C.; Wang, Z.-W. Digestive tract morphology and food habits in six species of rodents. Folia Zool. 2003, 52, 51–56. [Google Scholar]
- Hamaker, B.R.; Tuncil, Y.E. A perspective on the complexity of dietary fiber structures and their potential effect on the gut microbiota. J. Mol. Biol. 2014, 426, 3838–3850. [Google Scholar] [CrossRef]
- Escobar, J.S.; Klotz, B.; Valdes, B.E.; Agudelo, G.M. The gut microbiota of Colombians differs from that of Americans, Europeans and Asians. BMC Microbiol. 2014, 14, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Dai, X.; Tian, Y.; Li, J.; Su, X.; Wang, X.; Zhao, S.; Liu, L.; Luo, Y.; Liu, D.; Zheng, H.; et al. Metatranscriptomic analyses of plant cell wall polysaccharide degradation by microorganisms in the cow rumen. Appl. Environ. Microbiol. 2015, 81, 1375–1386. [Google Scholar] [CrossRef] [Green Version]
- Semova, I.; Carten, J.D.; Stombaugh, J.; Mackey, L.C.; Knight, R.; Farber, S.A.; Rawls, J.F. Microbiota regulate intestinal absorption and metabolism of fatty acids in the zebrafish. Cell Host Microbe 2012, 12, 277–288. [Google Scholar] [CrossRef] [Green Version]
- Qin, J.; Li, R.; Raes, J.; Arumugam, M.; Burgdorf, K.S.; Manichanh, C.; Nielsen, T.; Pons, N.; Levenez, F.; Yamada, T.; et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 2010, 464, 59–65. [Google Scholar] [CrossRef] [Green Version]
- El Kaoutari, A.; Armougom, F.; Gordon, J.I.; Raoult, D.; Henrissat, B. The abundance and variety of carbohydrate-active enzymes in the human gut microbiota. Nat. Rev. Microbiol. 2013, 11, 497–504. [Google Scholar] [CrossRef]
- Ley, R.E.; Lozupone, C.A.; Hamady, M.; Knight, R.; Gordon, J.I. Worlds within worlds: Evolution of the vertebrate gut microbiota. Nat. Rev. Microbiol. 2008, 6, 776–788. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muegge, B.D.; Kuczynski, J.; Knights, D.; Clemente, J.C.; González, A.; Fontana, L.; Henrissat, B.; Knight, R.; Gordon, J.I. Diet drives convergence in gut microbiome functions across mammalian phylogeny and within humans. Science 2011, 332, 970–974. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lozupone, C.A.; Stombaugh, J.I.; Gordon, J.I.; Jansson, J.K.; Knight, R. Diversity, stability and resilience of the human gut microbiota. Nature 2012, 489, 220–230. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bäuerl, C.; Collado, M.C.; Zúñiga, M.; Blas, E.; Martínez, G.P. Changes in cecal microbiota and mucosal gene expression revealed new aspects of epizootic rabbit enteropathy. PLoS ONE 2014, 9, e105707. [Google Scholar] [CrossRef] [Green Version]
- Ley, R.E.; Peterson, D.A.; Gordon, J.I. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell 2006, 124, 837–848. [Google Scholar] [CrossRef] [Green Version]
- Niu, Q.; Li, P.; Hao, S.; Zhang, Y.; Kim, S.W.; Li, H.; Ma, X.; Gao, S.; He, L.; Wu, W.; et al. Dynamic distribution of the gut microbiota and the relationship with apparent crude fiber digestibility and growth stages in pigs. Sci. Rep. 2015, 5, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Schnorr, S.L.; Candela, M.; Rampelli, S.; Centanni, M.; Consolandi, C.; Basaglia, G.; Turroni, S.; Biagi, E.; Peano, C.; Severgnini, M.; et al. Gut microbiome of the Hadza hunter-gatherers. Nat. Commun. 2014, 5, 1–12. [Google Scholar] [CrossRef]
- Tasse, L.; Bercovici, J.; Pizzut-Serin, S.; Robe, P.; Tap, J.; Klopp, C.; Cantarel, B.L.; Coutinho, P.M.; Henrissat, B.; Leclerc, M.; et al. Functional metagenomics to mine the human gut microbiome for dietary fiber catabolic enzymes. Genome Res. 2010, 20, 1605–1612. [Google Scholar] [CrossRef] [Green Version]
- O’Keefe, S.J.D.; Li, J.V.; Lahti, L.; Ou, J.; Carbonero, F.; Mohammed, K.; Posma, J.M.; Kinross, J.; Wahl, E.; Ruder, E.; et al. Fat, fibre and cancer risk in African Americans and rural Africans. Nat. Commun. 2015, 6, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Dodd, D.; Mackie, R.I.; Cann, I.K. Xylan degradation, a metabolic property shared by rumen and human colonic Bacteroidetes. Mol. Microbiol. 2011, 79, 292–304. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xue, Z.; Zhang, W.; Wang, L.; Hou, R.; Zhang, M.; Fei, L.; Zhang, X.; Huang, H.; Bridgewater, L.C.; Jiang, Y.; et al. The bamboo-eating giant panda harbors a carnivore-like gut microbiota, with excessive seasonal variations. MBio 2015, 6, e00022-15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ahn, J.; Sinha, R.; Pei, Z.; Dominianni, C.; Wu, J.; Shi, J.; Goedert, J.J.; Hayes, R.B.; Yang, L. Human gut microbiome and risk for colorectal cancer. J. Natl. Cancer Inst. 2013, 105, 1907–1911. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Evans, N.J.; Brown, J.M.; Murray, R.D.; Getty, B.; Birtles, R.J.; Hart, C.A.; Carter, S.D. Characterization of novel bovine gastrointestinal tract Treponema isolates and comparison with bovine digital dermatitis treponemes. Appl. Environ. Microbiol. 2011, 77, 138–147. [Google Scholar] [CrossRef] [PubMed] [Green Version]



| Group | Myospalax baileyi | Rattus norvegicus | ANOVA (df = 1) | Sample Size | |
|---|---|---|---|---|---|
| F | P | n1, n2 | |||
| OTUs | 1889.30 ± 262.01 | 2008.65 ± 294.44 | 1.046 | 0.317 | 11, 12 |
| Chao 1 | 4010.14 ± 516.92 | 4380.74 ± 575.65 | 2.620 | 0.120 | 11, 12 |
| Shannon | 9.136 ± 0.702 | 9.266 ± 0.655 | 0.211 | 0.650 | 11, 12 |
| Simpson | 0.987 ± 0.013 | 0.989 ± 0.009 | 0.365 | 0.552 | 11, 12 |
| Good’s coverage | 0.829 ± 0.023 | 0.814 ± 0.027 | 1.925 | 0.179 | 11, 12 |
| Phylum | Myospalax baileyi | Rattus norvegicus | ANOVA (df = 1) | Sample Size | |
|---|---|---|---|---|---|
| F | P | n1, n2 | |||
| Bacteroidetes | 41.130 ± 9.828 | 44.094 ± 13.429 | 0.359 | 0.555 | 11, 12 |
| Firmicutes | 31.672 ± 8.266 | 36.113 ± 11.663 | 1.090 | 0.308 | 11, 12 |
| Proteobacteria | 5.963 ± 1.469 | 4.889 ± 1.686 | 2.630 | 0.120 | 11, 12 |
| Verrucomicrobia | 4.197 ± 7.290 | 4.783 ± 6.962 | 0.039 | 0.846 | 11, 12 |
| Actinobacteria | 3.931 ± 1.078 | 2.474 ± 0.780 | 13.961 | 0.001 ** | 11, 12 |
| Planctomycetes | 0.637 ± 0.221 | 0.714 ± 0.306 | 0.460 | 0.505 | 11, 12 |
| Acidobacteria | 0.541 ± 0.175 | 0.426 ± 0.095 | 3.902 | 0.062 | 11, 12 |
| Chloroflexi | 0.274 ± 0.100 | 0.248 ± 0.078 | 0.486 | 0.493 | 11 12 |
| TM7 | 0.249 ± 0.230 | 0.277 ± 0.447 | 0.033 | 0.857 | 11, 12 |
| Tenericutes | 0.174 ± 0.087 | 0.731 ± 0.316 | 31.842 | 0.000 *** | 11, 12 |
| Gemmatimonadetes | 0.148 ± 0.051 | 0.119 ± 0.048 | 3.437 | 0.078 | 11, 12 |
| Genus | Myospalax baileyi | Rattus norvegicus | ANOVA (df = 1) | Sample Size | |
|---|---|---|---|---|---|
| F | P | n1, n2 | |||
| Akkermansia | 3.976 ± 7.267 | 4.498 ± 6.876 | 0.031 | 0.861 | 11, 12 |
| Lactobacillus | 3.746 ± 5.568 | 4.458 ± 3.520 | 0.137 | 0.715 | 11, 12 |
| Prevotella | 1.275 ± 1.563 | 20.617 ± 14.757 | 18.630 | 0.000 *** | 11, 12 |
| Oscillospira | 0.731 ± 0.452 | 1.962 ± 1.277 | 9.138 | 0.006 ** | 11, 12 |
| Coprococcus | 0.457 ± 0.530 | 0.205 ± 0.124 | 2.552 | 0.125 | 11, 12 |
| Clostridium | 0.387 ± 0.391 | 0.238 ± 0.119 | 1.586 | 0.222 | 11, 12 |
| Alicyclobacillus | 0.343 ± 0.102 | 0.285 ± 0.158 | 1.065 | 0.314 | 11, 12 |
| Altererythrobacter | 0.283 ± 0.084 | 0.188 ± 0.072 | 8.406 | 0.009 ** | 11, 12 |
| Desulfovibrio | 0.236 ± 0.213 | 0.115 ± 0.115 | 2.952 | 0.100 | 11, 12 |
| Pseudonocardia | 0.186 ± 0.063 | 0.137 ± 0.039 | 5.080 | 0.035 * | 11, 12 |
| DA101 | 0.173 ± 0.062 | 0.136 ± 0.044 | 2.725 | 0.114 | 11, 12 |
| Pseudomonas | 0.166 ± 0.064 | 0.133 ± 0.056 | 1.698 | 0.207 | 11, 12 |
| Rubrobacter | 0.144 ± 0.057 | 0.108 ± 0.050 | 2.574 | 0.124 | 11, 12 |
| Kaistobacter | 0.143 ± 0.043 | 0.125 ± 0.057 | 0.699 | 0.413 | 11, 12 |
| CF231 | 0.131 ± 0.138 | 2.764 ± 2.560 | 11.543 | 0.003 ** | 11, 12 |
| Ruminococcus | 0.027 ± 0.015 | 0.177 ± 0.082 | 34.716 | 0.000 *** | 11, 12 |
| Bacteroides | 0.018 ± 0.019 | 1.228 ± 0.916 | 19.109 | 0.000 *** | 11 12 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, S.-D.; Lin, G.-H.; Han, J.-R.; Lin, Y.-W.; Wang, F.-Q.; Lu, D.-C.; Xie, J.-X.; Zhao, J.-X. Digestive Tract Morphology and Gut Microbiota Jointly Determine an Efficient Digestive Strategy in Subterranean Rodents: Plateau Zokor. Animals 2022, 12, 2155. https://doi.org/10.3390/ani12162155
Zhang S-D, Lin G-H, Han J-R, Lin Y-W, Wang F-Q, Lu D-C, Xie J-X, Zhao J-X. Digestive Tract Morphology and Gut Microbiota Jointly Determine an Efficient Digestive Strategy in Subterranean Rodents: Plateau Zokor. Animals. 2022; 12(16):2155. https://doi.org/10.3390/ani12162155
Chicago/Turabian StyleZhang, Shou-Dong, Gong-Hua Lin, Ji-Ru Han, Yu-Wei Lin, Feng-Qing Wang, De-Chen Lu, Jiu-Xiang Xie, and Jin-Xin Zhao. 2022. "Digestive Tract Morphology and Gut Microbiota Jointly Determine an Efficient Digestive Strategy in Subterranean Rodents: Plateau Zokor" Animals 12, no. 16: 2155. https://doi.org/10.3390/ani12162155
APA StyleZhang, S.-D., Lin, G.-H., Han, J.-R., Lin, Y.-W., Wang, F.-Q., Lu, D.-C., Xie, J.-X., & Zhao, J.-X. (2022). Digestive Tract Morphology and Gut Microbiota Jointly Determine an Efficient Digestive Strategy in Subterranean Rodents: Plateau Zokor. Animals, 12(16), 2155. https://doi.org/10.3390/ani12162155








