Developmental Alterations of Colonic microRNA Profiles Imply Potential Biological Functions in Kid Goats
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Animals and Colon Tissue Collection
2.2. RNA Isolation, Library Construction and Sequencing
2.3. miRNA Data Analyses
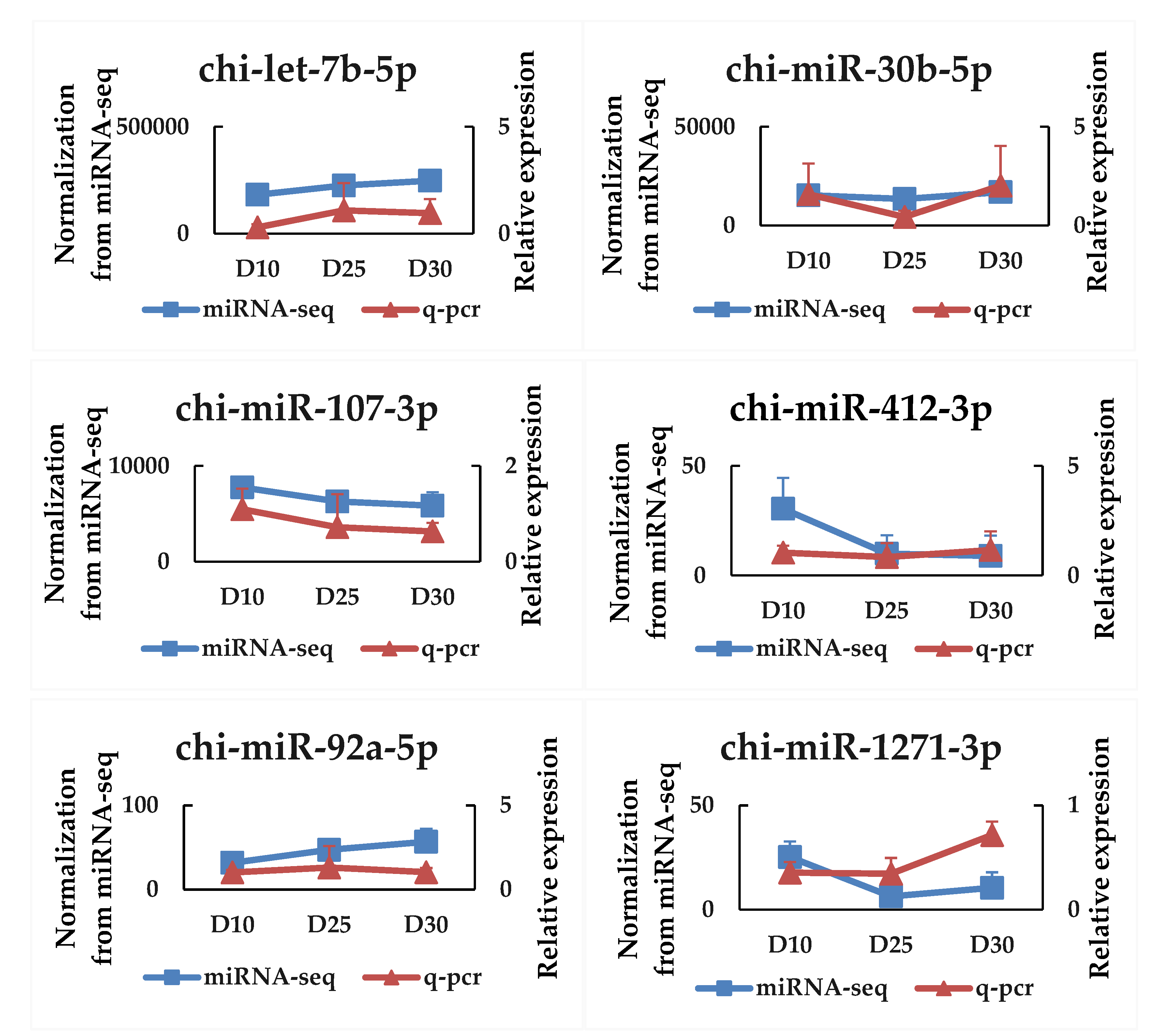
2.4. qPCR Validation of miRNA
2.5. Statistical Analyses
3. Results
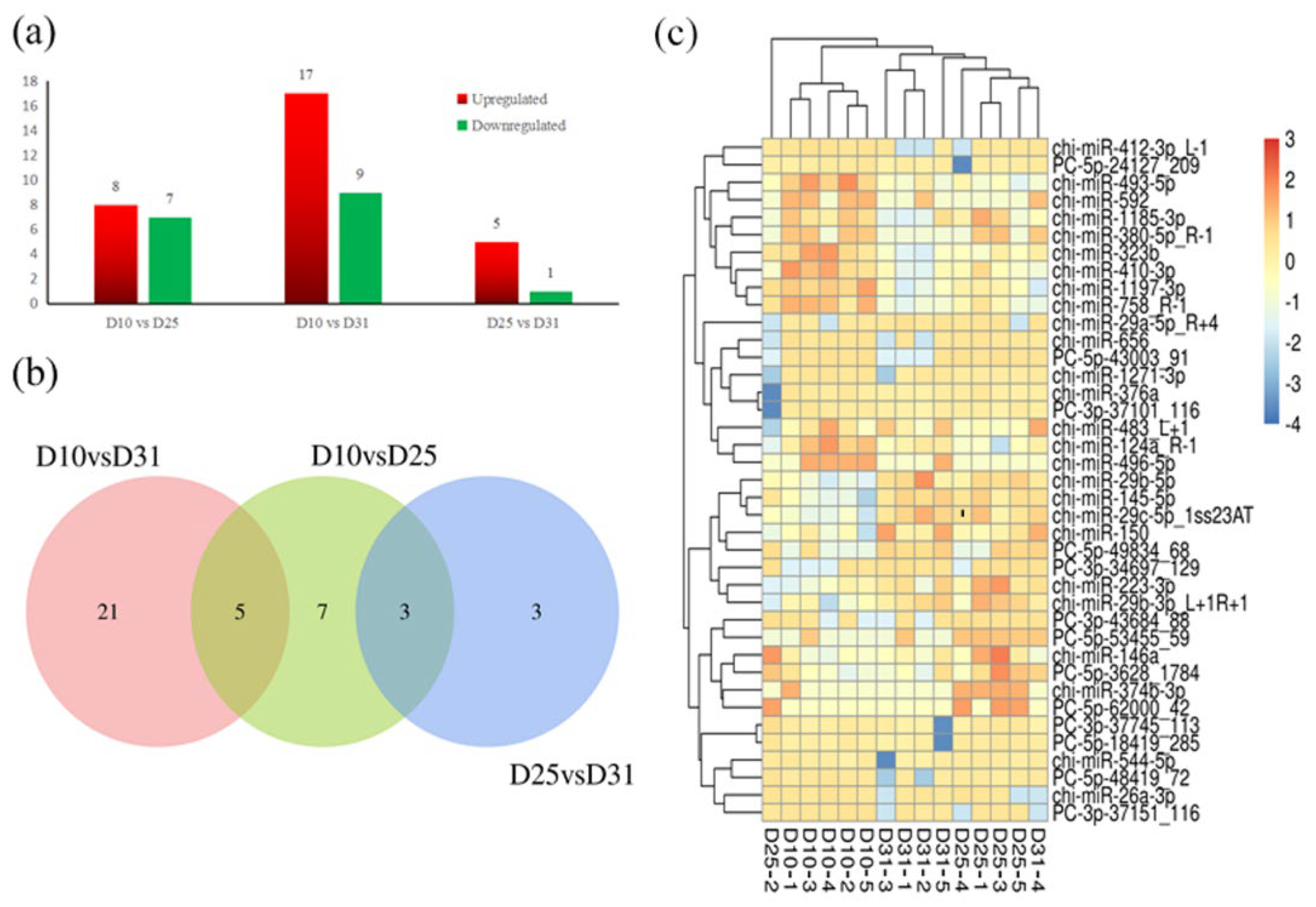
3.1. miRNAs Expression Profile in the Colon Tissues
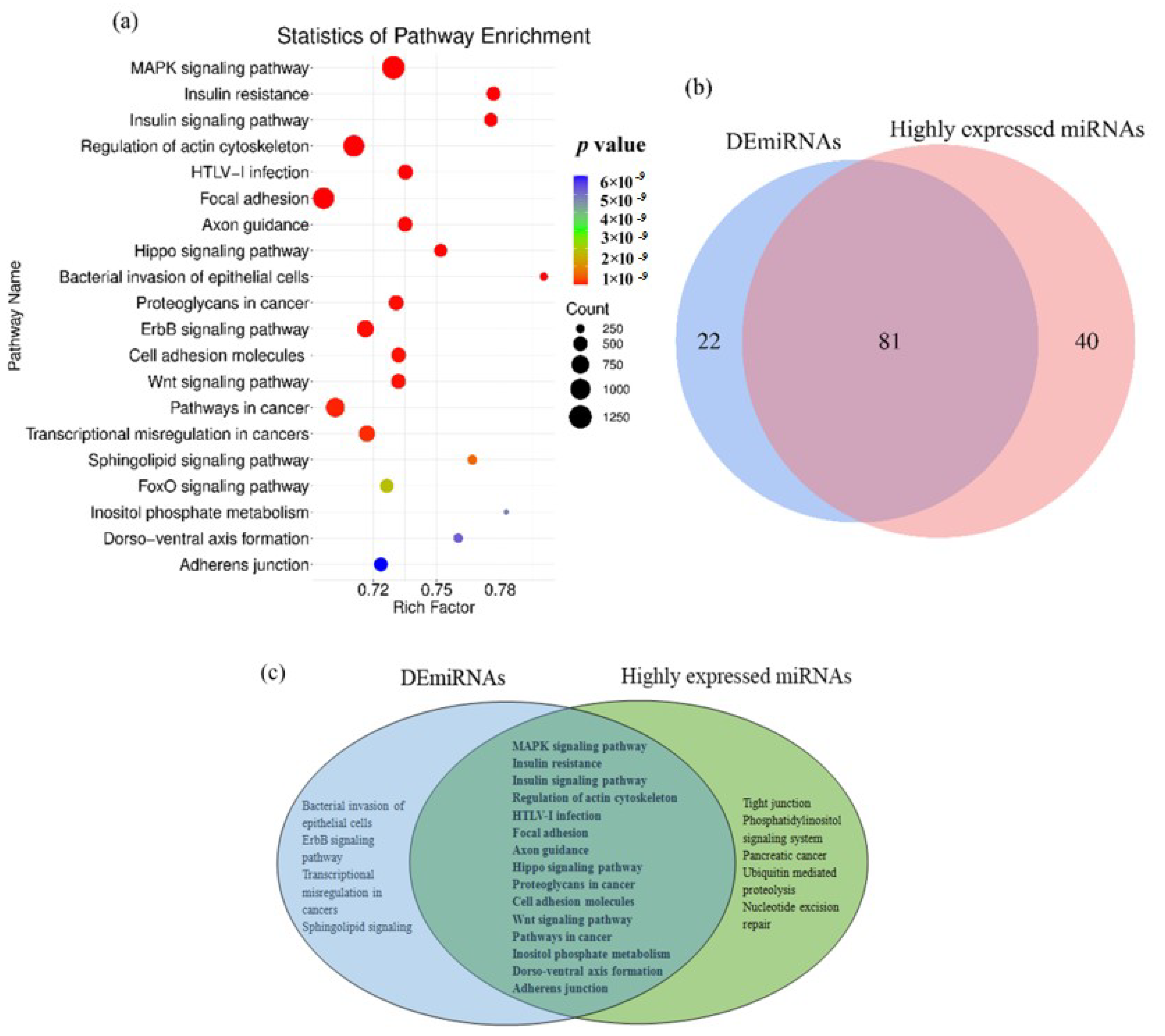
3.2. Pathway Analyses of the DEmiRNAs and HEmiRNAs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Graham, C.; Simmons, N.L. Functional organization of the bovine rumen epithelium. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2005, 288, R173–R181. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stumpff, F.; Georgi, M.-I.; Mundhenk, L.; Rabbani, I.; Fromm, M.; Martens, H.; Günzel, D. Sheep rumen and omasum primary cultures and source epithelia: Barrier function aligns with expression of tight junction proteins. J. Exp. Biol. 2011, 214, 2871–2882. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Plaizier, J.C.; Khafipour, E.; Li, S.; Gozho, G.N.; Krause, D.O. Subacute ruminal acidosis (SARA), endotoxins and health consequences. Anim. Feed Sci. Technol. 2012, 172, 9–21. [Google Scholar] [CrossRef]
- Hou, L.; Ji, Z.; Wang, G.; Wang, J.; Chao, T.; Wang, J. Identification and characterization of microRNAs in the intestinal tissues of sheep (Ovis aries). PLoS ONE 2018, 13, e0193371. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kostic, A.D.; Howitt, M.R.; Garrett, W.S. Exploring host-microbiota interactions in animal models and humans. Genes Dev. 2013, 27, 701–718. [Google Scholar] [CrossRef] [Green Version]
- Jiao, J.; Lu, Q.; Forster, R.; Zhou, C.; Wang, M.; Kang, J.; Tan, Z. Age and feeding system (supplemental feeding versus grazing) modulates colonic bacterial succession and host mucosal immune maturation in goats. J. Anim. Sci. 2016, 94, 2506–2518. [Google Scholar] [CrossRef] [Green Version]
- Kim, Y.S.; Ho, S.B. Intestinal Goblet Cells and Mucins in Health and Disease: Recent Insights and Progress. Curr. Gastroenterol. Rep. 2010, 12, 319–330. [Google Scholar] [CrossRef] [Green Version]
- Peterson, L.W.; Artis, D. Intestinal epithelial cells: Regulators of barrier function and immune homeostasis. Nat. Rev. Immunol. 2014, 14, 141–153. [Google Scholar] [CrossRef]
- Guilloteau, P.; Zabielski, R.; Blum, J.W. Gastrointestinal tract and digestion in the young ruminant: Ontogenesis, adaptations, consequences and manipulations. J. Physiol. Pharmacol. 2009, 60, 37–46. [Google Scholar]
- Li, B.; Zhang, K.; Li, C.; Wang, X.; Chen, Y.; Yang, Y. Characterization and Comparison of Microbiota in the Gastrointestinal Tracts of the Goat (Capra hircus) During Preweaning Development. Front. Microbiol. 2019, 10, 2125. [Google Scholar] [CrossRef] [Green Version]
- Lara-Villoslada, F.; Debras, E.; Nieto, A.; Concha, A.; Gálvez, J.; López-Huertas, E.; Boza, J.; Obled, C.; Xaus, J. Oligosaccharides isolated from goat milk reduce intestinal inflammation in a rat model of dextran sodium sulfate-induced colitis. Clin. Nutr. 2006, 25, 477–488. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.; Kittana, H.; Shu, J.; Kachman, S.D.; Cui, J.; Ramer-Tait, A.E.; Zempleni, J. Dietary Depletion of Milk Exosomes and Their MicroRNA Cargos Elicits a Depletion of miR-200a-3p and Elevated Intestinal Inflammation and Chemokine (C-X-C Motif) Ligand 9 Expression in Mdr1a-/- Mice. Curr. Dev. Nutr. 2019, 3, nzz122. [Google Scholar] [CrossRef] [PubMed]
- Zhou, F.; Paz, H.A.; Sadri, M.; Cui, J.; Kachman, S.D.; Fernando, S.C.; Zempleni, J. Dietary bovine milk exosomes elicit changes in bacterial communities in C57BL/6 mice. Am. J. Physiol. Liver Physiol. 2019, 317, G618–G624. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Bian, G.; Sun, D.; Zhu, W.; Mao, S. Starter Feeding Supplementation Alters Colonic Mucosal Bacterial Communities and Modulates Mucosal Immune Homeostasis in Newborn Lambs. Front. Microbiol. 2017, 8, 429. [Google Scholar] [CrossRef] [Green Version]
- Liang, G.; Malmuthuge, N.; McFadden, T.B.; Bao, H.; Griebel, P.J.; Stothard, P.; Guan, L.L. Potential Regulatory Role of MicroRNAs in the Development of Bovine Gastrointestinal Tract during Early Life. PLoS ONE 2014, 9, e92592. [Google Scholar] [CrossRef] [Green Version]
- Do, D.N.; Dudemaine, P.L.; Fomenky, B.E.; Ibeagha-Awemu, E.M. Integration of miRNA weighted gene co-expression network and miRNA-mRNA co-expression analyses reveals potential regulatory functions of miRNAs in calf rumen development. Genomics 2019, 111, 849–859. [Google Scholar] [CrossRef]
- Zhong, T.; Wang, C.; Hu, J.; Chen, X.; Niu, L.; Zhan, S.; Wang, L.; Guo, J.; Cao, J.; Li, L.; et al. Comparison of MicroRNA Transcriptomes Reveals the Association between MiR-148a-3p Expression and Rumen Development in Goats. Animals 2020, 10, 1951. [Google Scholar] [CrossRef]
- Sun, H.-Z.; Chen, Y.; Guan, L.L. MicroRNA expression profiles across blood and different tissues in cattle. Sci. Data 2019, 6, 190013. [Google Scholar] [CrossRef]
- Coutinho, L.; Matukumalli, L.K.; Sonstegard, T.S.; Van Tassell, C.; Gasbarre, L.C.; Capuco, A.V.; Smith, T.P.L. Discovery and profiling of bovine microRNAs from immune-related and embryonic tissues. Physiol. Genomics 2007, 29, 35–43. [Google Scholar] [CrossRef]
- Zhang, L.; Liang, R.; Zhang, G.; Zhai, Z.; Deng, Y.; Li, J.; Cui, S. Analysis of the microRNA expression profiles in feline kidney cell line infected with feline panleukopenia virus. Infect. Genet. Evol. 2019, 75, 103945. [Google Scholar] [CrossRef]
- Li, C.; Xiong, T.; Zhou, M.; Wan, L.; Xi, S.; Liu, Q.; Chen, Y.; Mao, H.; Liu, S.; Chen, B. Characterization of microRNAs during Embryonic Skeletal Muscle Development in the Shan Ma Duck. Animals 2020, 10, 1417. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Xia, Y.; Gu, Y.; Zhang, K.; Lang, Q.; Chen, L.; Guan, J.; Luo, Z.; Chen, H.; Li, Y.; et al. MicroRNAome of Porcine Pre- and Postnatal Development. PLoS ONE 2010, 5, e11541. [Google Scholar] [CrossRef] [PubMed]
- Meyer, C.; Grey, F.; Kreklywich, C.N.; Andoh, T.F.; Tirabassi, R.S.; Orloff, S.L.; Streblow, D.N. Cytomegalovirus MicroRNA Expression Is Tissue Specific and Is Associated with Persistence. J. Virol. 2011, 85, 378–389. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, Z.; Liu, X.; Feng, T.; Chang, Y. Novel and Conserved Micrornas in Dalian Purple Urchin (Strongylocentrotus Nudus) Identified by Next Generation Sequencing. Int. J. Biol. Sci. 2011, 7, 180–192. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kozomara, A.; Birgaoanu, M.; Griffiths-Jones, S. miRBase: From microRNA sequences to function. Nucleic Acids Res. 2019, 47, D155–D162. [Google Scholar] [CrossRef]
- Yan, Q.; Wang, K.; Han, X.; Tan, Z. The Regulatory Mechanism of Feeding a Diet High in Rice Grain on the Growth and microRNA Expression Profiles of the Spleen, Taking Goats as an Artiodactyl Model. Biology 2021, 10, 832. [Google Scholar] [CrossRef]
- Li, X.; Shahid, M.Q.; Wu, J.; Wang, L.; Liu, X.; Lu, Y. Comparative Small RNA Analysis of Pollen Development in Autotetraploid and Diploid Rice. Int. J. Mol. Sci. 2016, 17, 499. [Google Scholar] [CrossRef]
- Ernst, J.; Nau, G.; Bar-Joseph, Z. Clustering short time series gene expression data. Bioinformatics 2005, 21, i159–i168. [Google Scholar] [CrossRef] [Green Version]
- Ernst, J.; Bar-Joseph, Z. STEM: A tool for the analysis of short time series gene expression data. BMC Bioinform. 2006, 7, 191. [Google Scholar] [CrossRef] [Green Version]
- Anders, S.; Huber, W. Differential expression analysis for sequence count data. Genome Biol. 2010, 11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Agarwal, V.; Bell, G.W.; Nam, J.-W.; Bartel, D.P. Predicting effective microRNA target sites in mammalian mRNAs. eLife 2015, 4, e05005. [Google Scholar] [CrossRef] [PubMed]
- Friedman, R.C.; Farh, K.K.-H.; Burge, C.B.; Bartel, D.P. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009, 19, 92–105. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nam, J.-W.; Rissland, O.S.; Koppstein, D.; Abreu-Goodger, C.; Jan, C.H.; Agarwal, V.; Yildirim, M.; Rodriguez, A.; Bartel, D.P. Global Analyses of the Effect of Different Cellular Contexts on MicroRNA Targeting. Mol. Cell 2014, 53, 1031–1043. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Enright, A.J.; John, B.; Gaul, U.; Tuschl, T.; Sander, C.; Marks, D.S. MicroRNA targets in Drosophila. Genome Biol. 2003, 5, R1. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Azzalini, A.; Bowman, A.W. A look at some data on the old faithful geyser. J. R. Stat. Soc. Ser. C Appl. Stat. 1990, 39, 357–365. [Google Scholar] [CrossRef]
- Carr, D.B.; Littlefield, R.J.; Nicholson, W.L.; Littlefield, J.S. Scatterplot matrix techniques for large-N. J. Am. Stat. Assoc. 1987, 82, 424–436. [Google Scholar] [CrossRef]
- Eugster, M.J.A.; Schlesinger, T. osmar: OpenStreetMap and R. R J. 2013, 5, 53–63. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Huang, Y.; Du, K.L.; Guo, P.Y.; Zhao, R.M.; Wang, B.; Zhao, X.L.; Zhang, C.Q. IL-16 regulates macrophage polarization as a target gene of mir-145-3p. Mol. Immunol. 2019, 107, 1–9. [Google Scholar] [CrossRef]
- Fourie, N.; Peace, R.M.; Abey, S.K.; Sherwin, L.; Rahim-Williams, B.; Smyser, P.A.; Wiley, J.W.; Henderson, W.A. Elevated circulating miR-150 and miR-342-3p in patients with irritable bowel syndrome. Exp. Mol. Pathol. 2014, 96, 422–425. [Google Scholar] [CrossRef] [Green Version]
- Seo, S.-H.; Jang, M.S.; Kim, D.-J.; Kim, S.-M.; Oh, S.-C.; Jung, C.-R.; Park, Y.; Ha, S.-J.; Jung, H.; Park, Y.-J.; et al. MicroRNA-150 controls differentiation of intraepithelial lymphocytes through TGF-beta receptor II regulation. J. Allergy Clin. Immunol. 2018, 141, 1382–1394.e14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhi, L.; Yu, Y.; Li, X.; Wang, D. Molecular Control of Innate Immune Response to Pseudomonas aeruginosa Infection by Intestinal let-7 in Caenorhabditis elegans. PLoS Pathog. 2017, 13, e1006152. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Izumiya, M.; Tsuchiya, N.; Okamoto, K.; Nakagama, H. Systematic exploration of cancer-associated microRNA through functional screening assays. Cancer Sci. 2011, 102, 1615–1621. [Google Scholar] [CrossRef] [PubMed]
- Ueno, K.; Hirata, H.; Majid, S.; Yamamura, S.; Shahryari, V.; Tabatabai, Z.L.; Hinoda, Y.; Dahiya, R. Tumor Suppressor MicroRNA-493 Decreases Cell Motility and Migration Ability in Human Bladder Cancer Cells by Downregulating RhoC and FZD4. Mol. Cancer Ther. 2011, 11, 244–253. [Google Scholar] [CrossRef] [Green Version]
- Faridi, A.; Afgar, A.; Mousavi, S.M.; Nasibi, S.; Mohammadi, M.A.; Farajli Abbasi, M.; Fasihi Harandi, M. Intestinal Expression of miR-130b, miR-410b, and miR-98a in Experimental Canine Echinococcosis by Stem-Loop RT-qPCR. Front. Vet. Sci. 2020, 7, 507. [Google Scholar] [CrossRef]
- Zhu, Y.; Wang, W.; Yuan, T.; Fu, L.; Zhou, L.; Lin, G.; Zhao, S.; Zhou, H.; Wu, G.; Wang, J. MicroRNA-29a mediates the impairment of intestinal epithelial integrity induced by intrauterine growth restriction in pig. Am. J. Physiol. Gastrointest. Liver Physiol. 2017, 312, G434–G442. [Google Scholar] [CrossRef]
| miRNA | Sequences | D10_1 (norm) | D10_2 (norm) | D10_3 (norm) | D10_4 (norm) | D10_5 (norm) | D25_1 (norm) | D25_2 (norm) | D25_3 (norm) | D25_4 (norm) | D25_5 (norm) | D31_1 (norm) | D31_2 (norm) | D31_3 (norm) | D31_4 (norm) | D31_5 (norm) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| chi-miR-145-5p | GTCCAGTTTTCCCAGGAATCCCT | 413,928.08 | 273,737.54 | 305,373.85 | 259,564.08 | 158,024.67 | 793,964.42 | 638,968.89 | 541,464.83 | 631,723.29 | 709,422.09 | 752,564.55 | 708,834.75 | 619,420.59 | 683,159.93 | 806,093.4 |
| chi-miR-150 | TCTCCCAACCCTTGTACCAGTG | 7238.49 | 7862.5 | 10,018.71 | 5326.91 | 3185.05 | 7577.22 | 4807.91 | 8794.09 | 8113.97 | 11,637.64 | 8922.71 | 9453.68 | 18,551.5 | 17,071.68 | 17,578.87 |
| chi-miR-493-5p | TTGTACATGGTAGGCTTTCATT | 4999.65 | 7253.76 | 6637.88 | 4949.41 | 4818.39 | 2541.94 | 3025.21 | 3008.33 | 3475.73 | 1959.88 | 2622.32 | 3930.38 | 3009.98 | 2527.48 | 2423.76 |
| chi-let-7a-5p | TGAGGTAGTAGGTTGTGTGGTT | 273,674.8 | 308,517.33 | 296,359.97 | 317,003.02 | 330,373.78 | 311,181.55 | 521,287.55 | 306,841.98 | 352,508.62 | 303,980.85 | 399,522.55 | 392,244.07 | 339,988.32 | 255,973.03 | 245,615.7 |
| chi-let-7b-5p | TGAGGTAGTAGGTTGTGTGGTT | 176,240.7 | 173,168.72 | 187,498.46 | 222,957.97 | 151,793.56 | 227,617.78 | 245,458.22 | 194,446.03 | 201,309.73 | 256,727.74 | 242,442.15 | 267,225.59 | 239,427.19 | 275,758.49 | 21,3294.5 |
| chi-let-7c-5p | TGAGGTAGTAGGTTGTATGGTT | 19,998.78 | 19,328.18 | 22,494.28 | 25,363.75 | 15,374.01 | 17,784.34 | 27,865.21 | 14,034.72 | 20,558.67 | 15,736.86 | 22,688.46 | 29,583.58 | 24,529.86 | 19,915.93 | 15,916.33 |
| chi-let-7d-5p | AGAGGTAGTAGGTTGCATAGTT | 17,727.73 | 20,104.59 | 18,721.97 | 21,742.38 | 19,750.87 | 19,055.95 | 18,598.87 | 19,642.49 | 19,593.06 | 21,274.85 | 22,562.2 | 20,098.75 | 19,962.66 | 19,492.2 | 16,406.29 |
| chi-let-7d-3p | CTATACGACCTGCTGCCTTTCT | 4844.61 | 7089.73 | 6341.19 | 6762.08 | 5883.03 | 3801.47 | 14,170.87 | 6507.76 | 5145.36 | 9214.98 | 5528.73 | 7017.13 | 6903.8 | 7994.49 | 5557.47 |
| chi-let-7e-5p | TGAGGTAGGAGGTTGTATAGTT | 32,213.65 | 40,306.38 | 40,948.79 | 39,775.16 | 33,621.36 | 32,349.5 | 34,276.18 | 31,642.95 | 39,923.47 | 33,233.69 | 41,543.94 | 50,541.23 | 36,741.77 | 29,093.51 | 27,127.68 |
| chi-let-7f-5p | TGAGGTAGTAGATTGTATAGTT | 31,245.66 | 31,577.41 | 24,545.72 | 22,108.92 | 40,557.5 | 31,878.85 | 68,705.39 | 37,339.47 | 31,208.06 | 27,579.35 | 28,455.72 | 29,908.99 | 24,931.13 | 17,577.23 | 23,287.28 |
| chi-let-7g-5p | TGAGGTAGTAGTTTGTACAGTT | 20,827.40 | 27,945.82 | 19,867.21 | 20,364.65 | 31,014.77 | 20,261.49 | 53,386.92 | 22,753.72 | 24,312.68 | 20,075.23 | 24,907.09 | 27,355.42 | 20,522.19 | 15,088.47 | 16,710.72 |
| chi-let-7i-5p | TGAGGTAGTAGTTTGTGCTGTT | 32,712.21 | 31,512.86 | 26,314.27 | 25,278.73 | 34,437.59 | 31,221.13 | 79,115.93 | 37,108.08 | 29,293.7 | 28,501.77 | 28,455.99 | 31,761.57 | 27,101.97 | 22,835.44 | 20,823.23 |
| chi-miR-29a-5p | ACTGATTTCTTTTGGTGTTCAGAGT | 6.20 | 6.38 | 4.92 | 0.00 | 11.53 | 30.49 | 0.00 | 26.32 | 14.12 | 0.00 | 47.35 | 12.27 | 9.38 | 28.05 | 28.19 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yan, Q.; Tian, L.; Chen, W.; Kang, J.; Tang, S.; Tan, Z. Developmental Alterations of Colonic microRNA Profiles Imply Potential Biological Functions in Kid Goats. Animals 2022, 12, 1533. https://doi.org/10.3390/ani12121533
Yan Q, Tian L, Chen W, Kang J, Tang S, Tan Z. Developmental Alterations of Colonic microRNA Profiles Imply Potential Biological Functions in Kid Goats. Animals. 2022; 12(12):1533. https://doi.org/10.3390/ani12121533
Chicago/Turabian StyleYan, Qiongxian, Lina Tian, Wenxun Chen, Jinhe Kang, Shaoxun Tang, and Zhiliang Tan. 2022. "Developmental Alterations of Colonic microRNA Profiles Imply Potential Biological Functions in Kid Goats" Animals 12, no. 12: 1533. https://doi.org/10.3390/ani12121533
APA StyleYan, Q., Tian, L., Chen, W., Kang, J., Tang, S., & Tan, Z. (2022). Developmental Alterations of Colonic microRNA Profiles Imply Potential Biological Functions in Kid Goats. Animals, 12(12), 1533. https://doi.org/10.3390/ani12121533






