Feeding a Saccharomyces cerevisiae Fermentation Product (Olimond BB) Does Not Alter the Fecal Microbiota of Thoroughbred Racehorses
Abstract
:Simple Summary
Abstract
1. Introduction
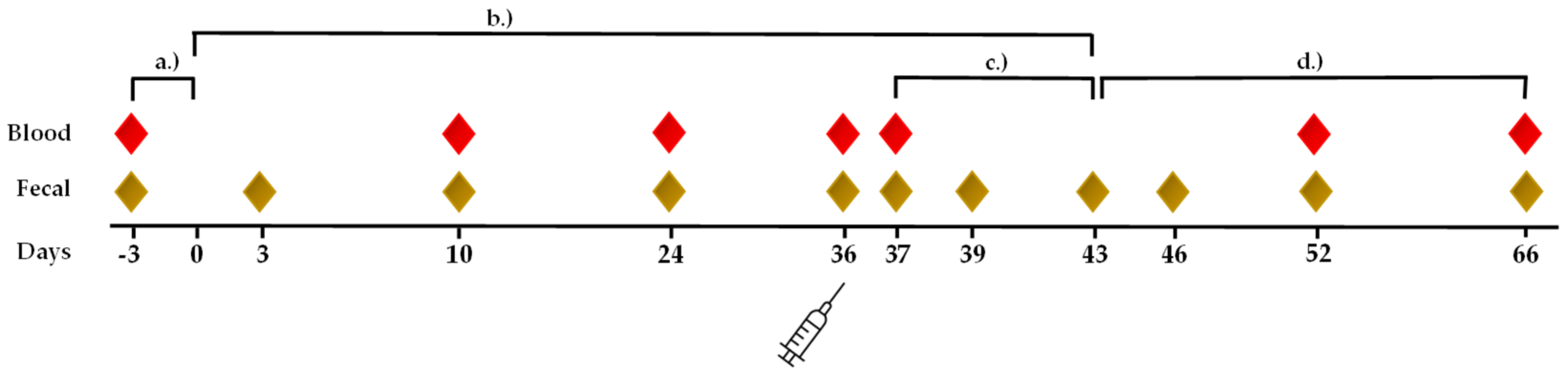
2. Materials and Methods
2.1. Animals
2.2. Supplementation
2.3. Blood Samples and Body Temperature
2.4. Blood Leukocyte Preparation and Subpopulation Determination
2.5. Flow Cytometric Determination of Lymphocyte Subpopulations and Reticulocytes
2.6. Vaccination and Determination of Influenza-Specific Antibodies
2.7. Fecal Samples for Microbiota Analysis
2.8. DNA Extraction
2.9. Statistical Analysis
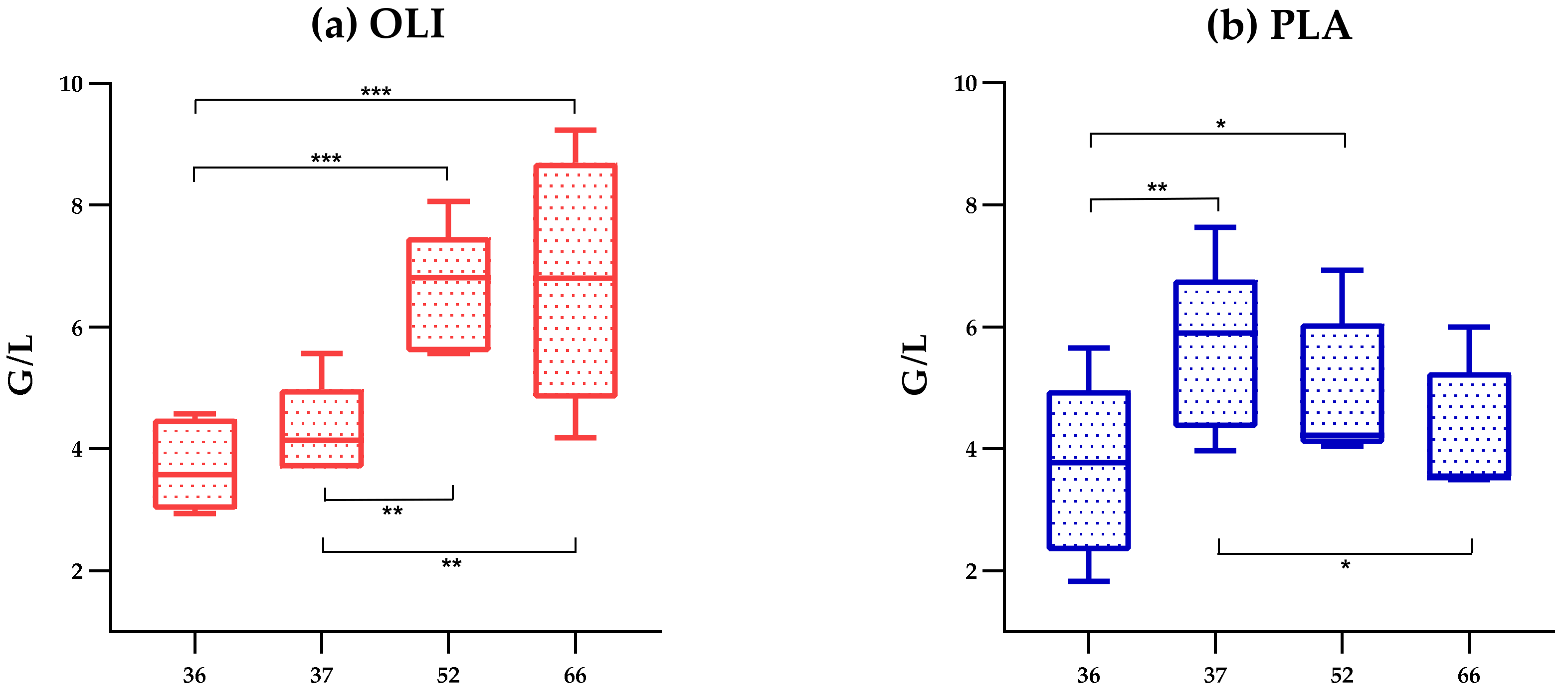
3. Results
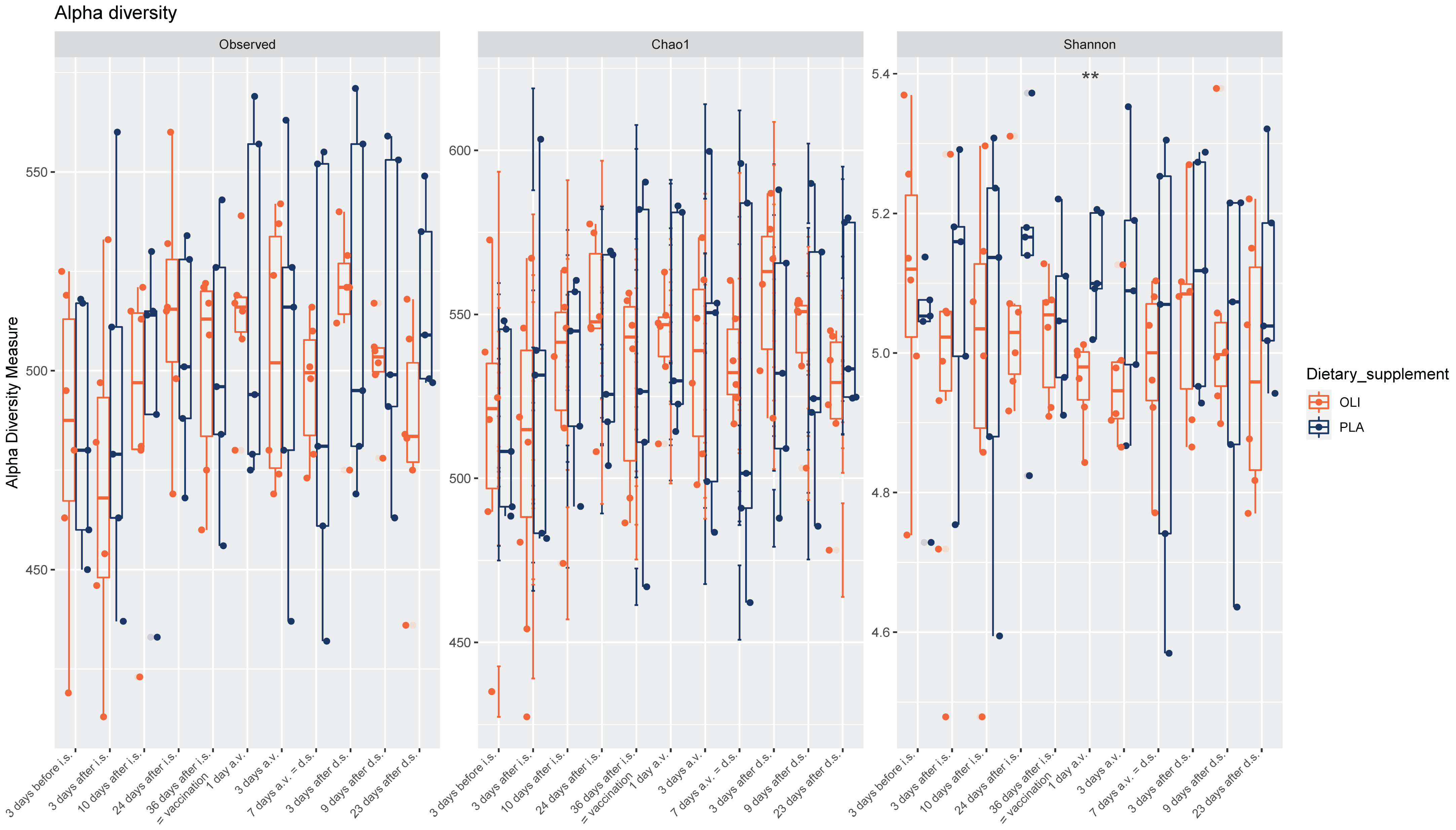
3.1. Fecal Microbiota Diversity
3.2. Heterogeneity of the Gut Microbial Community of the Individual Horse
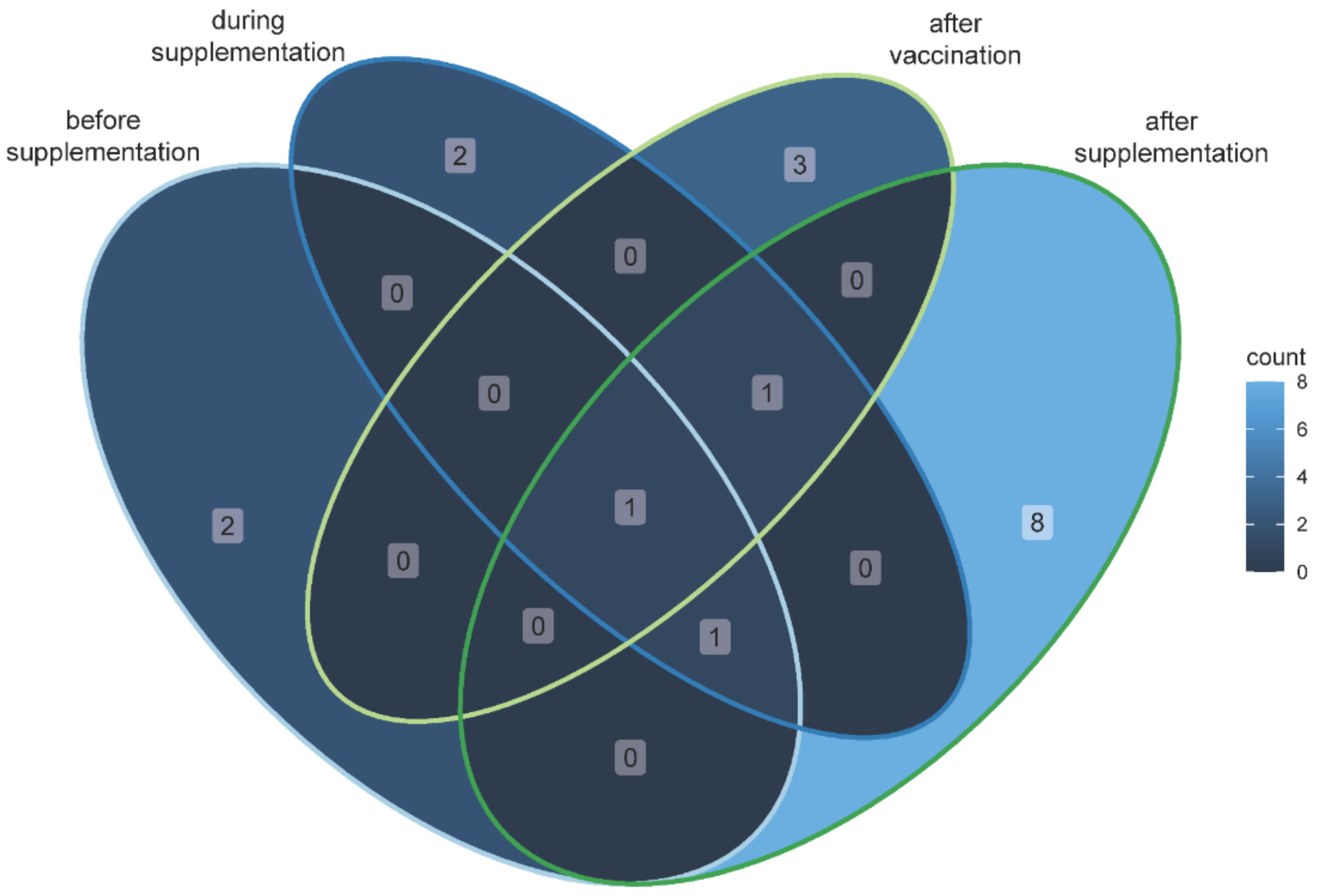
3.3. Identification of Specific Taxa Associated with the Feeding Measures
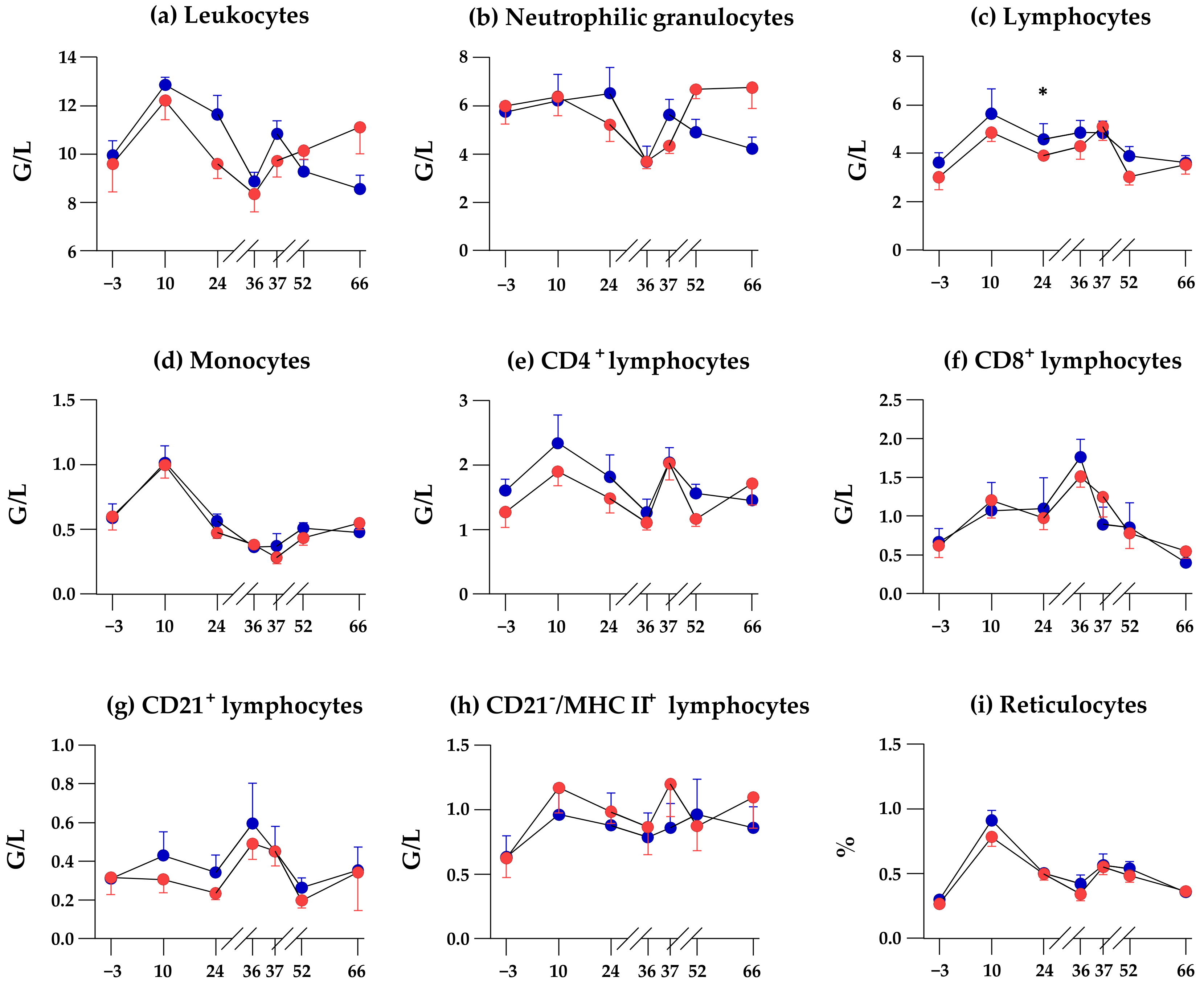
3.4. SCFP Feeding Does Not Alter Time-Dependend Changes of Blood Leukocyte Subpopulations
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kauter, A.; Epping, L.; Semmler, T.; Antao, E.-M.; Kannapin, D.; Stoeckle, S.D.; Gehlen, H.; Lübke-Becker, A.; Günther, S.; Wieler, L.H.; et al. The gut microbiome of horses: Current research on equine enteral microbiota and future perspectives. Anim. Microbiome 2019, 1, 14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McDermott, A.J.; Huffnagle, G.B. The microbiome and regulation of mucosal immunity. Immunology 2014, 142, 24–31. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, L.; Morley, P.S.; Traub-Dargatz, J.L.; Salman, M.D.; Gentry-Weeks, C. Factors associated with Salmonella shedding among equine colic patients at a veterinary teaching hospital. J. Am. Vet. Med. Assoc. 2001, 218, 740–748. [Google Scholar] [CrossRef] [PubMed]
- Belkaid, Y.; Harrison, O. Homeostatic Immunity and the Microbiota. Immunity 2017, 46, 562–576. [Google Scholar] [CrossRef] [Green Version]
- Tun, H.M.; Li, S.; Yoon, I.; Meale, S.J.; Azevedo, P.A.; Khafipour, E.; Plaizier, J.C. Saccharomyces cerevisiae fermentation products (SCFP) stabilize the ruminal microbiota of lactating dairy cows during periods of a depressed rumen pH. BMC Vet. Res. 2020, 16, 237. [Google Scholar] [CrossRef]
- Xiao, J.; Alugongo, G.; Chung, R.; Dong, S.; Li, S.; Yoon, I.; Wu, Z.; Cao, Z. Effects of Saccharomyces cerevisiae fermentation products on dairy calves: Ruminal fermentation, gastrointestinal morphology, and microbial community. J. Dairy Sci. 2016, 99, 5401–5412. [Google Scholar] [CrossRef] [Green Version]
- Alugongo, G.; Xiao, J.; Chung, Y.; Dong, S.; Li, S.; Yoon, I.; Wu, Z.; Cao, Z. Effects of Saccharomyces cerevisiae fermentation products on dairy calves: Performance and health. J. Dairy Sci. 2017, 100, 1189–1199. [Google Scholar] [CrossRef] [Green Version]
- Lin, C.-Y.; Alexander, C.; Steelman, A.J.; Warzecha, C.M.; de Godoy, M.R.C.; Swanson, K.S. Effects of a Saccharomyces cerevisiae fermentation product on fecal characteristics, nutrient digestibility, fecal fermentative end-products, fecal microbial populations, immune function, and diet palatability in adult Dogsj. Anim. Sci. 2019, 97, 1586–1599. [Google Scholar] [CrossRef]
- Matheus, L.F.D.O.; Risolia, L.W.; Ernandes, M.C.; de Souza, J.M.; Oba, P.M.; Vendramini, T.H.A.; Pedrinelli, V.; Henríquez, L.B.F.; Massoco, C.D.O.; Pontieri, C.F.F.; et al. Effects of Saccharomyces cerevisiae cell wall addition on feed digestibility, fecal fermentation and microbiota and immunological parameters in adult cats. BMC Vet. Res. 2021, 17, 351. [Google Scholar] [CrossRef]
- Daly, K.; Proudman, C.J.; Duncan, S.H.; Flint, H.J.; Dyer, J.; Shirazi-Beechey, S.P. Alterations in microbiota and fermentation products in equine large intestine in response to dietary variation and intestinal disease. Br. J. Nutr. 2011, 107, 989–995. [Google Scholar] [CrossRef] [Green Version]
- Dougal, K.; De La Fuente, G.; Harris, P.A.; Girdwood, S.E.; Pinloche, E.; Geor, R.; Nielsen, B.D.; Ii, H.C.S.; Elzinga, S.; Newbold, C.J. Characterisation of the Faecal Bacterial Community in Adult and Elderly Horses Fed a High Fibre, High Oil or High Starch Diet Using 454 Pyrosequencing. PLoS ONE 2014, 9, e87424. [Google Scholar] [CrossRef] [Green Version]
- Salem, S.E.; Maddox, T.W.; Berg, A.; Antczak, P.; Ketley, J.M.; Williams, N.J.; Archer, D.C. Variation in faecal microbiota in a group of horses managed at pasture over a 12-month period. Sci. Rep. 2018, 8, 8510. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schoster, A.; Guardabassi, L.; Staempfli, H.R.; Abrahams, M.; Jalali, M.; Weese, J.S. The longitudinal effect of a multi-strain probiotic on the intestinal bacterial microbiota of neonatal foals. Equine Vet. J. 2015, 48, 689–696. [Google Scholar] [CrossRef] [PubMed]
- Grimm, P.; Combes, S.; Pascal, G.; Cauquil, L.; Julliand, V. Dietary composition and yeast/microalgae combination supplementation modulate the microbial ecosystem in the caecum, colon and faeces of horses. Br. J. Nutr. 2020, 123, 372–382. [Google Scholar] [CrossRef] [Green Version]
- Cooke, C.G.; Gibb, Z.; E Harnett, J. The Safety, Tolerability and Efficacy of Probiotic Bacteria for Equine Use. J. Equine Vet. Sci. 2021, 99, 103407. [Google Scholar] [CrossRef]
- Gerstner, K.; Liesegang, A. Effect of a montmorillonite-bentonite-based product on faecal parameters of horses. J. Anim. Physiol. Anim. Nutr. 2018, 102, 43–46. [Google Scholar] [CrossRef] [Green Version]
- Valigura, H.C.; Leatherwood, J.L.; Martinez, R.E.; Norton, S.A.; White-Springer, S.H. Dietary supplementation of a Saccharomyces cerevisiae fermentation product attenuates exercise-induced stress markers in young horses. J. Anim. Sci. 2021, 99, skab199. [Google Scholar] [CrossRef]
- Martinez, R.E.; Leatherwood, J.L.; Arnold, C.E.; Glass, K.G.; Walter, K.W.; Valigura, H.C.; Norton, S.A.; White-Springer, S.H. Responses to an intra-articular lipopolysaccharide challenge following dietary supplementation of Saccharomyces cerevisiae fermentation product in young horses. J. Anim. Sci. 2021, 99, skab272. [Google Scholar] [CrossRef]
- Lucassen, A.; Finkler-Schade, C.; Schuberth, H.-J. A Saccharomyces cerevisiae Fermentation Product (Olimond BB) Alters the Early Response after Influenza Vaccination in Racehorses. Animals 2021, 11, 2726. [Google Scholar] [CrossRef]
- Lynn, D.J.; Pulendran, B. The potential of the microbiota to influence vaccine responses. J. Leukoc. Biol. 2017, 103, 225–231. [Google Scholar] [CrossRef] [Green Version]
- Jamieson, A.M. Influence of the microbiome on response to vaccination. Hum. Vaccines Immunother. 2015, 11, 2329–2331. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lynn, D.J.; Benson, S.C.; Lynn, M.A.; Pulendran, B. Modulation of immune responses to vaccination by the microbiota: Implications and potential mechanisms. Nat. Rev. Immunol. 2021, 22, 33–46. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, Q.N.; Himes, J.E.; Martinez, D.R.; Permar, S.R. The Impact of the Gut Microbiota on Humoral Immunity to Pathogens and Vaccination in Early Infancy. PLoS Pathog. 2016, 12, e1005997. [Google Scholar] [CrossRef]
- Borey, M.; Blanc, F.; Lemonnier, G.; Leplat, J.-J.; Jardet, D.; Rossignol, M.-N.; Ravon, L.; Billon, Y.; Bernard, M.; Estellé, J.; et al. Links between fecal microbiota and the response to vaccination against influenza A virus in pigs. NPJ Vaccines 2021, 6, 92. [Google Scholar] [CrossRef]
- Mir, R.A.; Schaut, R.G.; Allen, H.K.; Looft, T.; Loving, C.L.; Kudva, I.T.; Sharma, V.K. Cattle intestinal microbiota shifts following Escherichia coli O157:H7 vaccination and colonization. PLoS ONE 2019, 14, e0226099. [Google Scholar] [CrossRef] [PubMed]
- Caporaso, J.G.; Lauber, C.L.; Walters, W.A.; Berg-Lyons, D.; Lozupone, C.A.; Turnbaugh, P.J.; Fierer, N.; Knight, R. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. USA 2011, 108, 4516–4522. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA Ribosomal RNA Gene Database Project: Improved Data Processing and Web-Based Tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naïve Bayesian Classifier for Rapid Assignment of rRNA Sequences into the New Bacterial Taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef] [Green Version]
- McMurdie, P.J.; Holmes, S. Phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef] [Green Version]
- Kassambara, A. Pipe-Friendly Framework for Basic Statistical Tests [R Package Rstatix Version 0.7.0]. Available online: https://mran.microsoft.com/web/packages/rstatix/index.html (accessed on 20 January 2022).
- Fox, J. John Fox and Sanford Weisberg, 3rd ed.; SAGE: Thousand Oaks, CA, USA, 2019. [Google Scholar]
- Oksanen, J.; Blanchet, F.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Minchin, P.; O’Hara, R.; Simpson, G.; Solymos, P. Vegan: Community Ecology Package. R Package Version 2.5-7. Available online: https://CRAN.R-project.org/package=vegan (accessed on 18 January 2022).
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [Green Version]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. EdgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B Methodol. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- LeClere, M.; Costa, M.C. Fecal microbiota in horses with asthma. J. Vet. Intern. Med. 2020, 34, 996–1006. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fernandes, K.A.; Rogers, C.W.; Gee, E.K.; Kittelmann, S.; Bolwell, C.F.; Bermingham, E.N.; Biggs, P.J.; Thomas, D.G. Resilience of Faecal Microbiota in Stabled Thoroughbred Horses Following Abrupt Dietary Transition between Freshly Cut Pasture and Three Forage-Based Diets. Animals 2021, 11, 2611. [Google Scholar] [CrossRef]
- Sorensen, R.J.; Drouillard, J.S.; Douthit, T.L.; Ran, Q.; Marthaler, D.G.; Kang, Q.; Vahl, C.I.; Lattimer, J.M. Effect of hay type on cecal and fecal microbiome and fermentation parameters in horses. J. Anim. Sci. 2020, 99, skaa407. [Google Scholar] [CrossRef]
- Uhr, G.T.; Dohnalová, L.; Thaiss, C.A. The Dimension of Time in Host-Microbiome Interactions. mSystems 2019, 4, e112846. [Google Scholar] [CrossRef] [Green Version]
- Fernandes, K.A.; Kittelmann, S.; Rogers, C.; Gee, E.; Bolwell, C.F.; Bermingham, E.; Thomas, D.G. Faecal Microbiota of Forage-Fed Horses in New Zealand and the Population Dynamics of Microbial Communities following Dietary Change. PLoS ONE 2014, 9, e112846. [Google Scholar] [CrossRef] [Green Version]
- Costa, M.C.; Stämpfli, H.R.; Arroyo, L.G.; Allen-Vercoe, E.; Gomes, R.G.; Weese, J.S. Changes in the equine fecal microbiota associated with the use of systemic antimicrobial drugs. BMC Vet. Res. 2015, 11, 19. [Google Scholar] [CrossRef] [Green Version]
- Steelman, S.M.; Chowdhary, B.P.; Dowd, S.; Suchodolski, J.; E Janečka, J. Pyrosequencing of 16S rRNA genes in fecal samples reveals high diversity of hindgut microflora in horses and potential links to chronic laminitis. BMC Vet. Res. 2012, 8, 231. [Google Scholar] [CrossRef] [Green Version]
- Costa, M.; Silva, G.; Ramos, R.; Staempfli, H.; Arroyo, L.; Kim, P.; Weese, J. Characterization and comparison of the bacterial microbiota in different gastrointestinal tract compartments in horses. Vet. J. 2015, 205, 74–80. [Google Scholar] [CrossRef] [PubMed]
- Górniak, W.; Cholewińska, P.; Szeligowska, N.; Wołoszyńska, M.; Soroko, M.; Czyż, K. Effect of Intense Exercise on the Level of Bacteroidetes and Firmicutes Phyla in the Digestive System of Thoroughbred Racehorses. Animals 2021, 11, 290. [Google Scholar] [CrossRef] [PubMed]
- Mach, N.; Ruet, A.; Clark, A.; Bars-Cortina, D.; Ramayo-Caldas, Y.; Crisci, E.; Pennarun, S.; Dhorne-Pollet, S.; Foury, A.; Moisan, M.-P.; et al. Priming for welfare: Gut microbiota is associated with equitation conditions and behavior in horse athletes. Sci. Rep. 2020, 10, 8311. [Google Scholar] [CrossRef]
- Montgomery, L.; Flesher, B.; Stahl, D. Transfer of Bacteroides succinogenes (Hungate) to Fibrobacter gen. nov. as Fibrobacter succinogenes comb. nov. and Description of Fibrobacter intestinalis sp. nov. Int. J. Syst. Bacteriol. 1988, 38, 430–435. [Google Scholar] [CrossRef] [Green Version]
- Ransom-Jones, E.; Jones, D.L.; McCarthy, A.J.; McDonald, J.E. The Fibrobacteres: An Important Phylum of Cellulose-Degrading Bacteria. Microb. Ecol. 2012, 63, 267–281. [Google Scholar] [CrossRef]
- Neumann, A.P.; McCormick, C.A.; Suen, G. Fibrobacter communities in the gastrointestinal tracts of diverse hindgut-fermenting herbivores are distinct from those of the rumen. Environ. Microbiol. 2017, 19, 3768–3783. [Google Scholar] [CrossRef]
- Costa, M.C.; Stampfli, H.R.; Allenvercoe, E.; Weese, J.S. Development of the faecal microbiota in foals. Equine Vet. J. 2015, 48, 681–688. [Google Scholar] [CrossRef]
- Murray, J.-A.M.D.; Brown, S.; O’Shaughnessy, P.; Monteiro, A.; Warren, H.; Hastie, P.M. Effect of Live Yeast Culture Supplementation on Fibrolytic and Saccharolytic Bacterial Populations in the Feces of Horses Fed a High-Fiber or High-Starch Diet. J. Equine Vet. Sci. 2017, 51, 41–45. [Google Scholar] [CrossRef] [Green Version]
- Elzinga, S.; Weese, J.S.; Adams, A.A. Comparison of the Fecal Microbiota in Horses with Equine Metabolic Syndrome and Metabolically Normal Controls Fed a Similar All-Forage Diet. J. Equine Vet. Sci. 2016, 44, 9–16. [Google Scholar] [CrossRef]
- Mohajeri, M.H.; La Fata, G.; E Steinert, R.; Weber, P. Relationship between the gut microbiome and brain function. Nutr. Rev. 2018, 76, 481–496. [Google Scholar] [CrossRef]
- Deters, E.L.; Stokes, R.S.; Genther-Schroeder, O.N.; Hansen, S.L. Effects of a Saccharomyces cerevisiae fermentation product in receiving diets of newly weaned beef steers I: Growth performance and antioxidant defense. J. Anim. Sci. 2018, 96, 3897–3905. [Google Scholar] [CrossRef] [PubMed]
- Cortés-Coronado, R.F.; Gómez-Rosales, S.; Angeles, M.D.L.; Casaubon-Huguenin, M.T.; Sørensen-Dalgaard, T. Influence of a yeast fermented product on the serum levels of the mannan-binding lectin and the antibodies against the Newcastle disease virus in Ross broilers. J. Appl. Poult. Res. 2017, 26, 38–49. [Google Scholar] [CrossRef]
- Vaishnava, S.; Yamamoto, M.; Severson, K.M.; Ruhn, K.A.; Yu, X.; Koren, O.; Ley, R.; Wakeland, E.K.; Hooper, L.V. The Antibacterial Lectin RegIIIγ Promotes the Spatial Segregation of Microbiota and Host in the Intestine. Science 2011, 334, 255–258. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, N.; Li, N.; Duan, X.; Niu, H. Interaction between the gut microbiome and mucosal immune system. Mil. Med. Res. 2017, 4, 14. [Google Scholar] [CrossRef]
- Okumura, R.; Takeda, K. Roles of intestinal epithelial cells in the maintenance of gut homeostasis. Exp. Mol. Med. 2017, 49, e338. [Google Scholar] [CrossRef] [Green Version]

| DF | SumsOfSqs | F.Model | R2 | Pr (>F) | |
|---|---|---|---|---|---|
| Dietary_supplement | 1 | 0.485 | 7.097 | 0.055 | 0.001 |
| Animal_No | 9 | 4.296 | 6.980 | 0.484 | 0.001 |
| Residuals | 60 | 4.103 | |||
| Total | 70 | 8.884 |
| Samples | Comparison | p < 0.05 | pFDR < 0.05 & abs (log2FC) > 1 |
|---|---|---|---|
| Day −3 | PLA vs. OLI | 53 | 4 |
| Day 3 | PLA vs. OLI | 39 | 0 |
| Day 10 | PLA vs. OLI | 35 | 1 |
| Day 24 | PLA vs. OLI | 31 | 3 |
| Day 36 1 | PLA vs. OLI | 37 | 2 |
| Day 37 2 | PLA vs. OLI | 49 | 0 |
| Day 39 3 | PLA vs. OLI | 47 | 3 |
| Day 43 4 | PLA vs. OLI | 54 | 2 |
| Day 46 5 | PLA vs. OLI | 43 | 8 |
| Day 52 6 | PLA vs. OLI | 30 | 0 |
| Day 66 7 | PLA vs. OLI | 49 | 1 |
| Point in Time | Group | Group * Point in Time 1 | |
|---|---|---|---|
| Pr (>F) | Pr (>F) | Pr (>F) | |
| Leukocytes | <0.0001 | - | 0.002 |
| Neutrophilic granulocytes | <0.0001 | - | 0.002 |
| Lymphocytes | <0.0001 | - | - |
| Monocytes | <0.0001 | - | - |
| CD4+ lymphocytes | <0.0001 | - | - |
| CD8+ lymphocytes | <0.0001 | - | - |
| CD21+ lymphocytes | 0.006 | - | - |
| CD21−/MHC II+ lymphocytes | - | - | - |
| Reticulocytes | <0.0001 | - | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lucassen, A.; Hankel, J.; Finkler-Schade, C.; Osbelt, L.; Strowig, T.; Visscher, C.; Schuberth, H.-J. Feeding a Saccharomyces cerevisiae Fermentation Product (Olimond BB) Does Not Alter the Fecal Microbiota of Thoroughbred Racehorses. Animals 2022, 12, 1496. https://doi.org/10.3390/ani12121496
Lucassen A, Hankel J, Finkler-Schade C, Osbelt L, Strowig T, Visscher C, Schuberth H-J. Feeding a Saccharomyces cerevisiae Fermentation Product (Olimond BB) Does Not Alter the Fecal Microbiota of Thoroughbred Racehorses. Animals. 2022; 12(12):1496. https://doi.org/10.3390/ani12121496
Chicago/Turabian StyleLucassen, Alexandra, Julia Hankel, Christa Finkler-Schade, Lisa Osbelt, Till Strowig, Christian Visscher, and Hans-Joachim Schuberth. 2022. "Feeding a Saccharomyces cerevisiae Fermentation Product (Olimond BB) Does Not Alter the Fecal Microbiota of Thoroughbred Racehorses" Animals 12, no. 12: 1496. https://doi.org/10.3390/ani12121496
APA StyleLucassen, A., Hankel, J., Finkler-Schade, C., Osbelt, L., Strowig, T., Visscher, C., & Schuberth, H.-J. (2022). Feeding a Saccharomyces cerevisiae Fermentation Product (Olimond BB) Does Not Alter the Fecal Microbiota of Thoroughbred Racehorses. Animals, 12(12), 1496. https://doi.org/10.3390/ani12121496






