Sulforaphane Enhanced Proliferation of Porcine Satellite Cells via Epigenetic Augmentation of SMAD7
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Porcine Skeletal Muscle Satellite Cell Culture
2.2. PSC Viability Assay
2.3. HDAC Activity Assay
2.4. MiRNAs Targeting Porcine SMAD7
2.5. Quantitative Real-Time PCR (qRT-PCR) of mRNA and miRNA
2.6. Western Blot
2.7. Bisulfite Sequencing
2.8. Chromatin Immunoprecipitation (ChIP) Assay
2.9. Statistical Analysis
3. Results
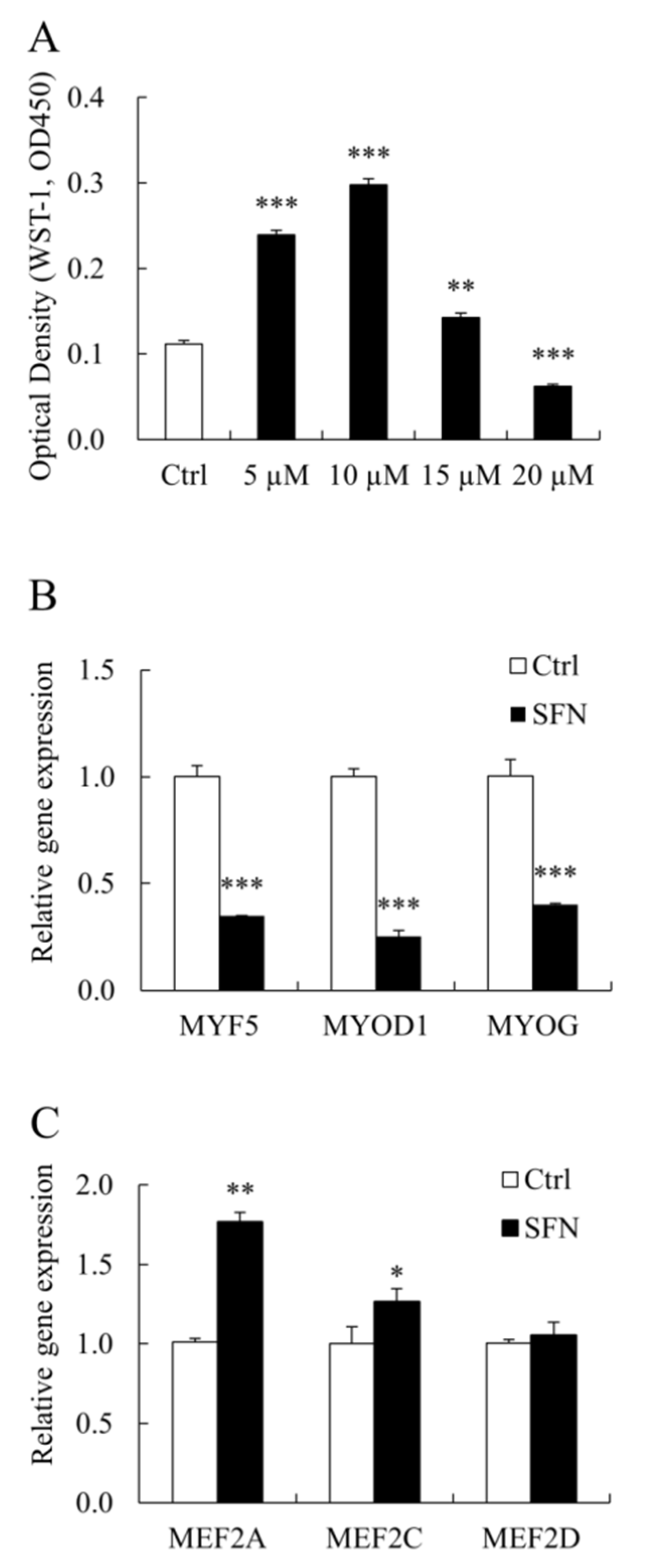
3.1. SFN Modulated PSC Proliferation and Myogenic Genes Expression
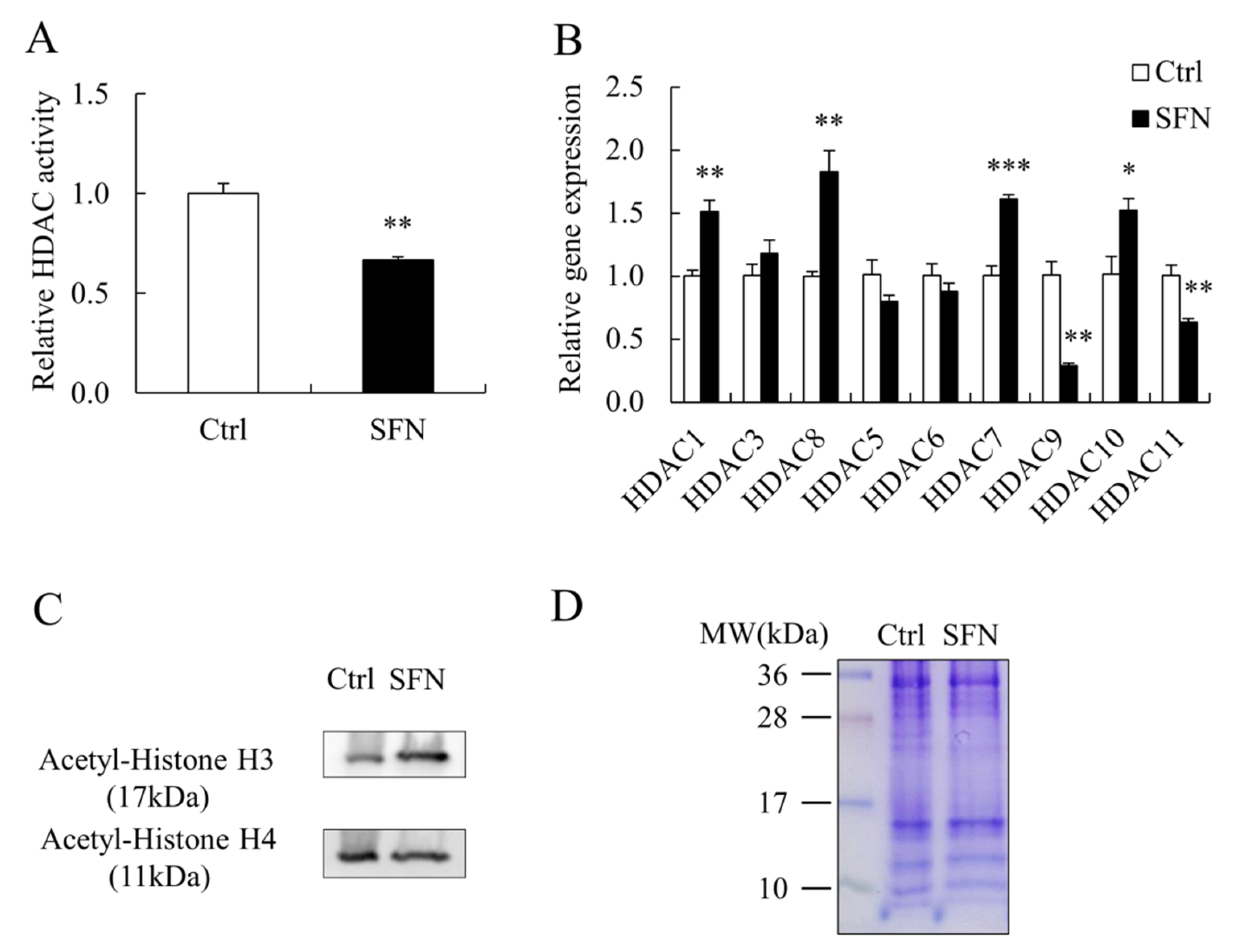
3.2. SFN Inhibited HDAC Activity and Elevated Global Histone Acetylation Level
3.3. SFN Increased SMAD7 Expression in PSCs
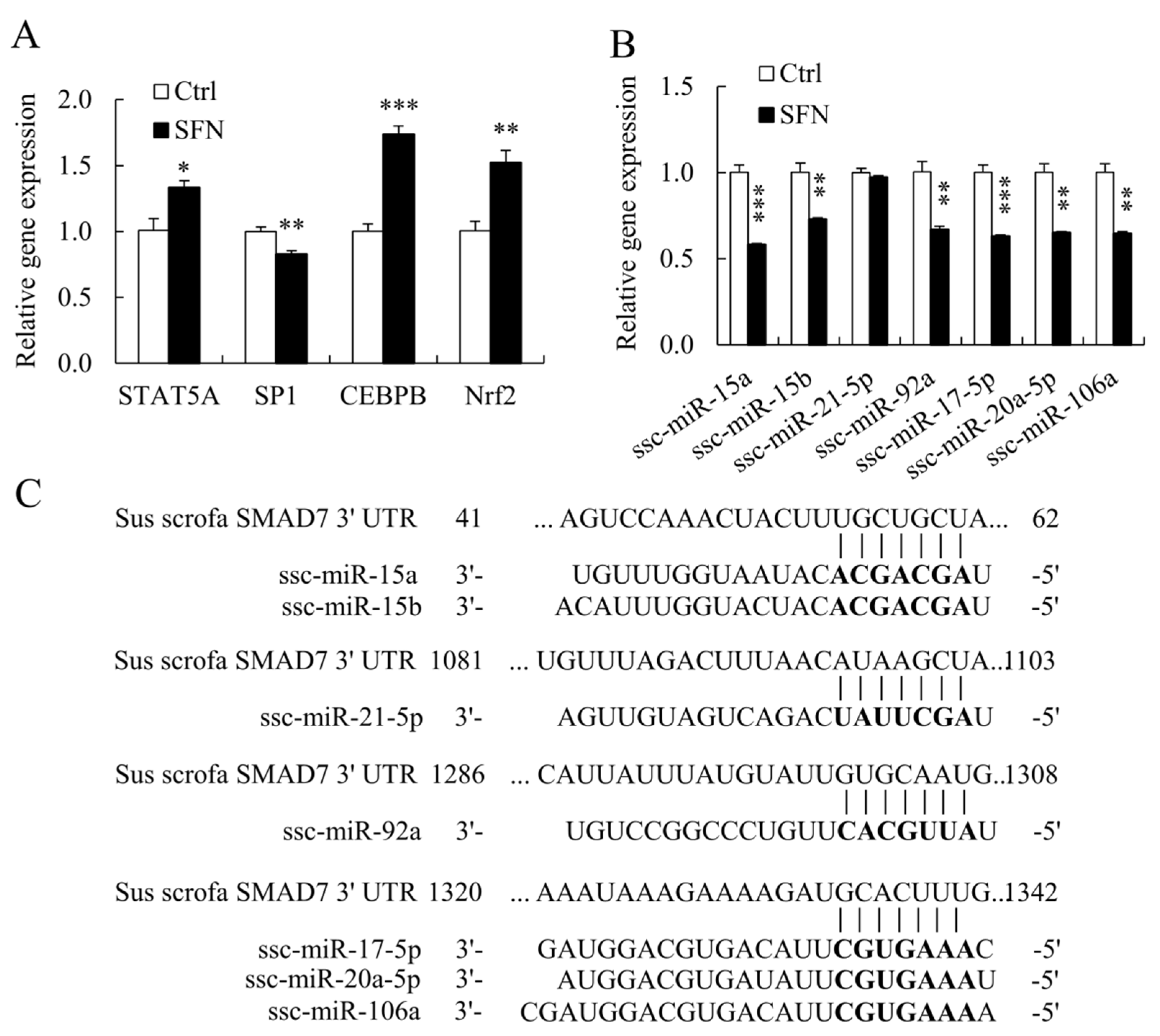
3.4. SFN Altered the Expression of TFs and miRNAs Involved in Regulating SMAD7
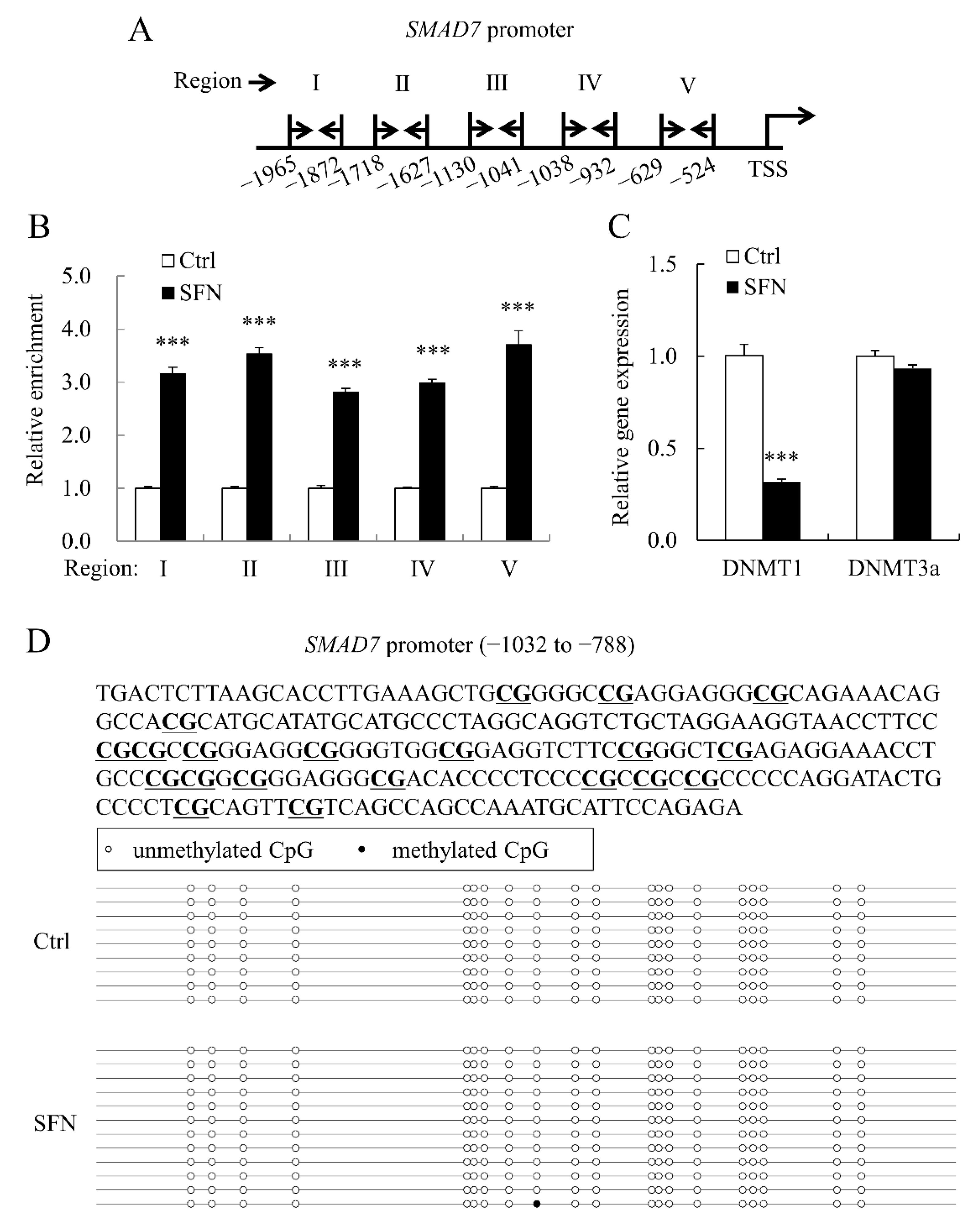
3.5. SFN Elevated Histone H4 Acetylation Level at SMAD7 Promoter
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mauro, A. Satellite cell of skeletal muscle fibers. J. Biophys. Biochem. Cytol. 1961, 9, 493–495. [Google Scholar] [CrossRef]
- Moss, F.P. The relationship between the dimensions of the fibres and the number of nuclei during normal growth of skeletal muscle in the domestic fowl. Am. J. Anat. 1968, 122, 555–563. [Google Scholar] [CrossRef] [PubMed]
- Molkentin, J.D.; Black, B.L.; Martin, J.F.; Olson, E.N. Cooperative activation of muscle gene expression by MEF2 and myogenic bHLH proteins. Cell 1995, 83, 1125–1136. [Google Scholar] [CrossRef] [Green Version]
- Fan, H.; Zhang, R.; Tesfaye, D.; Tholen, E.; Looft, C.; Hölker, M.; Schellander, K.; Cinar, M.U. Sulforaphane causes a major epigenetic repression of myostatin in porcine satellite cells. Epigenetics 2012, 7, 1379–1390. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hayashi, H.; Abdollah, S.; Qiu, Y.; Cai, J.; Xu, Y.Y.; Grinnell, B.W.; Richardson, M.A.; Topper, J.N.; Gimbrone, M.A., Jr.; Wrana, J.L.; et al. The MAD-related protein Smad7 associates with the TGFbeta receptor and functions as an antagonist of TGFbeta signaling. Cell 1997, 89, 1165–1173. [Google Scholar] [CrossRef] [Green Version]
- Winbanks, C.E.; Murphy, K.T.; Bernardo, B.C.; Qian, H.; Liu, Y.; Sepulveda, P.V.; Beyer, C.; Hagg, A.; Thomson, R.E.; Chen, J.L.; et al. Smad7 gene delivery prevents muscle wasting associated with cancer cachexia in mice. Sci. Transl. Med. 2016, 8, 348ra398. [Google Scholar] [CrossRef] [Green Version]
- Cohen, T.V.; Kollias, H.D.; Liu, N.; Ward, C.W.; Wagner, K.R. Genetic disruption of Smad7 impairs skeletal muscle growth and regeneration. J. Physiol. 2015, 593, 2479–2497. [Google Scholar] [CrossRef] [Green Version]
- Miyake, T.; Alli, N.S.; McDermott, J.C. Nuclear function of Smad7 promotes myogenesis. Mol. Cell. Biol. 2010, 30, 722–735. [Google Scholar] [CrossRef] [Green Version]
- Grönroos, E.; Hellman, U.; Heldin, C.H.; Ericsson, J. Control of Smad7 stability by competition between acetylation and ubiquitination. Mol. Cell 2002, 10, 483–493. [Google Scholar] [CrossRef]
- Wei, L.; Zhao, S.; Wang, G.; Zhang, S.; Luo, W.; Qin, Z.; Bi, X.; Tan, Y.; Meng, M.; Qin, J.; et al. SMAD7 methylation as a novel marker in atherosclerosis. Biochem. Biophys. Res. Commun. 2018, 496, 700–705. [Google Scholar] [CrossRef]
- Smith, A.L.; Iwanaga, R.; Drasin, D.J.; Micalizzi, D.S.; Vartuli, R.L.; Tan, A.C.; Ford, H.L. The miR-106b-25 cluster targets Smad7, activates TGF-beta signaling, and induces EMT and tumor initiating cell characteristics downstream of Six1 in human breast cancer. Oncogene 2012, 31, 5162–5171. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Myzak, M.C.; Karplus, P.A.; Chung, F.L.; Dashwood, R.H. A novel mechanism of chemoprotection by sulforaphane: Inhibition of histone deacetylase. Cancer Res. 2004, 64, 5767–5774. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Komine, S.; Miura, I.; Miyashita, N.; Oh, S.; Tokinoya, K.; Shoda, J.; Ohmori, H. Effect of a sulforaphane supplement on muscle soreness and damage induced by eccentric exercise in young adults: A pilot study. Physiol. Rep. 2021, 9, e15130. [Google Scholar] [CrossRef]
- Bose, C.; Alves, I.; Singh, P.; Palade, P.T.; Carvalho, E.; Borsheim, E.; Jun, S.R.; Cheema, A.; Boerma, M.; Awasthi, S.; et al. Sulforaphane prevents age-associated cardiac and muscular dysfunction through Nrf2 signaling. Aging Cell 2020, 19, e13261. [Google Scholar] [CrossRef] [PubMed]
- Son, Y.H.; Jang, E.J.; Kim, Y.W.; Lee, J.H. Sulforaphane prevents dexamethasone-induced muscle atrophy via regulation of the Akt/Foxo1 axis in C2C12 myotubes. Biomed. Pharmacother. 2017, 95, 1486–1492. [Google Scholar] [CrossRef]
- Moon, J.Y.; Kim, D.J.; Kim, H.S. Sulforaphane ameliorates serum starvation-induced muscle atrophy via activation of the Nrf2 pathway in cultured C2C12 cells. Cell Biol. Int. 2020, 44, 1831–1839. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Proll, M.J.; Salilew-Wondim, D.; Zhang, R.; Tesfaye, D.; Fan, H.; Cinar, M.U.; Grosse-Brinkhaus, C.; Tholen, E.; Islam, M.A.; et al. LPS-induced expression of CD14 in the TRIF pathway is epigenetically regulated by sulforaphane in porcine pulmonary alveolar macrophages. Innate Immun. 2016, 22, 682–695. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Grosse-Brinkhaus, C.; Heidt, H.; Uddin, M.J.; Cinar, M.U.; Tesfaye, D.; Tholen, E.; Looft, C.; Schellander, K.; Neuhoff, C. Polymorphisms and expression analysis of SOX-6 in relation to porcine growth, carcass, and meat quality traits. Meat Sci. 2015, 107, 26–32. [Google Scholar] [CrossRef]
- Wimmers, K.; Murani, E.; Ngu, N.T.; Schellander, K.; Ponsuksili, S. Structural and functional genomics to elucidate the genetic background of microstructural and biophysical muscle properties in the pig. J. Anim. Breed Genet. 2007, 124 (Suppl. S1), 27–34. [Google Scholar] [CrossRef]
- Lewis, B.P.; Burge, C.B.; Bartel, D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 2005, 120, 15–20. [Google Scholar] [CrossRef] [Green Version]
- Kruger, J.; Rehmsmeier, M. RNAhybrid: microRNA target prediction easy, fast and flexible. Nucleic Acids Res. 2006, 34, W451–W454. [Google Scholar] [CrossRef] [PubMed]
- Miranda, K.C.; Huynh, T.; Tay, Y.; Ang, Y.S.; Tam, W.L.; Thomson, A.M.; Lim, B.; Rigoutsos, I. A pattern-based method for the identification of MicroRNA binding sites and their corresponding heteroduplexes. Cell 2006, 126, 1203–1217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koressaar, T.; Remm, M. Enhancements and modifications of primer design program Primer3. Bioinformatics 2007, 23, 1289–1291. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Li, L.C.; Dahiya, R. MethPrimer: Designing primers for methylation PCRs. Bioinformatics 2002, 18, 1427–1431. [Google Scholar] [CrossRef] [Green Version]
- Bock, C.; Reither, S.; Mikeska, T.; Paulsen, M.; Walter, J.; Lengauer, T. BiQ Analyzer: Visualization and quality control for DNA methylation data from bisulfite sequencing. Bioinformatics 2005, 21, 4067–4068. [Google Scholar] [CrossRef]
- Messeguer, X.; Escudero, R.; Farre, D.; Nunez, O.; Martinez, J.; Alba, M.M. PROMO: Detection of known transcription regulatory elements using species-tailored searches. Bioinformatics 2002, 18, 333–334. [Google Scholar] [CrossRef]
- Heinemeyer, T.; Wingender, E.; Reuter, I.; Hermjakob, H.; Kel, A.E.; Kel, O.V.; Ignatieva, E.V.; Ananko, E.A.; Podkolodnaya, O.A.; Kolpakov, F.A.; et al. Databases on transcriptional regulation: TRANSFAC, TRRD and COMPEL. Nucleic Acids Res. 1998, 26, 362–367. [Google Scholar] [CrossRef] [Green Version]
- Chang, Y.; Liu, C.; Yang, J.; Liu, G.; Feng, F.; Tang, J.; Hu, L.; Li, L.; Jiang, F.; Chen, C.; et al. miR-20a triggers metastasis of gallbladder carcinoma. J. Hepatol. 2013, 59, 518–527. [Google Scholar] [CrossRef]
- Zanichelli, F.; Capasso, S.; Cipollaro, M.; Pagnotta, E.; Carteni, M.; Casale, F.; Iori, R.; Galderisi, U. Dose-dependent effects of R-sulforaphane isothiocyanate on the biology of human mesenchymal stem cells, at dietary amounts, it promotes cell proliferation and reduces senescence and apoptosis, while at anti-cancer drug doses, it has a cytotoxic effect. Age 2012, 34, 281–293. [Google Scholar] [CrossRef] [Green Version]
- Cusella-De Angelis, M.G.; Lyons, G.; Sonnino, C.; De Angelis, L.; Vivarelli, E.; Farmer, K.; Wright, W.E.; Molinaro, M.; Bouche, M.; Buckingham, M.; et al. MyoD, myogenin independent differentiation of primordial myoblasts in mouse somites. J. Cell Biol. 1992, 116, 1243–1255. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Whitman, S.A.; Long, M.; Wondrak, G.T.; Zheng, H.; Zhang, D.D. Nrf2 modulates contractile and metabolic properties of skeletal muscle in streptozotocin-induced diabetic atrophy. Exp. Cell Res. 2013, 319, 2673–2683. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, J.; McKinsey, T.A.; Nicol, R.L.; Olson, E.N. Signal-dependent activation of the MEF2 transcription factor by dissociation from histone deacetylases. Proc. Natl. Acad. Sci. USA 2000, 97, 4070–4075. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adler, K.; Molina, P.E.; Simon, L. Epigenomic mechanisms of alcohol-induced impaired differentiation of skeletal muscle stem cells; role of Class IIA histone deacetylases. Physiol. Genom. 2019, 51, 471–479. [Google Scholar] [CrossRef]
- Rajendran, P.; Kidane, A.I.; Yu, T.W.; Dashwood, W.M.; Bisson, W.H.; Lohr, C.V.; Ho, E.; Williams, D.E.; Dashwood, R.H. HDAC turnover, CtIP acetylation and dysregulated DNA damage signaling in colon cancer cells treated with sulforaphane and related dietary isothiocyanates. Epigenetics 2013, 8, 612–623. [Google Scholar] [CrossRef]
- Abbaoui, B.; Telu, K.H.; Lucas, C.R.; Thomas-Ahner, J.M.; Schwartz, S.J.; Clinton, S.K.; Freitas, M.A.; Mortazavi, A. The impact of cruciferous vegetable isothiocyanates on histone acetylation and histone phosphorylation in bladder cancer. J. Proteom. 2017, 156, 94–103. [Google Scholar] [CrossRef] [Green Version]
- Carlson, M.E.; Hsu, M.; Conboy, I.M. Imbalance between pSmad3 and Notch induces CDK inhibitors in old muscle stem cells. Nature 2008, 454, 528–532. [Google Scholar] [CrossRef] [Green Version]
- Cho, J.S.; Moon, Y.M.; Park, I.H.; Um, J.Y.; Moon, J.H.; Park, S.J.; Lee, S.H.; Kang, H.J.; Lee, H.M. Epigenetic regulation of myofibroblast differentiation and extracellular matrix production in nasal polyp-derived fibroblasts. Clin. Exp. Allergy 2012, 42, 872–882. [Google Scholar] [CrossRef]
- Salot, S.; Gude, R. MTA1-mediated transcriptional repression of SMAD7 in breast cancer cell lines. Eur. J. Cancer 2013, 49, 492–499. [Google Scholar] [CrossRef]
- Song, M.K.; Lee, J.H.; Ryoo, I.G.; Lee, S.H.; Ku, S.K.; Kwak, M.K. Bardoxolone ameliorates TGF-beta1-associated renal fibrosis through Nrf2/Smad7 elevation. Free Radic. Biol. Med. 2019, 138, 33–42. [Google Scholar] [CrossRef]



| Purpose | Gene | Sequence (5′—3′) | Size (bp) | GenBank Accession Number |
|---|---|---|---|---|
| qRT-PCR for mRNA expression | MYF5 | F: AGACGCCTCAAGAAGGTCAA | NM_001278775 | |
| R: TCCTGCAGGCTCTCAATGTA | 128 | |||
| MYOD1 | F: TGCAAACGCAAGACCACTAA | NM_001002824 | ||
| R: GCTGATTCGGGTTGCTAGAC | 127 | |||
| MYOG | F: CAGTGAATGCAGTTCCCACA | NM_001012406 | ||
| R: GGTGAGGGAGTGCAGATTGT | 130 | |||
| MEF2A | F: TGATGCGGAATCATAAAATCG | NM_001099698 | ||
| R: GCACCAGTAGTTCCAACCAAA | 358 | |||
| MEF2C | F: CGAGATACCCACAACACACG | NM_001044540 | ||
| R: CGCTTGACTGAGGGACTTTC | 175 | |||
| MEF2D | F: TCACTGCAGTTCAGCAATCC | XM_021089672 | ||
| R: AGGCCAGGAGACACACTGTT | 128 | |||
| HDAC1 | F: GGAAATCTATCGCCCTCACA | XM_013999116 | ||
| R: AAACACCGGACAGTCCTCAC | 157 | |||
| HDAC3 | F: CAACCAGGTGGTGGACTTCT | NM_001243827 | ||
| R: GCAGAGGGATGTTGAAGCTC | 152 | |||
| HDAC5 | F: AGATGCACTCCTCCAGTGCT | XM_021066892 | ||
| R: GGATGATGGCAAATCCATTC | 102 | |||
| HDAC6 | F: ATGGACGGGTATTGCATGTT | XM_003360315 | ||
| R: GCGGTGGATGGAGAAATAGA | 168 | |||
| HDAC7 | F: CGTCCCCTACAGAACTCTCG | XM_021092604 | ||
| R: TCAGGTTGGGCTCAGAGACT | 146 | |||
| HDAC8 | F: GGTGACGTGTCTGATGTTGG | XM_021080459 | ||
| R: AGCTCCCAGCTGTAAGACCA | 165 | |||
| HDAC9 | F: AACTGAAGCAACCAGGCAGT | XM_021102482 | ||
| R: CCCAACTTGTCCCAGTGAGT | 149 | |||
| HDAC10 | F: TCCATCCGAGTACCTTCCAC | XM_021091335 | ||
| R: GGCTGCTATGGCCACACTAT | 179 | |||
| HDAC11 | F: GACAAGCGCGTGTACATCAT | XM_021069384 | ||
| R: AGGTTCCTCTCCACCTTCGT | 143 | |||
| TGFB1 | F: CGTGCTAATGGTGGAAAGCG | XM_021093503 | ||
| R: AGAGCAATACAGGTTCCGGC | 122 | |||
| SMAD2 | F: GCAATCTTTGTGCAGAGCCC | NM_001256148 | ||
| R: ACACGGCTTCAAAACCCTGA | 157 | |||
| SMAD3 | F: GCTGGACGACTACAGCCATT | NM_214137 | ||
| R: TGTGGTTCATCTGGTGGTCG | 140 | |||
| SMAD7 | F: CCAACTGCAGACTGTCCAGA | XM_005659454 | ||
| R: CAGGCTCCAGAAGAAGTTGG | 106 | |||
| STAT5A | F: GAGGTGCTGAAGAAGCATCA | NM_214290 | ||
| R: GGCTTCAGATTCCACAGGTT | 200 | |||
| SP1 | F: TGCAGCAGAATTGAGTCACC | XM_005652567 | ||
| R: ACTGCTGCCACTTTGTTCCT | 180 | |||
| CEBPB | F: GCTTGAACAAGTTCCG | NM_001199889 | ||
| R: CAAGAAGACCGTGGATAAGC | 209 | |||
| Nrf2 | F: GTGCCTATAAGTCCCGGTCA | XM_013984303 | ||
| R: ATGCAGAGCTTTTGCCCTTA | 108 | |||
| DNMT1 | F: GCGGGACCTACCAAACAT | NM_001032355 | ||
| R: TTCCACGCAGGAGCAGAC | 133 | |||
| DNMT3a | F: CTGAGAAGCCCAAGGTCAAG | NM_001097437 | ||
| R: CAGCAGATGGTGCAGTAGGA | 238 | |||
| HPRT1 | F: AACCTTGCTTTCCTTGGTCA | NM_001032376 | ||
| R: TCAAGGGCATAGCCTACCAC | 150 | |||
| qRT-PCR for miRNA expression | ssc-miR-15a | F: TAGCAGCACATAATGGTTTGT | - | MIMAT0007753 |
| ssc-miR-15b | F: TAGCAGCACATCATGGTTTACA | - | MIMAT0002125 | |
| ssc-miR-17-5p | F: CAAAGTGCTTACAGTGCAGGTAG | - | MIMAT0007755 | |
| ssc-miR-20a-5p | F: TAAAGTGCTTATAGTGCAGGTA | - | MIMAT0002129 | |
| ssc-miR-21-5p | F: TAGCTTATCAGACTGATGTTGA | - | MIMAT0002165 | |
| ssc-miR-92a | F: TATTGCACTTGTCCCGGCCTGT | - | MIMAT0013908 | |
| ssc-miR-106a | F: AAAAGTGCTTACAGTGCAGGTAGC | - | MIMAT0002118 | |
| Bisulfite sequencing PCR | SMAD7-Bis | F: TGATTTTTAAGTATTTTGAAAGTTG | ||
| R: TCTCTAAAATACATTTAACTAACTAAC | 245 | |||
| ChIP-qPCR | Region I | F: TATGCCTCATGCACAGCACC | ||
| R: CCCATGCACAGGGAAAGACA | 93 | |||
| Region II | F: ATTGCAGCCTCTGTGGCTTA | |||
| R: GACCTAGGGATGCCAAGCAG | 91 | |||
| Region III | F: TGGTCCTTTGCCCTACCAAC | |||
| R: ATCCCCTTAGCCTGCGTTTT | 89 | |||
| Region IV | F: ACTCTCTGACTCTTAAGCACCT | |||
| R: AGGTTACCTTCCTAGCAGACCT | 106 | |||
| Region V | F: CGGTCCAGTCCGGTGTAAAT | |||
| R: CGTTTTGCCTTAAAGGCCCTG | 105 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, R.; Neuhoff, C.; Yang, Q.; Cinar, M.U.; Uddin, M.J.; Tholen, E.; Schellander, K.; Tesfaye, D. Sulforaphane Enhanced Proliferation of Porcine Satellite Cells via Epigenetic Augmentation of SMAD7. Animals 2022, 12, 1365. https://doi.org/10.3390/ani12111365
Zhang R, Neuhoff C, Yang Q, Cinar MU, Uddin MJ, Tholen E, Schellander K, Tesfaye D. Sulforaphane Enhanced Proliferation of Porcine Satellite Cells via Epigenetic Augmentation of SMAD7. Animals. 2022; 12(11):1365. https://doi.org/10.3390/ani12111365
Chicago/Turabian StyleZhang, Rui, Christiane Neuhoff, Qin Yang, Mehmet U. Cinar, Muhammad J. Uddin, Ernst Tholen, Karl Schellander, and Dawit Tesfaye. 2022. "Sulforaphane Enhanced Proliferation of Porcine Satellite Cells via Epigenetic Augmentation of SMAD7" Animals 12, no. 11: 1365. https://doi.org/10.3390/ani12111365
APA StyleZhang, R., Neuhoff, C., Yang, Q., Cinar, M. U., Uddin, M. J., Tholen, E., Schellander, K., & Tesfaye, D. (2022). Sulforaphane Enhanced Proliferation of Porcine Satellite Cells via Epigenetic Augmentation of SMAD7. Animals, 12(11), 1365. https://doi.org/10.3390/ani12111365







