Genetic Evaluation and Population Structure of Jiangsu Native Pigs in China Revealed by SINE Insertion Polymorphisms
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. DNA Extraction and SINE-RIPs Genotyping
2.3. Statistical Analyses
3. Results
3.1. Evaluation of the SINE-RIPs in Jiangsu Pig Populations
3.2. Genetic Variability
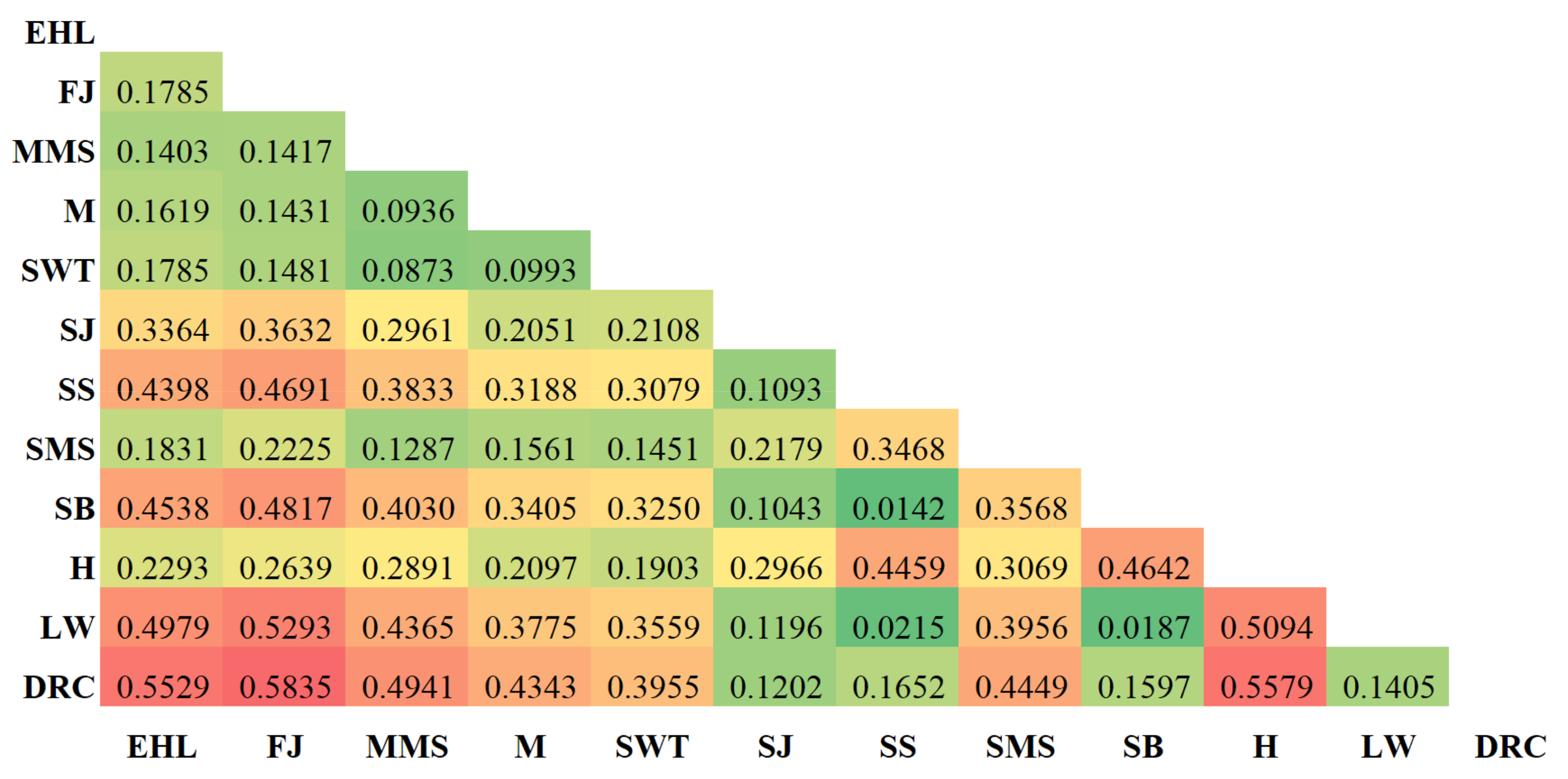
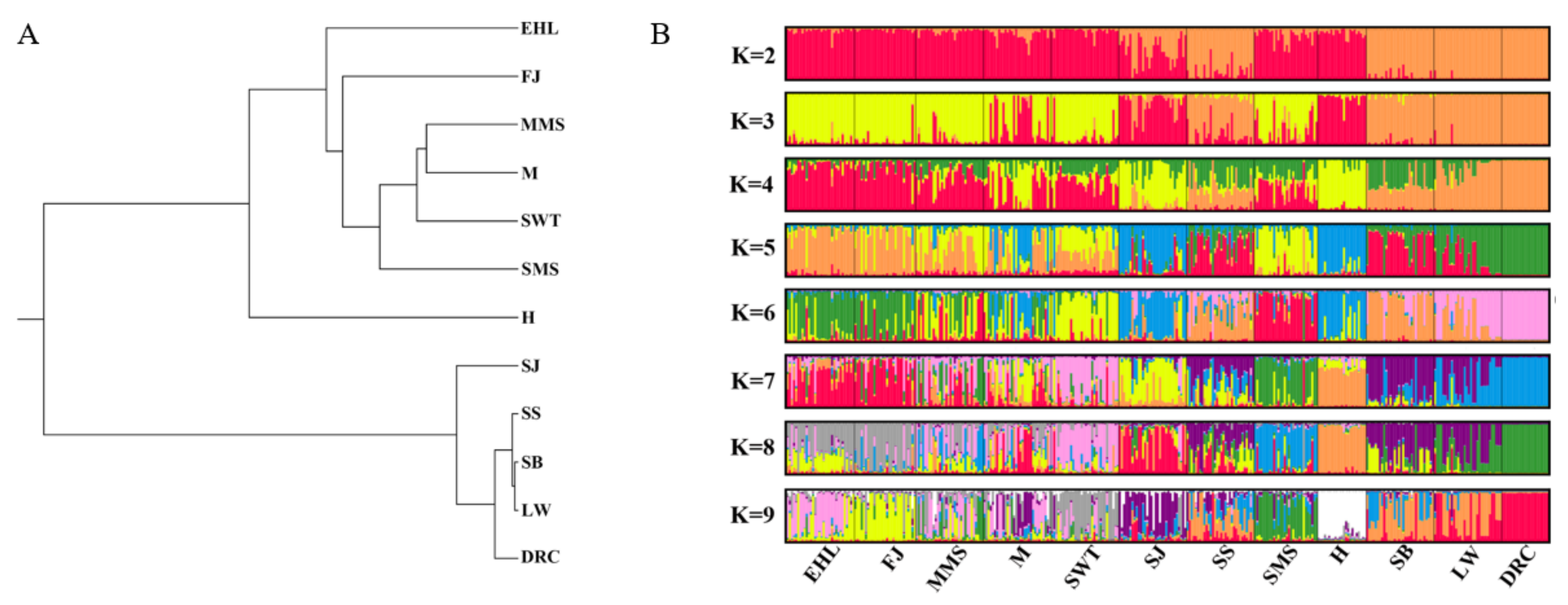
3.3. Genetic Distance and Population Structure
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wang, Z.; Chen, Q.; Yang, Y.; Liao, R.; Zhao, J.; Zhang, Z.; Chen, Z.; Zhang, X.; Xue, M.; Yang, H.; et al. Genetic diversity and population structure of six Chinese indigenous pig breeds in the Taihu Lake region revealed by sequencing data. Anim. Genet. 2015, 46, 697–701. [Google Scholar] [CrossRef] [PubMed]
- Cesar, A.S.M.; Silveira, A.C.P.; Freitas, P.F.A.; Guimaraes, E.C.; Batista, D.F.A.; Torido, L.C.; Meirelles, F.V.; Antunes, R.C. Influence of Chinese breeds on pork quality of commercial pig lines. Genet. Mol. Res. 2010, 9, 727–733. [Google Scholar] [CrossRef] [PubMed]
- Jing, R. The evolution of conversation and breeding of Jiangsu native pig breeds. Swine Ind. Sci. 2009, 26, 96–99. [Google Scholar]
- Chang, Q.; Zhou, K.Y.; Wang, Y.Q.; Zhang, Z.K.; Cao, X. RAPD analysis of genetic diversity and phylogenetic relationship of the Taihu pigs. Yi Chuan Xue Bao 1999, 26, 480–488. [Google Scholar] [PubMed]
- Fan, B.; Wang, Z.G.; Li, Y.J.; Zhao, X.L.; Liu, B.; Zhao, S.H.; Yu, M.; Li, M.H.; Chen, S.L.; Xiong, T.A.; et al. Genetic variation analysis within and among Chinese indigenous swine populations using microsatellite markers. Anim. Genet. 2002, 33, 422–427. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Chen, Q.; Liao, R.; Zhang, Z.; Zhang, X.; Liu, X.; Zhu, M.; Zhang, W.; Xue, M.; Yang, H.; et al. Genome-wide genetic variation discovery in Chinese Taihu pig breeds using next generation sequencing. Anim. Genet. 2017, 48, 38–47. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Q.B.; Sun, H.; Zhang, Z.; Xu, Z.; Olasege, B.S.; Ma, P.P.; Zhang, X.Z.; Wang, Q.S.; Pan, Y.C. Exploring the Structure of Haplotype Blocks and Genetic Diversity in Chinese Indigenous Pig Populations for Conservation Purpose. Evol. Bioinform. Online 2019, 15, 1176934318825082. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Q.B.; Lopez-Cortegano, E.; Oyelami, F.O.; Zhang, Z.; Ma, P.P.; Wang, Q.S.; Pan, Y.C. Conservation priorities analysis of chinese indigenous pig breeds in the Taihu lake region. Front. Genet. 2021, 12, 558873. [Google Scholar] [CrossRef]
- Wessler, S.R.; Bureau, T.E.; White, S.E. LTR-retrotransposons and MITEs: Important players in the evolution of plant genomes. Curr. Opin. Genet. Dev. 1995, 5, 814–821. [Google Scholar] [CrossRef]
- Finnegan, D.J. Eukaryotic transposable elements and genome evolution. Trends Genet. 1989, 5, 103–107. [Google Scholar] [CrossRef]
- Platt, R.N.; Vandewege, M.W.; Ray, D.A. Mammalian transposable elements and their impacts on genome evolution. Chromosome Res. 2018, 26, 25–43. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kalendar, R.; Schulman, A.H. Transposon-based tagging: IRAP, REMAP, and iPBS. Methods Mol. Biol. 2014, 1115, 233–255. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kalendar, R.; Muterko, A.; Boronnikova, S. Retrotransposable Elements: DNA Fingerprinting and the Assessment of Genetic Diversity. Methods Mol. Biol. 2021, 2222, 263–286. [Google Scholar] [CrossRef] [PubMed]
- Hancks, D.C.; Kazazian, H.H., Jr. Active human retrotransposons: Variation and disease. Curr. Opin. Genet. Dev. 2012, 22, 191–203. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Halo, J.V.; Pendleton, A.L.; Shen, F.; Doucet, A.J.; Derrien, T.; Hitte, C.; Kirby, L.E.; Myers, B.; Sliwerska, E.; Emery, S.; et al. Long-read assembly of a Great Dane genome highlights the contribution of GC-rich sequence and mobile elements to canine genomes. Proc. Natl. Acad. Sci. USA 2021, 118, e2016274118. [Google Scholar] [CrossRef] [PubMed]
- Shedlock, A.M.; Okada, N. SINE insertions: Powerful tools for molecular systematics. Bioessays 2000, 22, 148–160. [Google Scholar] [CrossRef]
- Kramerov, D.A.; Vassetzky, N.S. Short retroposons in eukaryotic genomes. Int. Rev. Cytol.-A Surv. Cell Biol. 2005, 247, 165–221. [Google Scholar] [CrossRef]
- Wenke, T.; Dobel, T.; Sorensen, T.R.; Junghans, H.; Weisshaar, B.; Schmidt, T. Targeted Identification of Short Interspersed Nuclear Element Families Shows Their Widespread Existence and Extreme Heterogeneity in Plant Genomes. Plant Cell 2011, 23, 3117–3128. [Google Scholar] [CrossRef] [Green Version]
- Kramerov, D.A.; Vassetzky, N.S. Origin and evolution of SINEs in eukaryotic genomes. Heredity 2011, 107, 487–495. [Google Scholar] [CrossRef]
- Seibt, K.M.; Wenke, T.; Wollrab, C.; Junghans, H.; Muders, K.; Dehmer, K.J.; Diekmann, K.; Schmidt, T. Development and application of SINE-based markers for genotyping of potato varieties. Theor. Appl. Genet. 2012, 125, 185–196. [Google Scholar] [CrossRef]
- Mao, H.; Wang, H. Distribution, Diversity, and Long-Term Retention of Grass Short Interspersed Nuclear Elements (SINEs). Genome Biol. Evol. 2017, 9, 2048–2056. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meng, H.; Feng, J.; Bai, T.; Jian, Z.; Chen, Y.; Wu, G. Genome-wide analysis of short interspersed nuclear elements provides insight into gene and genome evolution in citrus. DNA Res. 2020, 27, dsaa004. [Google Scholar] [CrossRef] [PubMed]
- Batzer, M.A.; Stoneking, M.; Alegria-Hartman, M.; Bazan, H.; Kass, D.H.; Shaikh, T.H.; Novick, G.E.; Ioannou, P.A.; Scheer, W.D.; Herrera, R.J. African origin of human-specific polymorphic Alu insertions. Proc. Natl. Acad. Sci. USA 1994, 91, 12288–12292. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, C.; Wang, W.; Wang, X.; Shen, D.; Wang, S.; Wang, Y.; Gao, B.; Wimmers, K.; Mao, J.; Li, K.; et al. Retrotransposons evolution and impact on lncRNA and protein coding genes in pigs. Mob. DNA 2019, 10, 19. [Google Scholar] [CrossRef]
- Chen, C.; D’Alessandro, E.; Murani, E.; Zheng, Y.; Giosa, D.; Yang, N.S.; Wang, X.Y.; Gao, B.; Li, K.; Wimmers, K.; et al. SINE jumping contributes to large-scale polymorphisms in the pig genomes. Mob. DNA 2021, 12, 17. [Google Scholar] [CrossRef]
- Chen, C.; Wang, X.; Zong, W.; D’Alessandro, E.; Giosa, D.; Guo, Y.; Mao, J.; Song, C. Genetic Diversity and Population Structures in Chinese Miniature Pigs Revealed by SINE Retrotransposon Insertion Polymorphisms, a New Type of Genetic Markers. Animals 2021, 11, 1136. [Google Scholar] [CrossRef]
- Zumbo, A.; Sutera, A.M.; Tardiolo, G.; D’Alessandro, E. Sicilian Black Pig: An Overview. Animals 2020, 10, 2326. [Google Scholar] [CrossRef]
- Yeh, F.; Yang, R.; Boyle, T. POPGENE Version 1.32 Microsoft Windows-Based Freeware for Populations Genetic Analysis; University of Alberta: Edmonton, AB, Canada, 1999. [Google Scholar]
- Botstein, D.; White, R.L.; Skolnick, M.; Davis, R.W. Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am. J. Hum. Genet. 1980, 32, 314–331. [Google Scholar]
- Earl, D.A.; Vonholdt, B.M. Structure Harvester: A website and program for visualizing structure output and implementing the Evanno method. Conserv. Genet. Resour. 2012, 4, 359–361. [Google Scholar] [CrossRef]
- Jakobsson, M.; Rosenberg, N.A. CLUMPP: A cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 2007, 23, 1801–1806. [Google Scholar] [CrossRef] [Green Version]
- Rosenberg, N.A. distruct: A program for the graphical display of population structure. Mol. Ecol. Notes 2004, 4, 137–138. [Google Scholar] [CrossRef]
- Ai, H.S.; Huang, L.S.; Ren, J. Genetic diversity, linkage disequilibrium and selection signatures in chinese and western pigs revealed by genome-wide snp markers. PLoS ONE 2013, 8, e56001. [Google Scholar] [CrossRef] [Green Version]
- Giuffra, E.; Kijas, J.M.; Amarger, V.; Carlborg, O.; Jeon, J.T.; Andersson, L. The origin of the domestic pig: Independent domestication and subsequent introgression. Genetics 2000, 154, 1785–1791. [Google Scholar] [CrossRef] [PubMed]
- Xu, P.; Ni, L.; Tao, Y.; Ma, Z.; Hu, T.; Zhao, X.; Yu, Z.; Lu, C.; Zhao, X.; Ren, J. Genome-wide association study for growth and fatness traits in Chinese Sujiang pigs. Anim. Genet. 2020, 51, 314–318. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Zhao, W.; Fu, Y.; Fang, X.; Ren, S.; Ren, J. Genome-wide detection of genetic loci and candidate genes for teat number and body conformation traits at birth in Chinese Sushan pigs. Anim. Genet. 2019, 50, 753–756. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Zhu, M.; Pan, Y.; Hou, Q.; Ni, L. Protection, development and utilization of local pig genetic resources in Jiangsu Province. Jiangsu Agric. Sci. 2018, 46, 179–181. (In Chinese) [Google Scholar]
- Zhang, X. Comparison On Partial Traits and Investigation On Breed Resourses OF Huai Bei Pig in Gan Yu Region; Nanjing Agricultural University: Nangjing, China, 2004. (In Chinese) [Google Scholar]



| SINE-RIPs | Insertion Frequency | No. of Populations Show Polymorphic | No. of Populations Show Hardy–Weinberg Disequilibrium | FIS | FST | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EHL | FJ | MMS | M | SWT | SJ | SS | SMS | H | SB | LW | DRC | |||||
| REF-815 | 1.00 | 0.02 | 0.47 | 0.25 | 0.19 | 0.00 | 0.00 | 0.22 | 0.00 | 0.00 | 0.00 | 0.00 | 5 | 0 | −0.0645 | 0.559 |
| REF-12270 | 0.48 | 0.36 | 0.53 | 0.45 | 0.23 | 0.02 | 0.64 | 0.10 | 0.00 | 0.52 | 0.45 | 0.00 | 10 | 1 | 0.0599 | 0.2325 |
| REF-13182 | 1.00 | 1.00 | 0.92 | 1.00 | 0.81 | 0.39 | 0.17 | 0.98 | 1.00 | 0.05 | 0.08 | 0.19 | 8 | 0 | −0.0894 | 0.6808 |
| REF-14427 | 0.58 | 0.41 | 0.11 | 0.16 | 0.00 | 0.03 | 0.06 | 0.02 | 0.96 | 0.02 | 0.00 | 0.00 | 9 | 2 | 0.3258 | 0.5339 |
| REF-16131 | 0.39 | 0.57 | 0.50 | 0.50 | 0.72 | 0.17 | 0.00 | 0.13 | 0.22 | 0.00 | 0.00 | 0.00 | 8 | 3 | −0.1121 | 0.3126 |
| REF-16684 | 0.08 | 0.40 | 0.08 | 0.05 | 0.06 | 0.00 | 0.00 | 0.30 | 0.00 | 0.00 | 0.00 | 0.00 | 6 | 0 | −0.1354 | 0.2129 |
| REF-18327 | 0.39 | 0.62 | 0.86 | 0.25 | 0.36 | 0.00 | 0.00 | 0.45 | 0.00 | 0.00 | 0.00 | 0.00 | 6 | 3 | 0.0704 | 0.4313 |
| REF-19717 | 0.05 | 0.14 | 0.33 | 0.13 | 0.56 | 0.03 | 0.00 | 0.32 | 0.13 | 0.00 | 0.00 | 0.00 | 8 | 0 | −0.0369 | 0.2385 |
| REF-21609 | 0.84 | 0.85 | 0.94 | 0.70 | 0.56 | 0.27 | 0.03 | 0.55 | 0.67 | 0.00 | 0.03 | 0.00 | 10 | 1 | −0.0914 | 0.4959 |
| REF-2929 | 0.98 | 1.00 | 0.98 | 0.83 | 0.61 | 0.25 | 0.16 | 1.00 | 0.37 | 0.25 | 0.16 | 0.00 | 9 | 0 | −0.1704 | 0.5683 |
| REF-3719 | 0.59 | 0.10 | 0.25 | 0.13 | 0.48 | 0.08 | 0.03 | 0.03 | 1.00 | 0.00 | 0.00 | 0.00 | 8 | 1 | −0.0223 | 0.5148 |
| REF-4531 | 0.52 | 0.67 | 0.14 | 0.05 | 0.14 | 0.06 | 0.11 | 0.18 | 0.00 | 0.19 | 0.03 | 0.00 | 10 | 1 | −0.1918 | 0.2789 |
| REF-5597 | 0.92 | 0.71 | 0.59 | 0.11 | 0.72 | 0.14 | 0.08 | 0.93 | 0.44 | 0.06 | 0.03 | 0.00 | 11 | 1 | −0.0636 | 0.5081 |
| REF-7445 | 0.66 | 0.38 | 0.48 | 0.48 | 0.33 | 0.30 | 0.03 | 0.35 | 0.44 | 0.06 | 0.00 | 0.00 | 10 | 1 | −0.1221 | 0.2137 |
| REF-8430 | 0.23 | 0.93 | 0.41 | 0.23 | 0.38 | 0.00 | 0.02 | 0.02 | 0.39 | 0.02 | 0.00 | 0.00 | 9 | 2 | −0.0053 | 0.4219 |
| REF-9435 | 0.84 | 0.86 | 0.47 | 0.83 | 0.28 | 0.31 | 0.05 | 0.58 | 0.61 | 0.03 | 0.00 | 0.00 | 10 | 2 | 0.1531 | 0.4416 |
| REF-10096 | 0.53 | 0.71 | 0.00 | 0.41 | 0.42 | 0.33 | 0.16 | 0.03 | 0.89 | 0.20 | 0.08 | 0.00 | 10 | 1 | −0.0309 | 0.358 |
| REF-11062 | 0.45 | 0.97 | 0.86 | 0.86 | 0.98 | 0.08 | 0.09 | 0.23 | 0.63 | 0.02 | 0.00 | 0.00 | 10 | 2 | 0.0896 | 0.6186 |
| No. of loci show non-polymorphic | 2 | 2 | 1 | 1 | 1 | 4 | 5 | 1 | 7 | 7 | 11 | 17 | N | N | N | N |
| Breed | Sample Size | He | Ho | Polymorphic Information Content (PIC) | Effective Number of Allele (Ne) | Fis |
|---|---|---|---|---|---|---|
| EHL | 32 | 0.3195 ± 0.1999 | 0.3108 ± 0.2347 | 0.2467 ± 0.1404 | 1.5687 ± 0.4102 | 0.0070 ± 0.2802 |
| FJ | 29 | 0.2984 ± 0.1894 | 0.3142 ± 0.2409 | 0.2339 ± 0.1347 | 1.5069 ± 0.3738 | −0.0261 ± 0.2812 |
| MMS | 32 | 0.3178 ± 0.1831 | 0.2969 ± 0.1661 | 0.2485 ± 0.1254 | 1.5515 ± 0.3936 | 0.0177 ± 0.1464 |
| M | 32 | 0.3042 ± 0.1535 | 0.2674 ± 0.1832 | 0.2438 ± 0.1064 | 1.4922 ± 0.3251 | 0.1108 ± 0.2999 |
| SWT | 32 | 0.3606 ± 0.1625 | 0.3854 ± 0.1896 | 0.2799 ± 0.1147 | 1.6265 ± 0.3317 | −0.0821 ± 0.1746 |
| SJ | 32 | 0.2039 ± 0.1828 | 0.2240 ± 0.2123 | 0.1653 ± 0.1360 | 1.3197 ± 0.3256 | 0.0744 ± 0.1656 |
| SS | 32 | 0.1242 ± 0.1314 | 0.1181 ± 0.1147 | 0.1069 ± 0.1022 | 1.1688 ± 0.2169 | −0.0142 ± 0.1425 |
| SMS | 30 | 0.2567 ± 0.1904 | 0.2704 ± 0.2351 | 0.2040 ± 0.1367 | 1.4238 ± 0.3691 | 0.0662 ± 0.3972 |
| SB | 32 | 0.1117 ± 0.1582 | 0.1215 ± 0.1790 | 0.0925 ± 0.1196 | 1.1688 ± 0.2783 | −0.0718 ± 0. 0988 |
| H | 23 | 0.2357 ± 0.2243 | 0.2464 ± 0.2400 | 0.1813 ± 0.1628 | 1.4123 ± 0.4214 | −0.0708 ± 0.1428 |
| LW | 32 | 0.0694 ± 0.1312 | 0.0677 ± 0.1190 | 0.0581 ± 0.0991 | 1.1039 ± 0.2387 | −0.0387 ± 0.1027 |
| DRC | 32 | 0.0172 ± 0.0730 | 0.0208 ± 0.0884 | 0.0143 ± 0.0592 | 1.0243 ± 0.1033 | −0.2308 ± 0.000 |
| Average | 0.2183 ± 0.1076 | 0.2203 ± 0.1076 | 0.1832 ± 0.0748 | 1.3640 ± 0.1936 | −0.0339 ± 0.0823 |
| ID | EHL | FJ | MMS | M | SWT | SJ | SS | SMS | SB | H | LW | DRC |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EHL | **** | 0.8110 | 0.8508 | 0.8289 | 0.7817 | 0.6502 | 0.5656 | 0.8236 | 0.5570 | 0.7769 | 0.5368 | 0.5171 |
| FJ | 0.2095 | **** | 0.8559 | 0.8586 | 0.8344 | 0.6288 | 0.5438 | 0.7854 | 0.5375 | 0.7482 | 0.5133 | 0.4998 |
| MMS | 0.1616 | 0.1556 | **** | 0.9089 | 0.9045 | 0.7114 | 0.6594 | 0.8843 | 0.6435 | 0.6950 | 0.6427 | 0.6243 |
| M | 0.1876 | 0.1525 | 0.0955 | **** | 0.8935 | 0.8297 | 0.7548 | 0.8596 | 0.7395 | 0.8119 | 0.7342 | 0.7217 |
| SWT | 0.2463 | 0.1811 | 0.1003 | 0.1126 | **** | 0.7990 | 0.7298 | 0.8543 | 0.7176 | 0.8058 | 0.7153 | 0.7167 |
| SJ | 0.4305 | 0.4640 | 0.3406 | 0.1867 | 0.2243 | **** | 0.9538 | 0.8384 | 0.9586 | 0.7704 | 0.9606 | 0.9718 |
| SS | 0.5699 | 0.6091 | 0.4165 | 0.2813 | 0.3150 | 0.0473 | **** | 0.7596 | 0.9962 | 0.6680 | 0.9958 | 0.9720 |
| SMS | 0.1941 | 0.2416 | 0.1229 | 0.1513 | 0.1575 | 0.1763 | 0.2750 | **** | 0.7589 | 0.7110 | 0.7571 | 0.7613 |
| SB | 0.5852 | 0.6209 | 0.4409 | 0.3018 | 0.3318 | 0.0423 | 0.0038 | 0.2758 | **** | 0.6515 | 0.9965 | 0.9755 |
| H | 0.2525 | 0.2900 | 0.3639 | 0.2084 | 0.2159 | 0.2608 | 0.4035 | 0.3410 | 0.4285 | **** | 0.6484 | 0.6674 |
| LW | 0.6221 | 0.6669 | 0.4421 | 0.3090 | 0.3351 | 0.0402 | 0.0042 | 0.2783 | 0.0035 | 0.4332 | **** | 0.9858 |
| DRC | 0.6596 | 0.6936 | 0.4711 | 0.3261 | 0.3331 | 0.0286 | 0.0284 | 0.2727 | 0.0249 | 0.4043 | 0.0143 | **** |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, X.; D’Alessandro, E.; Chi, C.; Moawad, A.S.; Zong, W.; Chen, C.; Song, C. Genetic Evaluation and Population Structure of Jiangsu Native Pigs in China Revealed by SINE Insertion Polymorphisms. Animals 2022, 12, 1345. https://doi.org/10.3390/ani12111345
Wang X, D’Alessandro E, Chi C, Moawad AS, Zong W, Chen C, Song C. Genetic Evaluation and Population Structure of Jiangsu Native Pigs in China Revealed by SINE Insertion Polymorphisms. Animals. 2022; 12(11):1345. https://doi.org/10.3390/ani12111345
Chicago/Turabian StyleWang, Xiaoyan, Enrico D’Alessandro, Chenglin Chi, Ali Shoaib Moawad, Wencheng Zong, Cai Chen, and Chengyi Song. 2022. "Genetic Evaluation and Population Structure of Jiangsu Native Pigs in China Revealed by SINE Insertion Polymorphisms" Animals 12, no. 11: 1345. https://doi.org/10.3390/ani12111345
APA StyleWang, X., D’Alessandro, E., Chi, C., Moawad, A. S., Zong, W., Chen, C., & Song, C. (2022). Genetic Evaluation and Population Structure of Jiangsu Native Pigs in China Revealed by SINE Insertion Polymorphisms. Animals, 12(11), 1345. https://doi.org/10.3390/ani12111345








