Simple Summary
Salmonella enterica serovar Gallinarum (S. enterica ser. Gallinarum) is a host-specific agent of fowl typhoid (FT). This is one of the most important bacterial infections in the poultry industry in both developing and developed countries, including South Korea. The use of antimicrobial drugs is the first choice for disease control. Antimicrobials, such as β-lactams, aminoglycosides, and fluoroquinolones, are frequently used to treat FT. However, the continuous use of antimicrobial drugs has led to the emergence and persistence of antimicrobial-resistant S. enterica ser. Gallinarum. In this study, we analyzed the antimicrobial susceptibility and epidemiological relationship of thirty isolates of S. enterica ser. Gallinarum isolated from poultry farms with an FT outbreak from 2013 to 2018 in South Korea. All the isolates showed a multi-drug resistant (MDR) phenotype. This study confirmed horizontal transmission and cross-contamination between farms within the same integrated poultry company or between farms belonging to different companies. The characterization of these isolates would be helpful to develop prevention and control strategies for the MDR S. enterica ser. Gallinarum infection in South Korea.
Abstract
Antimicrobial resistance and pulsed-field gel electrophoresis (PFGE) genotypes of collected S. enterica ser. Gallinarum isolates were investigated to examine the epidemiological relationship between field outbreak isolates of S. enterica ser. Gallinarum. Thirty S. enterica ser. Gallinarum isolates collected from poultry farms with FT outbreaks from 2013 to 2018 in South Korea were analyzed. All isolates were resistant to at least 3 of the 18 antimicrobials tested and exhibited an MDR phenotype. All isolates showed resistance to streptomycin, sulfisoxazole, and colistin. One isolate was resistant to 9 antimicrobials. The antimicrobial resistance profile, streptomycin-sulfisoxazole-colistin-nalidixic acid-ciprofloxacin-gentamicin (18/30, 60.0%), was the most prevalent. PFGE types were classified into 10 groups with a 100% correlation cutoff in dendrograms for 30 field isolates. The dominant PFGE types were 1 (8/30, 26.7%), 4 (7/30, 23.3%), and 9 (5/30, 16.7%). Interestingly some isolates collected from the same and different companies had the same PFGE type. We reported a high MDR rate in S. enterica ser. Gallinarum isolates. The present study highlights the occurrence of horizontal spread and cyclic contamination of MDR S. enterica ser. Gallinarum within the same company. Furthermore, we showed cross-contamination between different companies. The characterization of these isolates would be helpful in the development of prevention and control strategies for MDR S. enterica ser. Gallinarum infection in South Korea.
1. Introduction
Salmonella enterica serovar Gallinarum (S. enterica ser. Gallinarum) is the causative agent of fowl typhoid (FT). This is a disease characterized by severe hepatomegaly and splenomegaly accompanied by a liver with bronzing aspects, anemia, and septicemia, with mortality rates of up to 80% in affected chickens of any age [1,2,3]. FT has been almost eradicated from commercial poultry in many developed countries, including North American, Canada, Australia, Japan, and most European countries. This was completed by isolating and removing infected flocks, as well as implementing biosecurity and hygiene measures. Nonetheless, FT remains a significant economic problem in many parts of the world, such as Africa, Asia, and Central and South America [4,5].
After the first occurrence of FT in the field in 1992, it spread quickly throughout South Korea because the majority of the population was comprised of brown layers, which are more vulnerable to FT [6]. Nevertheless, producers would not replace brown layers with white chickens due to the preferences of Korean customers. Since then, FT has become one of the most serious bacterial diseases in the poultry industry. After suffering substantial economic losses due to this disease, South Korea opted to implement a nationwide vaccination program with a live attenuated strain S. enterica ser. Gallinarum 9R (SG9R) for commercial layers in 2001, rather than an eradication policy, to reduce the economic impact of FT. Subsequently, the number of reported cases decreased significantly. However, the vaccine has many shortcomings [7,8,9]; therefore, the use of antimicrobial drugs remains the first choice for disease control.
Antimicrobials, such as β-lactams, aminoglycosides, and fluoroquinolones, are frequently used to treat systemic bacterial infections, including those caused by S. enterica ser. Gallinarum in commercial chicken farms in South Korea. Fluoroquinolones, in particular, are extensively utilized due to the advantages of oral administration and strong efficacy against a wide range of Gram-negative bacteria [10]. However, the continuous use of antimicrobial drugs has led to the emergence and persistence of antimicrobial-resistant S. enterica ser. Gallinarum [10,11,12]. In recent years, many multidrug-resistant (MDR) S. enterica ser. Gallinarum isolates have been reported [11,13,14].
During a disease outbreak, a detailed epidemiological investigation would directly influence measures that should be adopted for the application of a successful control plan. Pulsed-field gel electrophoresis (PFGE) is an efficient and reliable method to identify the sources of infection or reservoirs of epidemic outbreaks, to understand the clonality among isolates and to determine transmission routes [15]. PFGE works by digesting the whole DNA using restriction enzymes to yield strain-specific fragment patterns. This method is based on genomic differences between isolates. It is these differences accumulated by genetic variation that cause slight detectable differences between DNA fingerprint patterns [16].
Here, we studied the antimicrobial susceptibility of thirty isolates of S. enterica ser. Gallinarum isolated from poultry farms with FT outbreaks from 2013 to 2018 in South Korea. We analyzed the epidemiological relationship of these isolates to determine whether there was horizontal transmission and cross-contamination of MDR isolates between farms belonging to the same or different integrated poultry companies.
2. Materials and Methods
2.1. Bacterial Isolates
The thirty isolates of S. enterica ser. Gallinarum used in this study were provided by the Department of Veterinary Infectious Diseases and Avian Diseases, College of Veterinary Medicine and Center for Poultry Diseases Control, Jeonbuk National University. The thirty S. enterica ser. Gallinarum isolates chosen were suspected of having an epidemiological relationship. This center has long-term cooperative relationships with main poultry companies in South Korea for disease monitoring. It should be emphasized that these isolates were representative isolates selected from the samples of FT outbreaks in farms of poultry companies from 2013 to 2018. These isolates were isolated from farms belonging to eight different integrated poultry companies in South Korea (Table 1). Among these isolates, some were isolated from the same farm of the same company, some were isolated from different farms of the same company, and some were isolated from different farms from different companies. To isolate Salmonella enterica, aseptically separated liver samples were directly streaked onto MacConkey Agar (Difco Laboratories, Detroit, MI, USA), and the plate was incubated for 24 h at 37 °C. Presumptive Salmonella colonies were further tested using a Salmonella latex test kit (Thermo Fisher Scientific, Oxoid Ltd., Basingstoke, UK). All positive isolates were serotyped per the Kauffmann–White scheme using slide agglutination with O and H antigen-specific sera (BD Difco, Sparks, MD, USA and Denka Seiken Co., Ltd., Tokyo, Japan). S. enterica ser. Gallinarum isolates were confirmed by the agglutination test using Salmonella O antiserum group D1, the motility test and biochemical tests. The isolates were also identified using VDx® Fowl Typhoid PCR (MEDIAN Diagnostics, Chuncheon, Gangwon-do, Korea). The S. enterica ser. Gallinarum live vaccine Nobilis SG9R (Intervet International, Boxmeer, The Netherlands) was used as a reference strain. All isolates were stored in Luria-Bertani (LB) medium containing 20% glycerol at −70 °C.

Table 1.
Isolates of S. enterica ser. Gallinarum used in the present study.
2.2. Antimicrobial Susceptibility Test
The KRNV5F Sensititre panel (TREK Diagnostic Systems, Incheon, Korea) was used to determine minimum inhibitory concentrations (MICs) according to the manufacturer’s instructions [17]. The antimicrobials used for MIC were amoxicillin/clavulanic acid (AUG2, 2/1–32/16 μg/mL), ampicillin (AMP, 2–64 μg/mL), cefoxitin (FOX, 1–32 μg/mL), ceftazidime (TAZ, 1–16 μg/mL), ceftiofur (XNL, 0.5–8 μg/mL), cefepime (FEP, 0.25–16 μg/mL), chloramphenicol (CHL, 2–64 μg/ mL), ciprofloxacin (CIP, 0.12–16 μg/mL), colistin (COL, 2–16 μg/mL), florfenicol (FFC, 2–64 μg/mL), gentamicin (GEN, 1–64 μg/mL), Kanamycin (K, 4–256 μg/mL), meropenem (MERO, 0.25–4 μg/mL), nalidixic acid (NAL, 2–128 μg/mL), sulfisoxazole (FIS, 16–256 μg/mL), streptomycin (STR, 16–128 μg/mL), tetracycline (TET, 2–128 μg/mL), and trimethoprim/sulfamethoxazole (SXT, 0.12/2.38–4/76 μg/mL). The quality control strain that was used was Escherichia coli ATCC 25922. The interpretive categories—susceptible, intermediate, or resistant—were used according to the Clinical and Laboratory Standard Institute (CLSI) guidelines [18], except for colistin, where an MIC value ≥4 μg/mL (resistant) was used [19]. Salmonella isolates, resistant to three or more antimicrobial classes, were defined as multidrug resistant (MDR).
2.3. PFGE and BioNumerics Analysis
S. enterica ser. Gallinarum isolates (n = 31, 30 field isolates and vaccine strain SG9R) were analyzed using PFGE, with some modifications, according to the Centers for Disease Control and Prevention procedures, as previously described [20]. In brief, a single colony of each isolate was streaked onto a MacConkey agar plate and incubated at 37 °C overnight. The bacteria were then suspended in PBS at a concentration of 0.6–0.8 optical density (OD). Genomic DNA samples were digested with 50U of XbaI (Thermo Fisher Scientific, Inchon, Korea) at 37 °C for 3 h after extraction with 1% SDS and 1 mg/mL proteinase K (Biosesang, Seoul, Korea). Using a CHEF-DR@ electrophoresis equipment (Bio-Rad, Hercules, CA, USA), the digested DNA was separated by electrophoresis in 0.5× TBE buffer at 14 °C for 18 h at a gradient of 6 V/cm, with a pulse angle of 120°. The pulse time was gradually increased from 2.16 to 63.8 s. The fragment sizes were determined using fragments from the Salmonella enterica serovar Braenderup H9812 reference standard (ATCC® BAA-664™). After electrophoresis, gels were stained using ethidium bromide solution. Then, the band patterns were photographed on a UV transilluminator (Bio Doc-It Imaging System, Upland, CA, USA). The gel images were analyzed by BioNumerics (version 5.10 for Windows, Applied Maths, Sint-Martens-Latem, Belgium). The dendrogram tree was constructed using Unweighted Pair Group Method with Arithmetic Mean (UPGMA) with Dice coefficient at an optimization setting of 1% and a position tolerance setting of 1%. The band of 100% similarity was regarded as the same PFGE type.
3. Results
3.1. Antimicrobial Susceptibility Test
The antimicrobial susceptibility test results of S. enterica ser. Gallinarum isolates is shown in Table 2. All isolates showed resistance to STR, FIS, and COL. The resistance rates to NAL, CIP, and GEN were 96.7%, 90.0%, and 66.7%, respectively. A 3.3% (1/30) resistance rate was observed in CHL, AMP, TET, and FFC. No isolates were resistant to the eight antimicrobials, including AUG2, FEP, FOX, SXT, TAZ, XNL, MERO, and K. Five antimicrobial resistance profiles were observed among the isolates. The antimicrobial resistance profile, STR-FIS-COL-NAL-CIP-GEN (18/30, 60.0%), was the most prevalent, followed by STR-FIS-COL-NAL-CIP (8/30, 27.0%). In addition, one isolate had a nine antimicrobial resistance pattern of STR-FIS-COL-NAL-CIP-CHL-AMP-TET-FFC. The distribution of MDR by year is shown in Table 3. All isolates were classified as MDR because they were resistant to ≥3 classes of antimicrobials. One isolate, A16-OTH-010 (3.3%), was resistant to eight antimicrobial classes. There were 26 isolates (86.7%) resistant to five antimicrobial classes. Two isolates were resistant to four antimicrobial classes. One isolate, A16-MRA-002 (3.3%), was resistant to three antimicrobial classes.

Table 2.
Antimicrobial resistance patterns of S. enterica ser. Gallinarum isolates (n = 30).

Table 3.
Multi-drug resistance of S. enterica ser. Gallinarum isolates obtained from 2013 to 2018.
3.2. PFGE and Bionomics Analysis
Thirty-one isolates of S. enterica ser. Gallinarum, including thirty field isolates collected from 2013–2018 and a vaccine strain (SG9R), were analyzed by PFGE after DNA digestion with XbaI (Figure 1). Eleven different PFGE types (1 to 11), exhibiting 100% similarity, were produced by enzyme digestion. The vaccine strain (SG9R) notably showed a different type (type 11). The dominant PFGE types were type 1 (8/30, 26.7%), 4 (7/30, 23.3%), and 9 (5/30, 16.7%). Isolates of the same PFGE type were observed in different companies (type 1 in companies A, B, and C; type 3 in companies D and E; type 4 in companies A, B, and G; type 6 in companies A and F; type 9 in companies D, E, and G). Conversely, isolates of different PFGE types were observed in the same companies (types 1, 4, 5, 6, 7 and 8 in company A; types 1 and 4 in company B; types 1 and 2 in company C; types 3 and 9 in companies D and E; types 6 and 10 in company F; types 4 and 9 in company G). There were five PFGE types in broiler chickens, namely types 1, 2, 4, 5, and 7; type 1 was the dominant type. There were four types of PFGE in layers, 6, 8, 9, and 10. There were three types of PFGE in native chickens, 1, 3, and 9. There was only one isolate taken from 2013, which had the PFGE type 1. The PFGE types from isolates in 2016 included 4, 5, 7, 8, and 9. The PFGE types from isolates in 2017 included 1, 2, 3, 4, 6, and 9. The PFGE types of isolates from 2018 included 1, 6, 8, and 10.
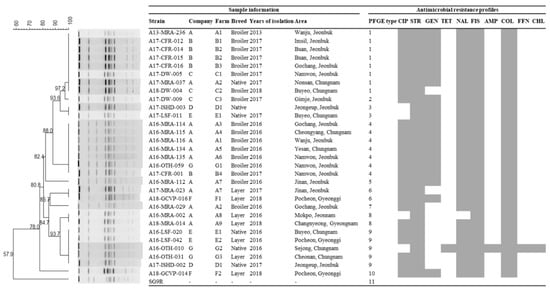
Figure 1.
A dendrogram based on the XbaI-pulsed field gel electrophoresis (PFGE) profiles of S. enterica ser. Gallinarum isolates from chicken. Corresponding antimicrobial susceptibility patterns to the 18 indicated antimicrobials. Gray squares represent resistance.
4. Discussion
In this study, thirty isolates from poultry farms with FT outbreaks from 2013 to 2018 were used. We used antimicrobial susceptibility tests to understand the drug resistance and PFGE to evaluate the genetic relationship of these isolates. We aimed to confirm whether there was horizontal transmission and cross-contamination between farms within the same or different companies.
A previous study has reported that the STR resistance rates of S. enterica ser. Gallinarum isolates from broilers and layers between 1999 and 2004 were 99.8% (55/56) and 81.8% (36/44), respectively [21]. In the present study, all isolates were STR resistant, which suggested that STR may be completely ineffective for the treatment of FT in poultry. Resistance to GEN, an aminoglycoside antibiotic such as STR, reached 64.5% in the present study. Compared with 56.6% in 2011, the GEN resistance rate has increased significantly [12]. Aminoglycosides, such as STR and GEN, are the most commonly used antimicrobials in poultry [10,22]. Therefore, this increased resistance may be related to their constant use. In addition, the use of antimicrobials as growth promoters remains common, which may have indirectly promoted the emergence of resistant strains of Salmonella.
It is noteworthy that the resistance to COL has also reached 100% in the present study. From 2014–2017, COL sales increased sharply, which suggests that the increased resistance of COL may be a consequence of its heavy use [11]. Infections caused by MDR gram-negative bacteria have increased dramatically, and polymyxins are often the only active antibiotics available [18]. In Europe, plasmid-mediated COL resistance in Enterobacteriaceae has spread widely in avian and pig farms. This has necessitated immediate international action to restrict or ban COL use in agriculture to prevent further resistance spread [23]. Mobilized colistin resistance (mcr) genes are plasmid-borne genes that confer resistance to COL. To date, mcr-1 to mcr-9 genes have been reported [24]. However, none of these genes were detected in our isolates (personal communication). Therefore, other resistance genes or mechanisms related to COL need to be studied. COL resistance can be transmitted to humans through the food chain, posing a threat to human health. Hence, it must be continuously monitored.
The resistance rate of S. enterica ser. Gallinarum isolates to CIP has increased from 0% in 1995 to 89.1% in 2002 [12]. The CIP resistance rate of our isolates reached 90%, which was consistent with the 2002 rate. Enrofloxacin (ENR) is metabolized to CIP; therefore, excessive use of ENR will correspondingly increase CIP resistance. The Food and Drug Administration (FDA) banned the use of ENR in the USA in 2005; however, it was not banned in chickens until May 2017 in South Korea [11,25]. Prior to this, ENR was continuously applied to poultry through drinking water, which may be the cause of increased resistance to CIP.
Between 1995 and 2001, 16.2% (22/136) of the isolates were MDR [12]. Of the isolates isolated between 2002 and 2007, 61.0% were MDR [10]. Of the isolates isolated in 2018, 60.7% (17/128) were MDR [11]. In this study, all isolates isolated between 2013 and 2018 were MDR, which suggested that the isolation of MDR isolate has become common and reflects the severity of the resistance of S. enterica ser. Gallinarum in Korea. This poses even greater hurdles for FT prevention and control in the poultry industry. The emergence of MDR isolates is usually caused by excessive and unreasonable simultaneous use of multiple antimicrobials. Here, it is important to note that there was one isolate that was resistant to nine different antimicrobials, which brings greater challenges to the use of antimicrobial treatment. Therefore, the monitoring of MDR isolates must be strengthened. Otherwise, more resistant super bacteria may appear. The universality and severity of MDR isolates once again emphasize the importance of the rational use of antimicrobials.
Many bacteria, including various Salmonella serotypes, have been successfully tested using PFGE. This genetic analysis can assess the genetic diversity of the entire genome. It is the gold standard method due to its accuracy and reproducibility [26,27,28,29]. First, isolates collected from different farms belonging to the same integrated company had the same PFGE type (for example, type 1 in B1, B2, and B3 farms belonging to company B; type 4 in A1, A3, A4, A5 and A6 farms belonging to company A). This indicated that the horizontal spread of MDR isolates existed between farms within the same company. This could be because the farms that belong to the same company share resources, including breeders, trucks for transport, veterinarians, chicks, and feed. Second, isolates collected from different farms belonging to different integrated companies had the same PFGE type (for example, type 1 in companies A, B, and C; type 4 in companies A, B, and G; type 9 in companies D, E, and G). This indicated that cross-contamination exists between different companies. Migratory birds may play a role in the dispersal of pathogenic and antimicrobial-resistant Salmonella in poultry [30]. Therefore, we hypothesize that migratory birds may be one of the causes of cross-contamination between companies. Third, some isolates had the same PFGE type and antimicrobial resistance profile (for example, type 1 isolates (A13-MRA-236, A17-CFR-012, A17-CFR-014, A17-CFR-015, A17-CFR-016, A17-DW-005, A17-MRA-037) with CIP-STR-GEN-NAL-FIS-COL; type 4 isolates (A16-MRA-114, A16-MRA-115, A16-MRA-116, A16-MRA-134, A16-MRA-135, A16-OTH-059, and A17-CFR-001 with CIP-STR-GEN-NAL-FIS-COL)). This further indicated that some isolates might come from the same clone.
Interestingly, the PFGE types (1, 2, 4, 5, and 7) of the isolates collected from broilers were different from the PFGE types (6, 8, 9, and 10) of the isolates collected from the layers. This indicated that there were different isolate genotypes infecting broilers and layers. One explanation might be that different breeds of chickens have different susceptibilities to S. enterica ser. Gallinarum [31]. Additionally, layers are older and have a longer exposure period than broilers. Their treatment regimens often differ as well. Further research and analysis are needed to explain this result.
Whole-genome sequencing (WGS) is an important tool for the analysis of resistant bacteria. WGS technology can be used to analyze drug resistance genes and their mutations. In addition to known drug resistance mechanisms, it can also be used to predict unknown potential drug resistance mechanisms [32,33]. To deeply study the resistance genes and resistance mechanisms of these currently isolated S. enterica ser. Gallinarum isolates, WGS has been listed as a further plan.
5. Conclusions
We reported a high MDR rate in S. enterica ser. Gallinarum isolates from farms of poultry companies from 2013 to 2018 in South Korea. The present study highlights the horizontal spread of MDR S. enterica ser. Gallinarum within and between the same and different companies. We believe the characterization of these isolates will be helpful in the development of prevention and control strategies.
Author Contributions
M.K. and S.-Y.C. contributed to the conception and design of experiments. Y.-J.L., J.-Y.P., Y.-R.C. and S.-W.K. contributed to the acquisition, analysis, and interpretation of data. K.S., J.-F.Z., M.K., B.W. and H.-K.J. drafted and/or revised the article. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Korea Institute of Planning and Evaluation for Technology in Food, Agriculture and Forestry (IPET) through Agriculture, Food and Rural Affairs Convergence Technologies Program for Educating Creative Global Leaders (716002-7, 320005-4) funded by Ministry of Agriculture, Food and Rural Affairs (MAFRA). And this work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korean government (MSIT) (No. 2020R1F1A1065136). And this paper was supported by the selection of a research-oriented professor of Jeonbuk National University in 2020.
Institutional Review Board Statement
The study was conducted according to the guidelines of the Animal Care and Ethics Committees of Jeonbuk National University and were approved by the National Association of Laboratory Animal Care (JBNU 2020-0162).
Data Availability Statement
The data presented in this study are available from the corresponding author on reasonable request.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Shehata, A.A.; Sultan, H.; Hafez, H.M.; Kruger, M. Safety and efficacy of a metabolic drift live attenuated Salmonella Gallinarum vaccine against fowl typhoid. Avian Dis. 2013, 57, 29–35. [Google Scholar] [CrossRef] [PubMed]
- Berhanu, G.; Fulasa, A. Pullorum Disease and Fowl Typhoid in Poultry: A Review. Br. J. Poult. Sci. 2020, 9, 48–56. [Google Scholar]
- Shivaprasad, H.L. Fowl typhoid and pullorum disease. Rev. Sci. Tech. 2000, 19, 405–424. [Google Scholar] [CrossRef] [PubMed]
- Jones, M.A.; Wigley, P.; Page, K.L.; Hulme, S.D.; Barrow, P.A. Salmonella enterica serovar Gallinarum requires the Salmonella pathogenicity island 2 type III secretion system but not the Salmonella pathogenicity island 1 type III secretion system for virulence in chickens. Infect. Immun. 2001, 69, 5471–5476. [Google Scholar] [CrossRef]
- Barrow, P.A.; Freitas Neto, O.C. Pullorum disease and fowl typhoid--new thoughts on old diseases: A review. Avian Pathol. 2011, 40, 1–13. [Google Scholar] [CrossRef]
- Kim, K.; Lee, H.; Mo, I.; Kim, S. Outbreak of fowl typhoid from chickens in Korea. RDA J. Agric. Sci. 1995, 37, 544–549. [Google Scholar]
- Bouzoubaa, K.; Nagaraja, K.V.; Kabbaj, F.Z.; Newman, J.A.; Pomeroy, B.S. Feasibility of Using Proteins from Salmonella Gallinarum Vs 9R Live Vaccine for the Prevention of Fowl Typhoid in Chickens. Avian Dis. 1989, 33, 385–391. [Google Scholar] [CrossRef]
- Silva, E.N.; Snoeyenbos, G.H.; Weinack, O.M.; Smyser, C.F. Studies on the Use of 9R Strain of Salmonella-Gallinarum as a Vaccine in Chickens. Avian Dis. 1981, 25, 38–52. [Google Scholar] [CrossRef]
- Van Immerseel, F.; Studholme, D.J.; Eeckhaut, V.; Heyndrickx, M.; Dewulf, J.; Dewaele, I.; Van Hoorebeke, S.; Haesebrouck, F.; Van Meirhaeghe, H.; Ducatelle, R.; et al. Salmonella Gallinarum field isolates from laying hens are related to the vaccine strain SG9R. Vaccine 2013, 31, 4940–4945. [Google Scholar] [CrossRef]
- Kang, M.S.; Kim, A.; Jung, B.Y.; Her, M.; Jeong, W.; Cho, Y.M.; Oh, J.Y.; Lee, Y.J.; Kwon, J.H.; Kwon, Y.K. Characterization of antimicrobial resistance of recent Salmonella enterica serovar Gallinarum isolates from chickens in South Korea. Avian Pathol. 2010, 39, 201–205. [Google Scholar] [CrossRef]
- Seo, K.W.; Kim, J.J.; Mo, I.P.; Lee, Y.J. Molecular characteristic of antimicrobial resistance of Salmonella Gallinarum isolates from chickens in Korea, 2014 to 2018. Poult. Sci. 2019, 98, 5416–5423. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.J.; Kim, K.S.; Kwon, Y.K.; Tak, R.B. Biochemical characteristics and antimicrobials susceptibility of Salmonella gallinarum isolated in Korea. J. Vet. Sci. 2003, 4, 161–166. [Google Scholar] [CrossRef] [PubMed]
- Parvej, M.S.; Nazir, K.H.; Rahman, M.B.; Jahan, M.; Khan, M.F.; Rahman, M. Prevalence and characterization of multi-drug resistant Salmonella enterica serovar Gallinarum biovar Pullorum and Gallinarum from chicken. Vet. World 2016, 9, 65–70. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Haque, A.; Akter, M.R.; Islam, S.K.S.; Alam, J.; Neogi, S.B.; Yamasaki, S.; Kabir, S.M.L. Salmonella Gallinarum in Small-Scale Commercial Layer Flocks: Occurrence, Molecular Diversity and Antibiogram. Vet. Sci. 2021, 8, 71. [Google Scholar] [CrossRef]
- Diaz-Torres, O.; Lugo-Melchor, O.Y.; de Anda, J.; Gradilla-Hernandez, M.S.; Amezquita-Lopez, B.A.; Meza-Rodriguez, D. Prevalence, Distribution, and Diversity of Salmonella Strains Isolated from a Subtropical Lake. Front. Microbiol. 2020, 11, 521146. [Google Scholar] [CrossRef] [PubMed]
- Zakaria, Z.; Hassan, L.; Sharif, Z.; Ahmad, N.; Ali, R.M.; Husin, S.A.; Hazis, N.; Sohaimi, N.F.M.; Bakar, S.A.; Garba, B. Analysis of Salmonella enterica serovar Enteritidis isolates from chickens and chicken meat products in Malaysia using PFGE, and MLST. BMC Vet. Res. 2020, 16, 393. [Google Scholar] [CrossRef]
- Wei, B.; Shang, K.; Cha, S.Y.; Zhang, J.F.; Jang, H.K.; Kang, M. Clonal dissemination of Salmonella enterica serovar albany with concurrent resistance to ampicillin, chloramphenicol, streptomycin, sulfisoxazole, tetracycline, and nalidixic acid in broiler chicken in Korea. Poult. Sci. 2021, 100, 101141. [Google Scholar] [CrossRef] [PubMed]
- Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing-26th Edition. CLSI Document M100-S26; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2016. [Google Scholar]
- Biswas, S.; Brunel, J.M.; Dubus, J.C.; Reynaud-Gaubert, M.; Rolain, J.M. Colistin: An update on the antibiotic of the 21st century. Expert Rev. Anti. Infect. Ther. 2012, 10, 917–934. [Google Scholar] [CrossRef]
- Shang, K.; Wei, B.; Kang, M. Distribution and dissemination of antimicrobial-resistant Salmonella in broiler farms with or without enrofloxacin use. BMC Vet. Res. 2018, 14, 257. [Google Scholar] [CrossRef] [PubMed]
- Bae, J.-C.; Kim, S.-G.; Kim, Y.-H.; Jo, M.-H.; Lee, Y.-J.; Park, C.-K. Antimicrobial resistance and pulsed-field gel electrophoresis (PFGE) patterns of Salmonella Gallinarum isolated from chicken. Korean J. Vet. Serv. 2009, 32, 155–163. [Google Scholar]
- Lee, S.K.; Chon, J.W.; Song, K.Y.; Hyeon, J.Y.; Moon, J.S.; Seo, K.H. Prevalence, characterization, and antimicrobial susceptibility of Salmonella Gallinarum isolated from eggs produced in conventional or organic farms in South Korea. Poult. Sci. 2013, 92, 2789–2797. [Google Scholar] [CrossRef] [PubMed]
- Kempf, I.; Fleury, M.A.; Drider, D.; Bruneau, M.; Sanders, P.; Chauvin, C.; Madec, J.Y.; Jouy, E. What do we know about resistance to colistin in Enterobacteriaceae in avian and pig production in Europe? Int. J. Antimicrob. Agents 2013, 42, 379–383. [Google Scholar] [CrossRef] [PubMed]
- Borowiak, M.; Baumann, B.; Fischer, J.; Thomas, K.; Deneke, C.; Hammerl, J.A.; Szabo, I.; Malorny, B. Development of a Novel mcr-6 to mcr-9 Multiplex PCR and Assessment of mcr-1 to mcr-9 Occurrence in Colistin-Resistant Salmonella enterica Isolates from Environment, Feed, Animals and Food (2011–2018) in Germany. Front. Microbiol. 2020, 11, 80. [Google Scholar] [CrossRef] [PubMed]
- Han, F.; Lestari, S.I.; Pu, S.; Ge, B. Prevalence and antimicrobial resistance among Campylobacter spp. in Louisiana retail chickens after the enrofloxacin ban. Foodborne Pathog. Dis. 2009, 6, 163–171. [Google Scholar] [CrossRef] [PubMed]
- Tenover, F.C.; Arbeit, R.D.; Goering, R.V.; Mickelsen, P.A.; Murray, B.E.; Persing, D.H.; Swaminathan, B. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: Criteria for bacterial strain typing. J. Clin. Microbiol. 1995, 33, 2233–2239. [Google Scholar] [CrossRef] [PubMed]
- Dominguez, J.E.; Vinas, M.R.; Herrera, M.; Moroni, M.; Gutkind, G.O.; Mercado, E.C.; Di Conza, J.A.; Chacana, P.A. Molecular characterization and antimicrobial resistance profiles of Salmonella Heidelberg isolates from poultry. Zoonoses Public Health 2021, 68, 309–315. [Google Scholar] [CrossRef] [PubMed]
- Oh, J.Y.; Kwon, Y.K.; Wei, B.; Jang, H.K.; Lim, S.K.; Kim, C.H.; Jung, S.C.; Kang, M.S. Epidemiological relationships of Campylobacter jejuni strains isolated from humans and chickens in South Korea. J. Microbiol. 2017, 55, 13–20. [Google Scholar] [CrossRef]
- Seo, K.W.; Lee, Y.J. Molecular characterization of fluoroquinolone-resistant Escherichia coli from broiler breeder farms. Poult. Sci. 2021, 100, 101250. [Google Scholar] [CrossRef] [PubMed]
- Wei, B.; Shang, K.; Cha, S.Y.; Zhang, J.F.; Kang, M.; Jang, H.K. Prevalence and potential risk of Salmonella enterica in migratory birds from South Korea. Vet. Microbiol. 2020, 249, 108829. [Google Scholar] [CrossRef] [PubMed]
- Smith, H.W. The Susceptibility of Different Breeds of Chickens to Experimental Salmonella-Gallinarum Infection. Poult. Sci. 1956, 35, 701–705. [Google Scholar] [CrossRef]
- Koser, C.U.; Ellington, M.J.; Peacock, S.J. Whole-genome sequencing to control antimicrobial resistance. Trends Genet. 2014, 30, 401–407. [Google Scholar] [CrossRef] [PubMed]
- Tyson, G.H.; McDermott, P.F.; Li, C.; Chen, Y.; Tadesse, D.A.; Mukherjee, S.; Bodeis-Jones, S.; Kabera, C.; Gaines, S.A.; Loneragan, G.H.; et al. WGS accurately predicts antimicrobial resistance in Escherichia coli. J. Antimicrob. Chemother. 2015, 70, 2763–2769. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).