Early Life Fecal Microbiota Transplantation in Neonatal Dairy Calves Promotes Growth Performance and Alleviates Inflammation and Oxidative Stress during Weaning
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Design
2.2. Donor Selection and Fecal Preparation
2.3. Pre-Trial Phase
2.4. Trial Phase: Study Design and Fecal Microbiota Inoculations
2.5. Animal Measurements and Sample Collection
2.6. Polymorphonuclear Leukocytes (PMNL) Isolation
2.7. Fecal DNA Isolation and Bioinformatic Analysis for 16S rRNA Gene-Based Composition Analysis
2.8. RNA Isolation, cDNA Synthesis, and Quantitative PCR (qPCR)
2.9. Statistical Analysis
3. Results
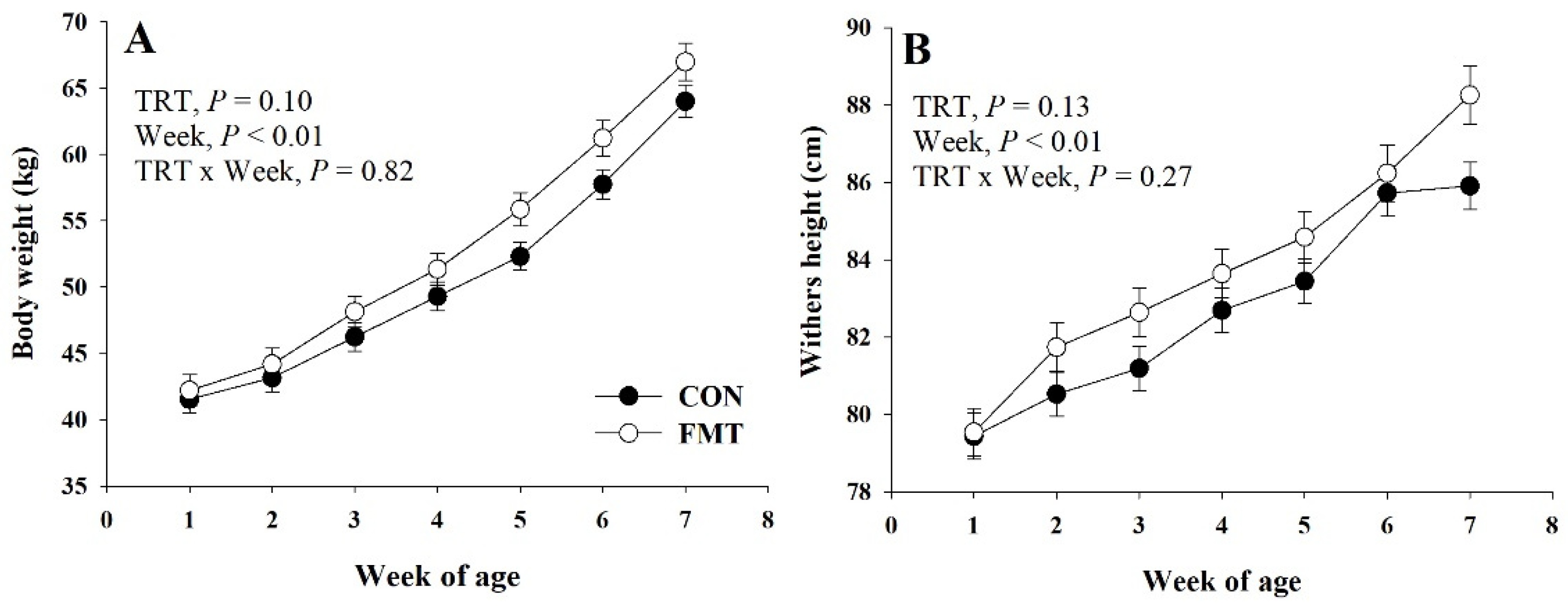
3.1. Growth Performance and Health
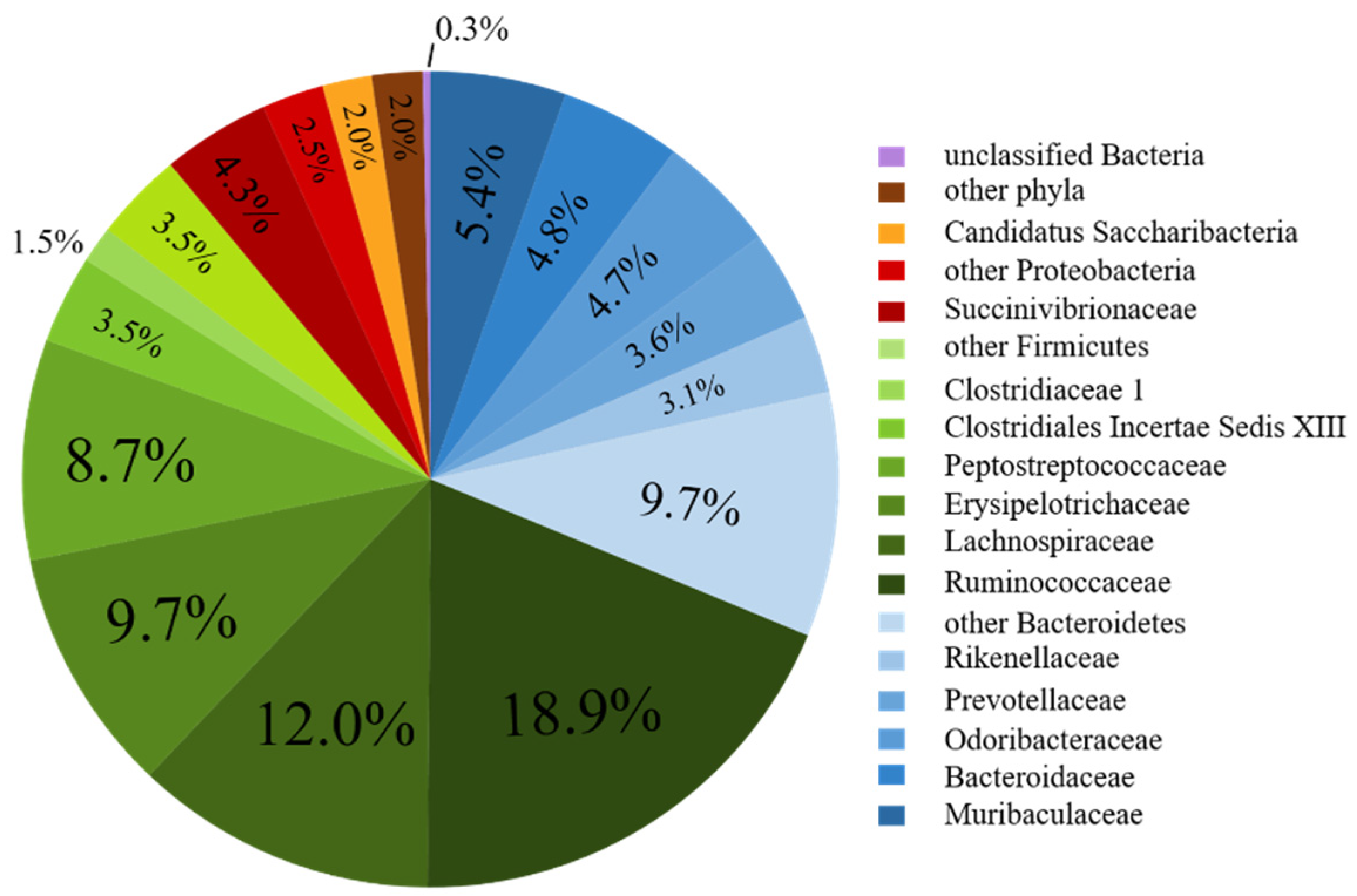
3.2. Bacterial Community Composition of the Fecal Donor
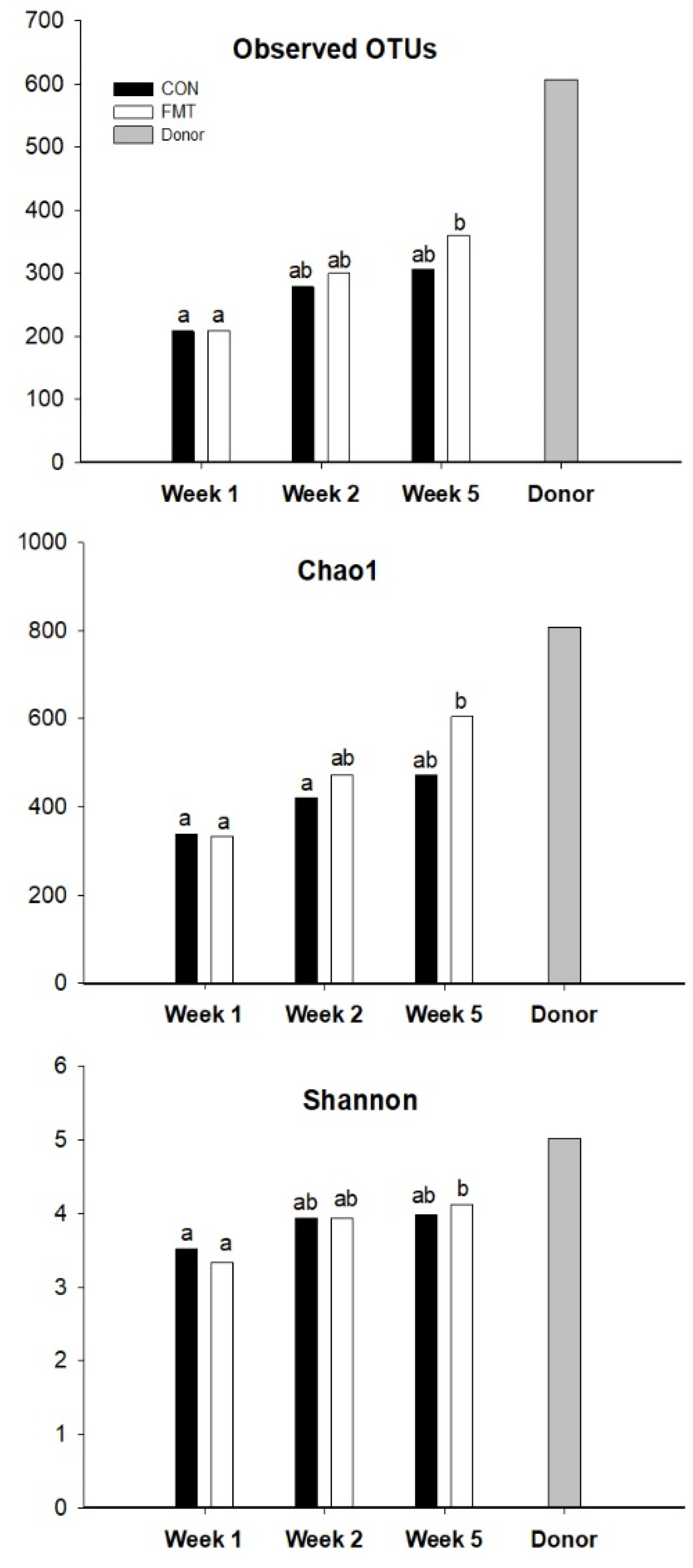
3.3. Fecal Microbiota Composition of the FMT Recipients
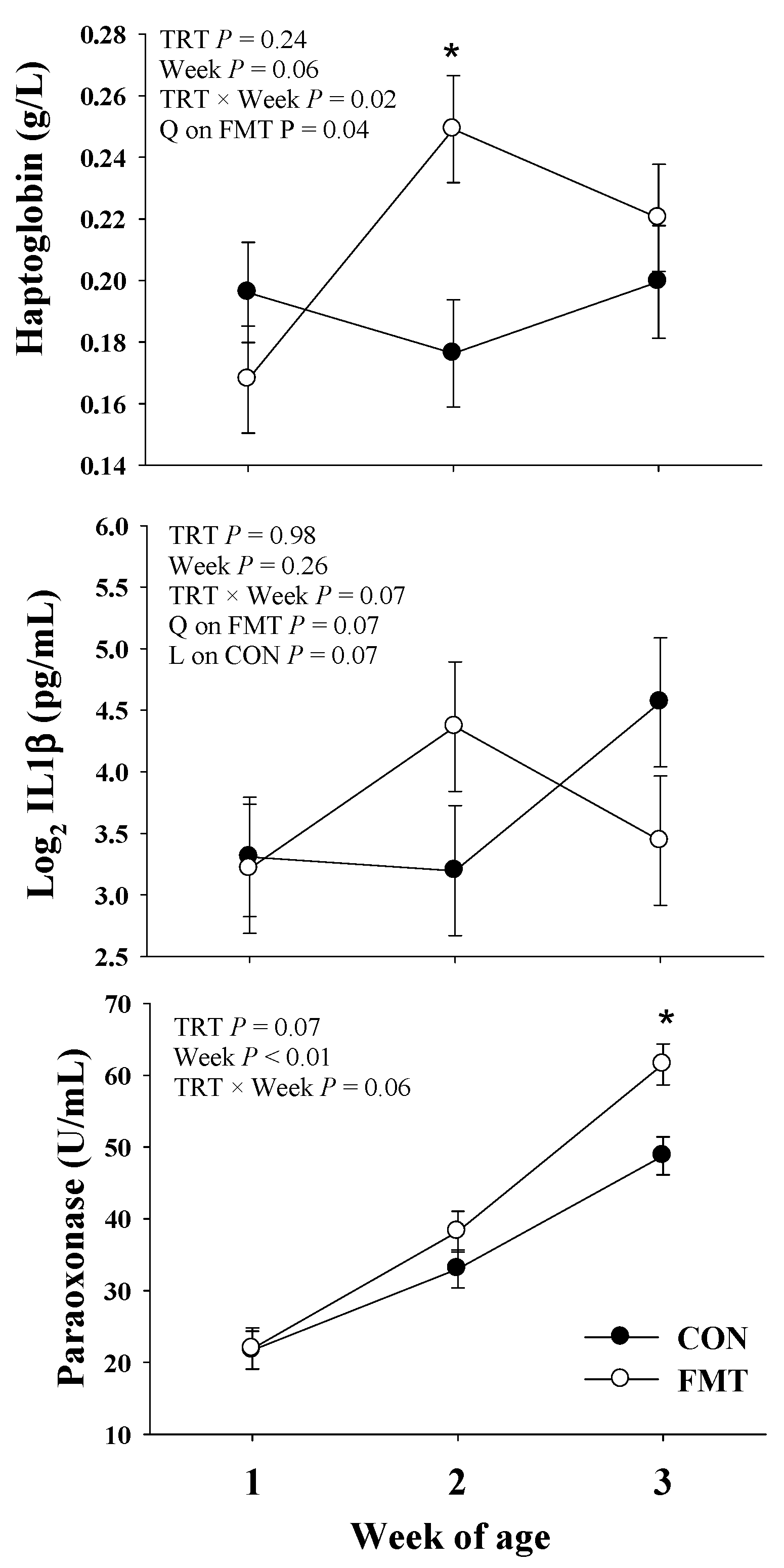
3.4. Blood Biomarkers
3.4.1. Energy Metabolism
3.4.2. Inflammation
3.4.3. Liver Function
3.4.4. Oxidative Stress
3.5. PMNL Gene Expression
4. Discussion
4.1. Gut Microbial Maturation Impacted by FMT Early in Life
4.2. Neonatal Liver Function and Inflammation
4.3. FMT Effects on Inflammation and Oxidative Stress during Weaning
4.4. PMNL Gene Expression
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Elgin, T.G.; Kern, S.L.; McElroy, S.J. Development of the Neonatal Intestinal Microbiome and Its Association with Necrotizing Enterocolitis. Clin. Ther. 2016. [Google Scholar] [CrossRef] [Green Version]
- Jiménez, E.; Marín, M.L.; Martín, R.; Odriozola, J.M.; Olivares, M.; Xaus, J.; Fernández, L.; Rodríguez, J.M. Is meconium from healthy newborns actually sterile? Res. Microbiol. 2008, 159, 187–193. [Google Scholar] [CrossRef]
- Jimenez, E.; Fernandez, L.; Marin, M.L.; Martin, R.; Odriozola, J.M.; Nueno-Palop, C.; Narbad, A.; Olivares, M.; Xaus, J.; Rodriguez, J.M. Isolation of commensal bacteria from umbilical cord blood of healthy neonates born by cesarean section. Curr. Microbiol. 2005, 51, 270–274. [Google Scholar] [CrossRef] [PubMed]
- Aagaard, K.; Ma, J.; Antony, K.M.; Ganu, R.; Petrosino, J.; Versalovic, J. The Placenta Harbors a Unique Microbiome. Sci. Transl. Med. 2014, 6, 237ra265. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Collado, M.C.; Rautava, S.; Aakko, J.; Isolauri, E.; Salminen, S. Human gut colonisation may be initiated in utero by distinct microbial communities in the placenta and amniotic fluid. Sci. Rep. 2016, 6, 23129. [Google Scholar] [CrossRef] [Green Version]
- Mayer, M.; Abenthum, A.; Matthes, J.M.; Kleeberger, D.; Ege, M.J.; Hölzel, C.; Bauer, J.; Schwaiger, K. Development and genetic influence of the rectal bacterial flora of newborn calves. Vet. Microbiol. 2012, 161, 179–185. [Google Scholar] [CrossRef]
- Karstrup, C.C.; Klitgaard, K.; Jensen, T.K.; Agerholm, J.S.; Pedersen, H.G. Presence of bacteria in the endometrium and placentomes of pregnant cows. Theriogenology 2017, 99, 41–47. [Google Scholar] [CrossRef]
- Malmuthuge, N.; Guan, L.L. Understanding the gut microbiome of dairy calves: Opportunities to improve early-life gut health1. J. Dairy Sci. 2017, 100, 5996–6005. [Google Scholar] [CrossRef]
- Dominguez-Bello, M.G.; De Jesus-Laboy, K.M.; Shen, N.; Cox, L.M.; Amir, A.; Gonzalez, A.; Bokulich, N.A.; Song, S.J.; Hoashi, M.; Rivera-Vinas, J.I.; et al. Partial restoration of the microbiota of cesarean-born infants via vaginal microbial transfer. Nat. Med. 2016. [Google Scholar] [CrossRef] [PubMed]
- Pyo, J.; Hare, K.; Pletts, S.; Inabu, Y.; Haines, D.; Sugino, T.; Guan, L.L.; Steele, M. Feeding colostrum or a 1:1 colostrum:milk mixture for 3 days postnatal increases small intestinal development and minimally influences plasma glucagon-like peptide-2 and serum insulin-like growth factor-1 concentrations in Holstein bull calves. J. Dairy Sci. 2020, 103, 4236–4251. [Google Scholar] [CrossRef]
- Le Doare, K.; Holder, B.; Bassett, A.; Pannaraj, P.S. Mother’s Milk: A Purposeful Contribution to the Development of the Infant Microbiota and Immunity. Front. Immunol. 2018, 9. [Google Scholar] [CrossRef] [Green Version]
- Francino, M.P. Antibiotics and the Human Gut Microbiome: Dysbioses and Accumulation of Resistances. Front. Microbiol. 2015, 6, 1543. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thames, C.H.; Pruden, A.; James, R.E.; Ray, P.P.; Knowlton, K.F. Excretion of antibiotic resistance genes by dairy calves fed milk replacers with varying doses of antibiotics. Front. Microbiol. 2012, 3, 139. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hansen, C.H.; Nielsen, D.S.; Kverka, M.; Zakostelska, Z.; Klimesova, K.; Hudcovic, T.; Tlaskalova-Hogenova, H.; Hansen, A.K. Patterns of early gut colonization shape future immune responses of the host. PLoS ONE 2012, 7, e34043. [Google Scholar] [CrossRef] [Green Version]
- Herfel, T.M.; Jacobi, S.K.; Lin, X.; Fellner, V.; Walker, D.C.; Jouni, Z.E.; Odle, J. Polydextrose enrichment of infant formula demonstrates prebiotic characteristics by altering intestinal microbiota, organic acid concentrations, and cytokine expression in suckling piglets. J. Nutr. 2011, 141, 2139–2145. [Google Scholar] [CrossRef] [Green Version]
- Vieira, A.T.; Teixeira, M.M.; Martins, F.S. The role of probiotics and prebiotics in inducing gut immunity. Front. Immunol. 2013, 4, 445. [Google Scholar] [CrossRef] [PubMed]
- de Groot, P.F.; Frissen, M.N.; de Clercq, N.C.; Nieuwdorp, M. Fecal microbiota transplantation in metabolic syndrome: History, present and future. Gut Microbes 2017, 8, 253–267. [Google Scholar] [CrossRef]
- Ponte, A.; Pinho, R.; Mota, M.; Silva, J.; Vieira, N.; Oliveira, R.; Pinto-Pais, T.; Fernandes, C.; Ribeiro, I.; Rodrigues, J.; et al. Initial experience with fecal microbiota transplantation in Clostridium difficile infection - transplant protocol and preliminary results. Rev. Esp. Enferm. Dig. 2015, 107, 402–407. [Google Scholar] [CrossRef] [Green Version]
- Suskind, D.L.; Brittnacher, M.J.; Wahbeh, G.; Shaffer, M.L.; Hayden, H.S.; Qin, X.; Singh, N.; Damman, C.J.; Hager, K.R.; Nielson, H.; et al. Fecal Microbial Transplant Effect on Clinical Outcomes and Fecal Microbiome in Active Crohn’s Disease. Inflamm. Bowel Dis. 2015, 21, 556–563. [Google Scholar] [CrossRef] [Green Version]
- Kelly, C.R.; Ihunnah, C.; Fischer, M.; Khoruts, A.; Surawicz, C.; Afzali, A.; Aroniadis, O.; Barto, A.; Borody, T.; Giovanelli, A.; et al. Fecal microbiota transplant for treatment of Clostridium difficile infection in immunocompromised patients. Am. J. Gastroenterol. 2014, 109, 1065–1071. [Google Scholar] [CrossRef] [Green Version]
- Fareed, S.; Sarode, N.; Stewart, F.J.; Malik, A.; Laghaie, E.; Khizer, S.; Yan, F.; Pratte, Z.; Lewis, J.; Immergluck, L.C. Applying fecal microbiota transplantation (FMT) to treat recurrent Clostridium difficile infections (rCDI) in children. PeerJ 2018, 6, e4663. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Li, X.; Shang, Q.; Gao, Z.; Hao, F.; Guo, H.; Guo, C. Fecal microbiota transplantation (FMT) could reverse the severity of experimental necrotizing enterocolitis (NEC) via oxidative stress modulation. Free Radic. Biol. Med. 2017, 108, 32–43. [Google Scholar] [CrossRef]
- Khanna, S.; Vazquez-Baeza, Y.; Gonzalez, A.; Weiss, S.; Schmidt, B.; Muniz-Pedrogo, D.A.; Rainey, J.F., 3rd; Kammer, P.; Nelson, H.; Sadowsky, M.; et al. Changes in microbial ecology after fecal microbiota transplantation for recurrent C. difficile infection affected by underlying inflammatory bowel disease. Microbiome 2017, 5, 55. [Google Scholar] [CrossRef]
- Vaughn, B.P.; Vatanen, T.; Allegretti, J.R.; Bai, A.; Xavier, R.J.; Korzenik, J.; Gevers, D.; Ting, A.; Robson, S.C.; Moss, A.C. Increased Intestinal Microbial Diversity Following Fecal Microbiota Transplant for Active Crohn’s Disease. Inflamm. Bowel Dis. 2016, 22, 2182–2190. [Google Scholar] [CrossRef] [Green Version]
- Hu, J.; Chen, L.; Tang, Y.; Xie, C.; Xu, B.; Shi, M.; Zheng, W.; Zhou, S.; Wang, X.; Liu, L.; et al. Standardized Preparation for Fecal Microbiota Transplantation in Pigs. Front. Microbiol. 2018, 9, 1328. [Google Scholar] [CrossRef] [PubMed]
- McCormack, U.M.; Curiao, T.; Wilkinson, T.; Metzler-Zebeli, B.U.; Reyer, H.; Ryan, T.; Calderon-Diaz, J.A.; Crispie, F.; Cotter, P.D.; Creevey, C.J.; et al. Fecal Microbiota Transplantation in Gestating Sows and Neonatal Offspring Alters Lifetime Intestinal Microbiota and Growth in Offspring. mSystems 2018, 3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, W.; Chen, D.; Yu, B.; He, J.; Huang, Z.; Zheng, P.; Mao, X.; Luo, Y.; Luo, J.; Wang, Q.; et al. Capsulized faecal microbiota transplantation ameliorates post-weaning diarrhoea by modulating the gut microbiota in piglets. Vet. Res. 2020, 51, 55. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Canibe, N.; O’Dea, M.; Abraham, S. Potential relevance of pig gut content transplantation for production and research. J. Anim. Sci. Biotechnol. 2019, 10, 55. [Google Scholar] [CrossRef] [PubMed]
- Kamada, N.; Seo, S.U.; Chen, G.Y.; Nunez, G. Role of the gut microbiota in immunity and inflammatory disease. Nat. Rev. Immunol. 2013, 13, 321–335. [Google Scholar] [CrossRef]
- Wang, J.W.; Kuo, C.H.; Kuo, F.C.; Wang, Y.K.; Hsu, W.H.; Yu, F.J.; Hu, H.M.; Hsu, P.I.; Wang, J.Y.; Wu, D.C. Fecal microbiota transplantation: Review and update. J. Formos. Med. Assoc. 2019, 118 (Suppl. 1), S23–S31. [Google Scholar] [CrossRef]
- Osorio, J.S.; Wallace, R.L.; Tomlinson, D.J.; Earleywine, T.J.; Socha, M.T.; Drackley, J.K. Effects of source of trace minerals and plane of nutrition on growth and health of transported neonatal dairy calves. J. Dairy Sci. 2012, 95, 5831–5844. [Google Scholar] [CrossRef] [PubMed]
- Walpole, S.C.; Prieto-Merino, D.; Edwards, P.; Cleland, J.; Stevens, G.; Roberts, I. The weight of nations: An estimation of adult human biomass. BMC Public Health 2012, 12, 439. [Google Scholar] [CrossRef] [Green Version]
- Trevisi, E.; Amadori, M.; Cogrossi, S.; Razzuoli, E.; Bertoni, G. Metabolic stress and inflammatory response in high-yielding, periparturient dairy cows. Res. Vet. Sci. 2012, 93, 695–704. [Google Scholar] [CrossRef] [PubMed]
- Jacometo, C.B.; Zhou, Z.; Luchini, D.; Trevisi, E.; Correa, M.N.; Loor, J.J. Maternal rumen-protected methionine supplementation and its effect on blood and liver biomarkers of energy metabolism, inflammation, and oxidative stress in neonatal Holstein calves. J. Dairy Sci. 2016, 99, 6753–6763. [Google Scholar] [CrossRef]
- Batistel, F.; Osorio, J.S.; Ferrari, A.; Trevisi, E.; Socha, M.T.; Loor, J.J. Immunometabolic Status during the Peripartum Period Is Enhanced with Supplemental Zn, Mn, and Cu from Amino Acid Complexes and Co from Co Glucoheptonate. PLoS ONE 2016, 11, e0155804. [Google Scholar] [CrossRef]
- Rosa, F.; Osorio, J.S. Short communication: Comparative gene expression analysis on the enrichment of polymorphonuclear leukocytes and gastrointestinal epithelial cells in fecal RNA from nondiarrheic neonatal dairy calves. J. Dairy Sci. 2019, 102, 7464–7468. [Google Scholar] [CrossRef] [PubMed]
- Opdahl, L.J.; Gonda, M.G.; St-Pierre, B. Identification of Uncultured Bacterial Species from Firmicutes, Bacteroidetes and CANDIDATUS Saccharibacteria as Candidate Cellulose Utilizers from the Rumen of Beef Cows. Microorganisms 2018, 6, 17. [Google Scholar] [CrossRef] [Green Version]
- Poudel, P.; Levesque, C.L.; Samuel, R.; St-Pierre, B. Dietary inclusion of Peptiva, a peptide-based feed additive, can accelerate the maturation of the fecal bacterial microbiome in weaned pigs. BMC Vet. Res. 2020, 16, 60. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef] [Green Version]
- Johnson, J.S.; Spakowicz, D.J.; Hong, B.Y.; Petersen, L.M.; Demkowicz, P.; Chen, L.; Leopold, S.R.; Hanson, B.M.; Agresta, H.O.; Gerstein, M.; et al. Evaluation of 16S rRNA gene sequencing for species and strain-level microbiome analysis. Nat. Commun. 2019, 10, 5029. [Google Scholar] [CrossRef] [Green Version]
- Kim, M.; Morrison, M.; Yu, Z. Evaluation of different partial 16S rRNA gene sequence regions for phylogenetic analysis of microbiomes. J. Microbiol. Methods 2011, 84, 81–87. [Google Scholar] [CrossRef]
- Edgar, R.C.; Haas, B.J.; Clemente, J.C.; Quince, C.; Knight, R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 2011, 27, 2194–2200. [Google Scholar] [CrossRef] [Green Version]
- Altschul, S.F.; Madden, T.L.; Schaffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [Green Version]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef] [Green Version]
- Parte, A.C. LPSN--list of prokaryotic names with standing in nomenclature. Nucleic Acids Res. 2014, 42, D613–D616. [Google Scholar] [CrossRef] [Green Version]
- Bionaz, M.; Loor, J.J. Identification of reference genes for quantitative real-time PCR in the bovine mammary gland during the lactation cycle. Physiol. Genom. 2007, 29, 312–319. [Google Scholar] [CrossRef] [PubMed]
- Moyes, K.M.; Drackley, J.K.; Morin, D.E.; Loor, J.J. Greater expression of TLR2, TLR4, and IL6 due to negative energy balance is associated with lower expression of HLA-DRA and HLA-A in bovine blood neutrophils after intramammary mastitis challenge with Streptococcus uberis. Funct. Integr. Genom. 2010, 10, 53–61. [Google Scholar] [CrossRef] [PubMed]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, RESEARCH0034. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pereira, G.Q.; Gomes, L.A.; Santos, I.S.; Alfieri, A.F.; Weese, J.S.; Costa, M.C. Fecal microbiota transplantation in puppies with canine parvovirus infection. J. Vet. Intern. Med. 2018, 32, 707–711. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.; Yan, H.; Diao, H.; Yu, B.; He, J.; Yu, J.; Zheng, P.; Mao, X.; Luo, Y.; Chen, D. Early Gut Microbiota Intervention Suppresses DSS-Induced Inflammatory Responses by Deactivating TLR/NLR Signalling in Pigs. Sci. Rep. 2017, 7, 3224. [Google Scholar] [CrossRef] [Green Version]
- Kim, H.S.; Whon, T.W.; Sung, H.; Jeong, Y.S.; Jung, E.S.; Shin, N.R.; Hyun, D.W.; Kim, P.S.; Lee, J.Y.; Lee, C.H.; et al. Longitudinal evaluation of fecal microbiota transplantation for ameliorating calf diarrhea and improving growth performance. Nat. Commun. 2021, 12, 161. [Google Scholar] [CrossRef]
- Hu, L.; Geng, S.; Li, Y.; Cheng, S.; Fu, X.; Yue, X.; Han, X. Exogenous Fecal Microbiota Transplantation from Local Adult Pigs to Crossbred Newborn Piglets. Front. Microbiol. 2018, 8, 2663. [Google Scholar] [CrossRef]
- Siegerstetter, S.-C.; Petri, R.M.; Magowan, E.; Lawlor, P.G.; Zebeli, Q.; O’Connell, N.E.; Metzler-Zebeli, B.U. Fecal Microbiota Transplant from Highly Feed-Efficient Donors Shows Little Effect on Age-Related Changes in Feed-Efficiency-Associated Fecal Microbiota from Chickens. Appl. Environ. Microbiol. 2018, 84, e02330-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gareau, M.G.; Sherman, P.M.; Walker, W.A. Probiotics and the gut microbiota in intestinal health and disease. Nat. Rev. Gastroenterol. Hepatol. 2010, 7, 503–514. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Borody, T.J.; Khoruts, A. Fecal microbiota transplantation and emerging applications. Nat. Rev. Gastroenterol. Hepatol. 2011, 9, 88–96. [Google Scholar] [CrossRef]
- Liu, S.X.; Li, Y.H.; Dai, W.K.; Li, X.S.; Qiu, C.Z.; Ruan, M.L.; Zou, B.; Dong, C.; Liu, Y.H.; He, J.Y.; et al. Fecal microbiota transplantation induces remission of infantile allergic colitis through gut microbiota re-establishment. World J. Gastroenterol. 2017, 23, 8570–8581. [Google Scholar] [CrossRef]
- Yeh, A.; Morowitz, M.J. Probiotics and fecal microbiota transplantation in surgical disorders. Semin. Colon Rectal Surg. 2018, 29, 37–43. [Google Scholar] [CrossRef]
- Prado, C.; Michels, M.; Ávila, P.; Burger, H.; Milioli, M.V.M.; Dal-Pizzol, F. The protective effects of fecal microbiota transplantation in an experimental model of necrotizing enterocolitis. J. Pediatric Surg. 2019, 54, 1578–1583. [Google Scholar] [CrossRef] [PubMed]
- Hennessy, M.; Indugu, N.; Vecchiarelli, B.; Redding, L.; Bender, J.; Pappalardo, C.; Leibstein, M.; Toth, J.; Stefanovski, D.; Katepalli, A.; et al. Short communication: Comparison of the fecal bacterial communities in diarrheic and nondiarrheic dairy calves from multiple farms in southeastern Pennsylvania. J. Dairy Sci. 2021, 104, 7225–7232. [Google Scholar] [CrossRef] [PubMed]
- Shin, N.R.; Whon, T.W.; Bae, J.W. Proteobacteria: Microbial signature of dysbiosis in gut microbiota. Trends Biotechnol. 2015, 33, 496–503. [Google Scholar] [CrossRef] [PubMed]
- King, C.H.; Desai, H.; Sylvetsky, A.C.; LoTempio, J.; Ayanyan, S.; Carrie, J.; Crandall, K.A.; Fochtman, B.C.; Gasparyan, L.; Gulzar, N.; et al. Baseline human gut microbiota profile in healthy people and standard reporting template. PLoS ONE 2019, 14, e0206484. [Google Scholar] [CrossRef] [Green Version]
- Kim, E.T.; Lee, S.J.; Kim, T.Y.; Lee, H.G.; Atikur, R.M.; Gu, B.H.; Kim, D.H.; Park, B.Y.; Son, J.K.; Kim, M.H. Dynamic Changes in Fecal Microbial Communities of Neonatal Dairy Calves by Aging and Diarrhea. Animals 2021, 11, 1113. [Google Scholar] [CrossRef]
- Bertoni, G.; Ferrari, A.; Gubbiotti, A.; Trevisi, E. Blood indices calves: Relationship with mother values and changes in the first days of life. Ital. J. Anim. Sci. 2009, 8, 595–597. [Google Scholar] [CrossRef]
- Orro, T.; Jacobsen, S.; LePage, J.-P.; Niewold, T.; Alasuutari, S.; Soveri, T. Temporal changes in serum concentrations of acute phase proteins in newborn dairy calves. Vet. J. 2008, 176, 182–187. [Google Scholar] [CrossRef]
- Ontsouka, E.C.; Albrecht, C.; Bruckmaier, R.M. Invited review: Growth-promoting effects of colostrum in calves based on interaction with intestinal cell surface receptors and receptor-like transporters. J. Dairy Sci. 2016, 99, 4111–4123. [Google Scholar] [CrossRef] [PubMed]
- Hulbert, L.E.; Moisa, S.J. Stress, immunity, and the management of calves. J. Dairy Sci. 2016, 99, 3199–3216. [Google Scholar] [CrossRef] [Green Version]
- Schaff, C.T.; Rohrbeck, D.; Steinhoff-Wagner, J.; Kanitz, E.; Sauerwein, H.; Bruckmaier, R.M.; Hammon, H.M. Effects of colostrum versus formula feeding on hepatic glucocorticoid and alpha(1)- and beta(2)-adrenergic receptors in neonatal calves and their effect on glucose and lipid metabolism. J. Dairy Sci. 2014, 97, 6344–6357. [Google Scholar] [CrossRef] [Green Version]
- Baldwin VI, R.L.; McLeod, K.R.; Klotz, J.L.; Heitmann, R.N. Rumen development, intestinal growth and hepatic metabolism in the pre- and postweaning ruminant. J. Dairy Sci. 2004, 87, E55–E65. [Google Scholar] [CrossRef] [Green Version]
- Trevisi, E.; Bertoni, G.; Lombardelli, R.; Minuti, A. Relation of inflammation and liver function with the plasma cortisol response to adrenocorticotropin in early lactating dairy cows. J. Dairy Sci. 2013, 96, 5712–5722. [Google Scholar] [CrossRef] [Green Version]
- Giordano, A.; Veronesi, M.C.; Rossi, G.; Pezzia, F.; Probo, M.; Giori, L.; Paltrinieri, S. Serum paraoxonase-1 activity in neonatal calves: Age related variations and comparison between healthy and sick animals. Vet. J. 2013, 197, 499–501. [Google Scholar] [CrossRef] [PubMed]
- Marcato, F.; van den Brand, H.; Kemp, B.; van Reenen, K. Evaluating Potential Biomarkers of Health and Performance in Veal Calves. Front. Vet. Sci. 2018, 5. [Google Scholar] [CrossRef]
- Silva, D.G.d.; Silva, P.R.L.e.; Silva, P.C.d.; Fagliari, J.J. Serum protein concentrations, including acute phase proteins, in calves experimentally infected with Salmonella Dublin. Pesqui. Veterinária Bras. 2011, 31, 551–554. [Google Scholar] [CrossRef]
- Rosa, F.; Busato, S.; Avaroma, F.C.; Linville, K.; Trevisi, E.; Osorio, J.S.; Bionaz, M. Transcriptional changes detected in fecal RNA of neonatal dairy calves undergoing a mild diarrhea are associated with inflammatory biomarkers. PLoS ONE 2018, 13, e0191599. [Google Scholar] [CrossRef] [Green Version]
- Connor, E.E.; Baldwin, R.L.t.; Li, C.J.; Li, R.W.; Chung, H. Gene expression in bovine rumen epithelium during weaning identifies molecular regulators of rumen development and growth. Funct. Integr. Genom. 2013, 13, 133–142. [Google Scholar] [CrossRef] [PubMed]
- Meale, S.J.; Li, S.C.; Azevedo, P.; Derakhshani, H.; DeVries, T.J.; Plaizier, J.C.; Steele, M.A.; Khafipour, E. Weaning age influences the severity of gastrointestinal microbiome shifts in dairy calves. Sci. Rep. 2017, 7, 198. [Google Scholar] [CrossRef]
- Kim, M.-H.; Yang, J.-Y.; Upadhaya, S.D.; Lee, H.-J.; Yun, C.-H.; Ha, J.K. The stress of weaning influences serum levels of acute-phase proteins, iron-binding proteins, inflammatory cytokines, cortisol, and leukocyte subsets in Holstein calves. J. Vet. Sci. 2011, 12, 151–157. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiang, Q.; Wu, X.; Pan, Y.; Wang, L.; Cui, C.; Guo, Y.; Zhu, L.; Peng, J.; Wei, H. Early-Life Intervention Using Fecal Microbiota Combined with Probiotics Promotes Gut Microbiota Maturation, Regulates Immune System Development, and Alleviates Weaning Stress in Piglets. Int. J. Mol. Sci. 2020, 21, 503. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mutinati, M.; Pantaleo, M.; Roncetti, M.; Piccinno, M.; Rizzo, A.; Sciorsci, R. Oxidative Stress in Neonatology. A Review. Reprod. Domest. Anim. 2014, 49, 7–16. [Google Scholar] [CrossRef] [PubMed]
- Ranade, R.; Talukder, S.; Muscatello, G.; Celi, P. Assessment of oxidative stress biomarkers in exhaled breath condensate and blood of dairy heifer calves from birth to weaning. Vet. J. 2014, 202, 583–587. [Google Scholar] [CrossRef]
- Kuriakose, T.; Kanneganti, T.D. ZBP1: Innate Sensor Regulating Cell Death and Inflammation. Trends Immunol. 2018, 39, 123–134. [Google Scholar] [CrossRef]
- Takaoka, A.; Wang, Z.; Choi, M.K.; Yanai, H.; Negishi, H.; Ban, T.; Lu, Y.; Miyagishi, M.; Kodama, T.; Honda, K.; et al. DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature 2007, 448, 501–505. [Google Scholar] [CrossRef] [PubMed]
- Jacometo, C.B.; Alharthi, A.S.; Zhou, Z.; Luchini, D.; Loor, J.J. Maternal supply of methionine during late pregnancy is associated with changes in immune function and abundance of microRNA and mRNA in Holstein calf polymorphonuclear leukocytes. J. Dairy Sci. 2018, 101, 8146–8158. [Google Scholar] [CrossRef] [PubMed]
- Pearson, J.S.; Giogha, C.; Muhlen, S.; Nachbur, U.; Pham, C.L.; Zhang, Y.; Hildebrand, J.M.; Oates, C.V.; Lung, T.W.; Ingle, D.; et al. EspL is a bacterial cysteine protease effector that cleaves RHIM proteins to block necroptosis and inflammation. Nat. Microbiol. 2017, 2, 16258. [Google Scholar] [CrossRef] [PubMed]
| Parameter | Group | SEM 2 | p-Value 3 | |||
|---|---|---|---|---|---|---|
| CON | FMT | TRT | Week | T × W | ||
| Growth parameters 1 | ||||||
| BW, kg | 50.6 | 52.8 | 0.91 | 0.10 | <0.01 | 0.82 |
| Birth BW, kg | 41.6 | 38.8 | 2.52 | 0.43 | -- | -- |
| Pre-weaning BW, kg | 57.7 | 61.2 | 1.34 | 0.06 | -- | -- |
| Final BW, kg | 64.0 | 67.0 | 1.40 | 0.12 | -- | -- |
| WH, cm | 82.6 | 83.8 | 0.49 | 0.13 | <0.01 | 0.34 |
| Birth WH, cm | 78.4 | 76.6 | 1.22 | 0.31 | -- | -- |
| Pre-weaning WH, cm | 85.2 | 86.4 | 0.64 | 0.21 | -- | -- |
| Final WH, cm | 85.9 | 87.9 | 0.61 | 0.03 | -- | -- |
| ADG, kg/d | 0.46 | 0.47 | 0.04 | 0.92 | <0.01 | 0.99 |
| Intakes and feed efficiency | ||||||
| Stater intake, g/d | 504 | 486 | 73 | 0.86 | <0.01 | 0.26 |
| Total intake, g/d | 1027 | 1009 | 72 | 0.86 | <0.01 | 0.23 |
| G:F, g/g | 0.40 | 0.40 | 0.03 | 0.96 | <0.01 | 0.76 |
| Health parameters | ||||||
| Fecal scores 4 | 1.86 | 2.16 | 0.17 | 0.25 | 0.28 | 0.86 |
| Scour days 5 | 3.3 | 5.5 | 2.1 | 0.48 | -- | -- |
| Rectal temperature, °C | 38.6 | 38.6 | 0.06 | 0.94 | 0.39 | 0.32 |
| OTU | Week 1 | Week 2 | Week 5 | Donor | |||
|---|---|---|---|---|---|---|---|
| CON | FMT | CON | FMT | CON | FMT | ||
| Firmicutes | 32.03 | 38.32 | 44.0 | 35.19 | 33.0 | 36.44 | 57.76 |
| Ruminococcaceae | 10.01 | 9.91 | 7.63 | 8.48 | 9.97 | 12.45 | 18.86 |
| Lachnospiraceae | 5.67 | 4.32 | 8.81 | 6.30 | 7.61 | 7.81 | 11.97 |
| Erysipelotrichaceae | 4.67 | 4.18 | 8.10 | 7.68 | 3.64 | 5.65 | 9.72 |
| Peptostreptococcaceae | 0.01 | 0.15 | 0.03 | 0.26 | 0.24 | 0.18 | 8.74 |
| Clostridiales Inc. Sedis XIII | 0.27 | 0.16 | 0.68 | 0.20 | 0.18 | 0.19 | 3.49 |
| Clostridiaceae1 | 0.09 | 0.12 | <0.01 | 0.06 | 0.85 | 0.05 | 1.46 |
| Lactobacillaceae | 1.03 | 1.73 | 2.15 | 0.58 | 1.50 | 0.49 | 0.14 |
| Streptococcaceae | 1.89 | 5.93 | 4.08 | 1.00 | 1.20 | 0.24 | 0.23 |
| Veillonellaceae # | 4.38 | 4.69 | 4.62 | 2.54 | 2.14 | 0.62 | 0 |
| Enterococcaceae # | 0.35 | 1.64 | 1.14 | 0.50 | 0.46 | 0.01 | 0 |
| Selenomonadaceae | 1.96 | 3.47 | 3.08 | 2.39 | 1.94 | 1.24 | 0.03 |
| Other Firmicutes * | 1.61 | 2.04 | 3.67 | 5.21 | 3.26 | 7.50 | 3.13 |
| Bacteroidetes | 30.01 | 24.94 | 34.70 | 34.27 | 35.22 | 33.29 | 31.25 |
| Muribaculaceae | 0.03 | 0.02 | 0.04 | 0.02 | 0.05 | 0.05 | 5.36 |
| Bacteroidaceae # | 5.48 | 0.56 | 5.96 | 2.72 | 2.09 | 1.62 | 4.79 |
| Odoribacteraceae # | 0.03 ab | 0.01 ab | 0.10 a | <0.01 a | 0.20 bc | 0.18 c | 4.71 |
| Prevotellaceae | 22.95 | 23.68 | 24.42 | 30.41 | 30.35 | 28.08 | 3.63 |
| Porphyromonodaceae | 0.66 | 0.20 | 1.39 | 0.31 | 0.43 | 0.22 | 0.18 |
| Rikenellaceae | 0.03 | 0.04 | 0.07 | 0.08 | 0.02 | 0.04 | 3.05 |
| Other Bacteroidetes * | 0.81 | 0.43 | 2.73 | 0.71 | 2.08 | 3.11 | 9.52 |
| Proteobacteria | 28.73 | 29.95 | 12.98 | 24.98 | 21.96 | 22.53 | 6.72 |
| Succinivibrionaceae# | 2.32 | 2.81 | 0.85 | 8.84 | 6.20 | 4.13 | 4.27 |
| Pasteurellaceae Pasteurellaceae # | 2.69 ab | 2.18 ab | 1.82a | 2.76 a | 2.52 ab | 0.04 b | 0 |
| Sutterellaceae | 12.49 | 9.17 | 5.32 | 8.39 | 9.23 | 8.26 | 0.20 |
| Enterobacteriaceae # | 10.61 | 14.59 | 3.63 | 2.70 | 2.21 | 0.79 | 0.20 |
| Other Proteobacteria * | 0.63 | 1.21 | 1.37 | 2.28 | 1.81 | 9.31 | 2.00 |
| Actinobacteria # | 2.27 ab | 2.27 a | 4.65a | 1.17 ab | 2.08 ab | 0.75 b | 0.36 |
| Verrucomicrobia | 2.72 | 1.88 | 0.03 | 0.22 | 1.35 | <0.01 | 0.01 |
| Campylobacterota | 3.74 | 2.31 | 2.37 | 1.33 | 2.06 | 0.06 | 0.01 |
| Other Bacteria * | 0.49 | 0.34 | 1.27 | 2.83 | 4.33 | 6.91 | 3.88 |
| Parameter 1 | Group | SEM 2 | p-Value 3 | |||
|---|---|---|---|---|---|---|
| CON | FMT | TRT | Week | T × W | ||
| Energy metabolism | ||||||
| Glucose, mmol/L | 6.23 | 5.69 | 0.34 | 0.33 | 0.04 | 0.28 |
| BHBA 4, mmol/L | 0.09 | 0.07 | 0.01 | 0.20 | 0.36 | 0.28 |
| Creatinine, µmol/L | 94.65 | 91.75 | 4.41 | 0.65 | 0.03 | 0.72 |
| Urea, mmol/L | 2.98 | 2.74 | 0.16 | 0.29 | 0.01 | 0.07 |
| Inflammation | ||||||
| Haptoglobin, g/L | 0.19 | 0.21 | 0.01 | 0.24 | 0.06 | 0.02 |
| Ceruloplasmin, µmol/L | 1.82 | 1.95 | 0.13 | 0.51 | 0.38 | 0.29 |
| IL1-β (log2) pg/mL | 3.69 | 3.67 | 0.39 | 0.98 | 0.26 | 0.07 |
| IL-6, (log2) pg/mL | 8.17 | 7.96 | 0.20 | 0.48 | <0.01 | 0.68 |
| Liver function | ||||||
| Albumin, g/L | 30.61 | 30.03 | 0.29 | 0.18 | <0.01 | 0.14 |
| Paraoxonase, U/mL | 34.50 | 40.56 | 2.19 | 0.07 | <0.01 | 0.06 |
| Total bilirubin, µmol/L | 4.67 | 6.30 | 0.97 | 0.26 | <0.01 | 0.13 |
| Cholesterol, mmol/L | 2.68 | 2.40 | 0.17 | 0.25 | <0.01 | 0.31 |
| GOT 5 (log2), U/L | 5.76 | 5.89 | 0.11 | 0.42 | <0.01 | 0.71 |
| GGT 6, U/L | 458.4 | 418.1 | 87.4 | 0.76 | <0.01 | 0.16 |
| Oxidative Stress | ||||||
| ROM 7, mg H2O2/100 mL | 12.06 | 12.09 | 0.60 | 0.98 | 0.23 | 0.20 |
| FRAP 8, mmol/L | 160.45 | 149.64 | 7.79 | 0.36 | 0.46 | 0.34 |
| Parameter 1 | Group | SEM 2 | p-Value 3 | |||||
|---|---|---|---|---|---|---|---|---|
| CON | FMT | |||||||
| 6 | 7 | 6 | 7 | TRT | Week | T × W | ||
| Energy metabolism | ||||||||
| Glucose, mmol/L | 5.61 | 4.93 | 5.63 | 5.12 | 0.25 | 0.78 | <0.01 | 0.34 |
| BHBA 4, mmol/L | 0.18 | 0.25 | 0.15 | 0.26 | 0.03 | 0.86 | 0.02 | 0.62 |
| Creatinine, µmol/L | 80.9 | 82.3 | 80.9 | 82.4 | 3.5 | 0.99 | 0.45 | 0.98 |
| Urea, mmol/L | 4.43 | 7.29 | 3.68 | 6.0 | 0.34 | 0.05 | <0.01 | 0.33 |
| Inflammation | ||||||||
| Haptoglobin (log2), g/L | 0.22 | 0.47 | 0.32 | 0.49 | 0.26 | 0.48 | 0.05 | 0.54 |
| Ceruloplasmin, µmol/L | 1.50 | 1.72 | 1.62 | 1.69 | 0.19 | 0.88 | 0.14 | 0.40 |
| IL1-β (log2) pg/mL | 27.5 | 12.0 | 11.6 | 4.6 | 0.27 | 0.01 | 0.01 | 0.87 |
| IL-6, (log2) pg/mL | 159.2 | 154.4 | 72.2 | 69.0 | 0.36 | 0.05 | 0.87 | 0.98 |
| Liver function | ||||||||
| Albumin, g/L | 32.4 | 33.2 | 32.7 | 33.3 | 0.50 | 0.83 | 0.09 | 0.84 |
| Paraoxonase, U/mL | 77.5 | 74.0 | 92.8 | 85.3 | 5.85 | 0.09 | 0.18 | 0.61 |
| Total bilirubin, µmol/L | 2.49 | 2.36 | 3.12 | 2.78 | 0.40 | 0.38 | 0.41 | 0.72 |
| Cholesterol, mmol/L | 3.66 | 2.69 | 3.89 | 3.08 | 0.24 | 0.37 | <0.01 | 0.69 |
| GOT 5 (log2), U/L | 102.4 a | 87.1 b | 82.4 b | 89.2 ab | 0.11 | 0.33 | 0.37 | 0.02 |
| GGT 6 (log2), U/L | 84.6 a | 43.7 bc | 44.5 b | 30.0 c | 0.29 | 0.07 | <0.01 | 0.01 |
| Oxidative Stress | ||||||||
| ROM 7, mg H2O2/100 mL | 10.2 b | 13.1 a | 10.2 a | 9.6 b | 1.11 | 0.25 | 0.07 | 0.01 |
| FRAP 8, mmol/L | 159.8 a | 137.7 b | 161.5 a | 161.0 a | 9.3 | 0.30 | 0.01 | 0.01 |
| Transcript 1 | Group | SEM 2 | p-Value 3 | |||
|---|---|---|---|---|---|---|
| CON | FMT | TRT | Week | T × W | ||
| ZBP1 | 1.09 | 1.85 | 0.23 | 0.03 | 0.40 | 0.64 |
| NFKB1 | 1.21 | 1.57 | 0.15 | 0.12 | 0.20 | 0.53 |
| STAT3 | −0.50 | −0.34 | 0.21 | 0.60 | 0.87 | 0.92 |
| SELL | 0.82 | 1.27 | 0.30 | 0.30 | 0.10 | 0.86 |
| TLR2 | 1.24 | 1.41 | 0.27 | 0.66 | 0.50 | 0.11 |
| TLR4 | 0.40 | 0.65 | 0.26 | 0.50 | 0.36 | 0.36 |
| IL1B | 0.62 | 0.74 | 0.40 | 0.84 | 0.37 | 0.45 |
| IL8 | 0.27 | 0.87 | 0.54 | 0.44 | 0.01 | 0.60 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rosa, F.; Michelotti, T.C.; St-Pierre, B.; Trevisi, E.; Osorio, J.S. Early Life Fecal Microbiota Transplantation in Neonatal Dairy Calves Promotes Growth Performance and Alleviates Inflammation and Oxidative Stress during Weaning. Animals 2021, 11, 2704. https://doi.org/10.3390/ani11092704
Rosa F, Michelotti TC, St-Pierre B, Trevisi E, Osorio JS. Early Life Fecal Microbiota Transplantation in Neonatal Dairy Calves Promotes Growth Performance and Alleviates Inflammation and Oxidative Stress during Weaning. Animals. 2021; 11(9):2704. https://doi.org/10.3390/ani11092704
Chicago/Turabian StyleRosa, Fernanda, Tainara Cristina Michelotti, Benoit St-Pierre, Erminio Trevisi, and Johan S. Osorio. 2021. "Early Life Fecal Microbiota Transplantation in Neonatal Dairy Calves Promotes Growth Performance and Alleviates Inflammation and Oxidative Stress during Weaning" Animals 11, no. 9: 2704. https://doi.org/10.3390/ani11092704
APA StyleRosa, F., Michelotti, T. C., St-Pierre, B., Trevisi, E., & Osorio, J. S. (2021). Early Life Fecal Microbiota Transplantation in Neonatal Dairy Calves Promotes Growth Performance and Alleviates Inflammation and Oxidative Stress during Weaning. Animals, 11(9), 2704. https://doi.org/10.3390/ani11092704









