Transcriptomic Response under Heat Stress in Chickens Revealed the Regulation of Genes and Alteration of Metabolism to Maintain Homeostasis
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animal Experiments
2.2. RNA Isolation and Sequencing
2.3. Quality Control, Mapping, Counting and Batch Correction
2.4. Differentially Expressed Genes Analysis
2.5. Gene Ontology, KEGG Pathway, and Network Analysis
2.6. Statistical Analysis
3. Results
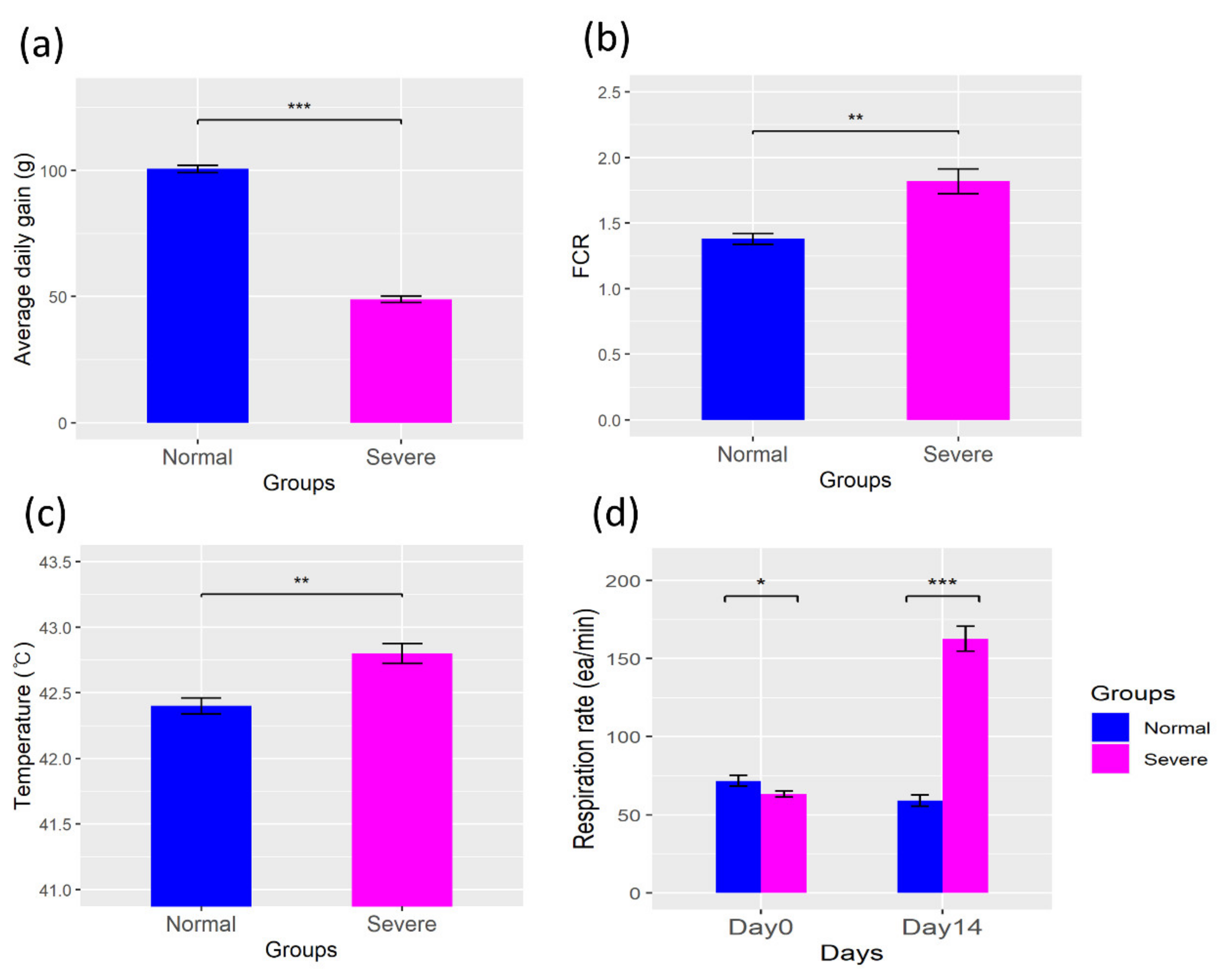
3.1. Physiologic Parameters
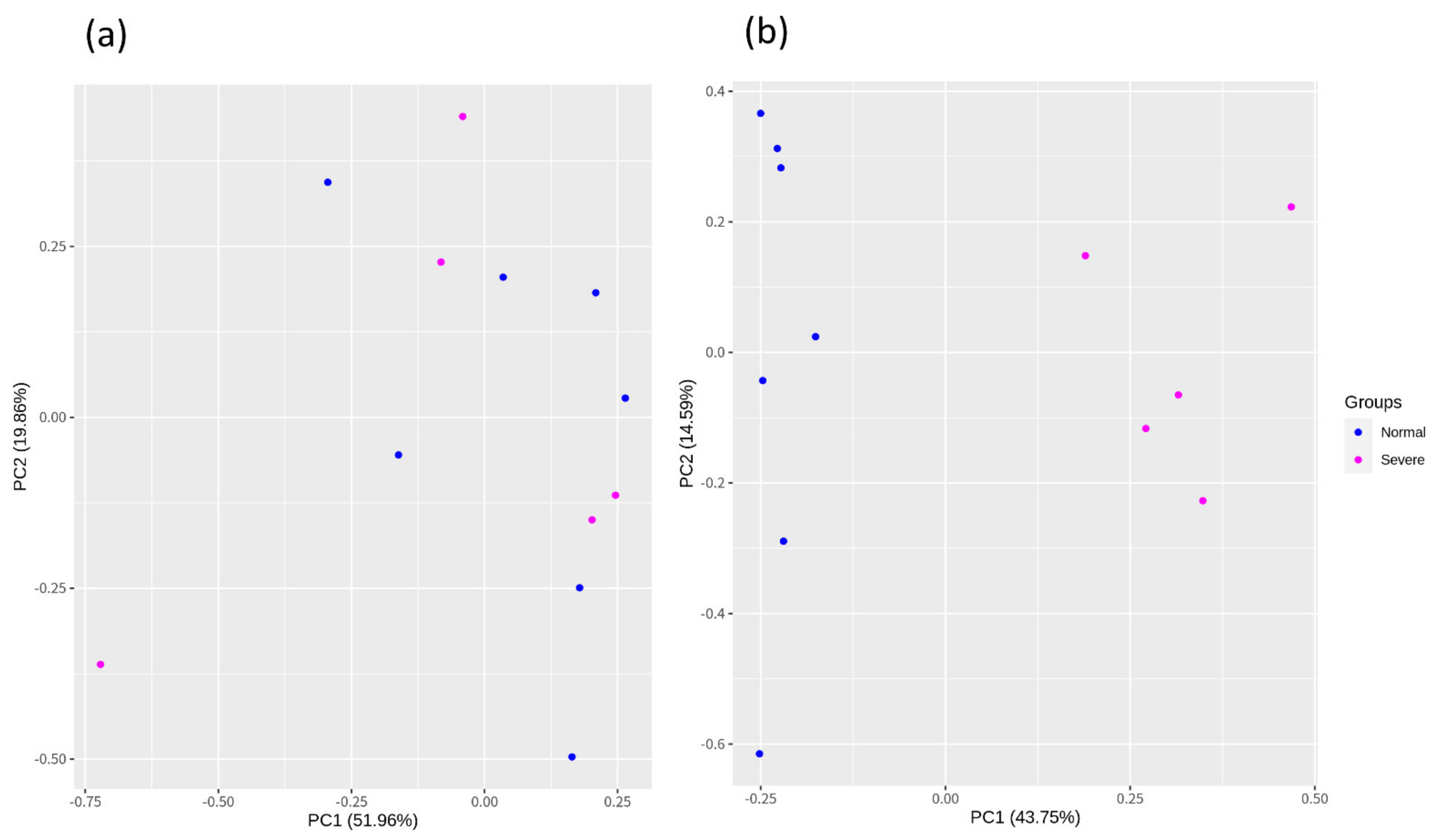
3.2. Construction of the Raw Reads, Mapping, and Batch Correction
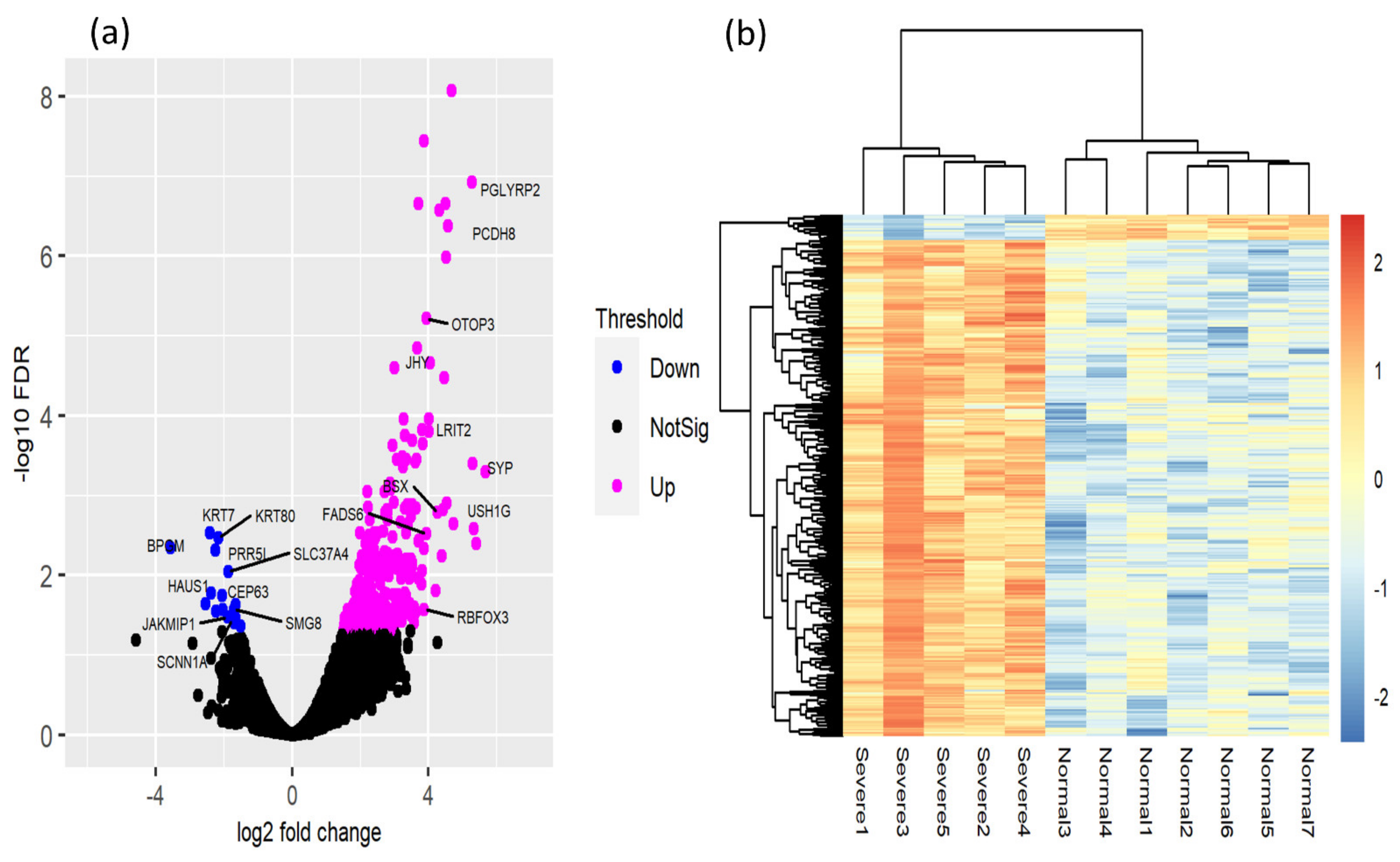
3.3. Identification of DEGs
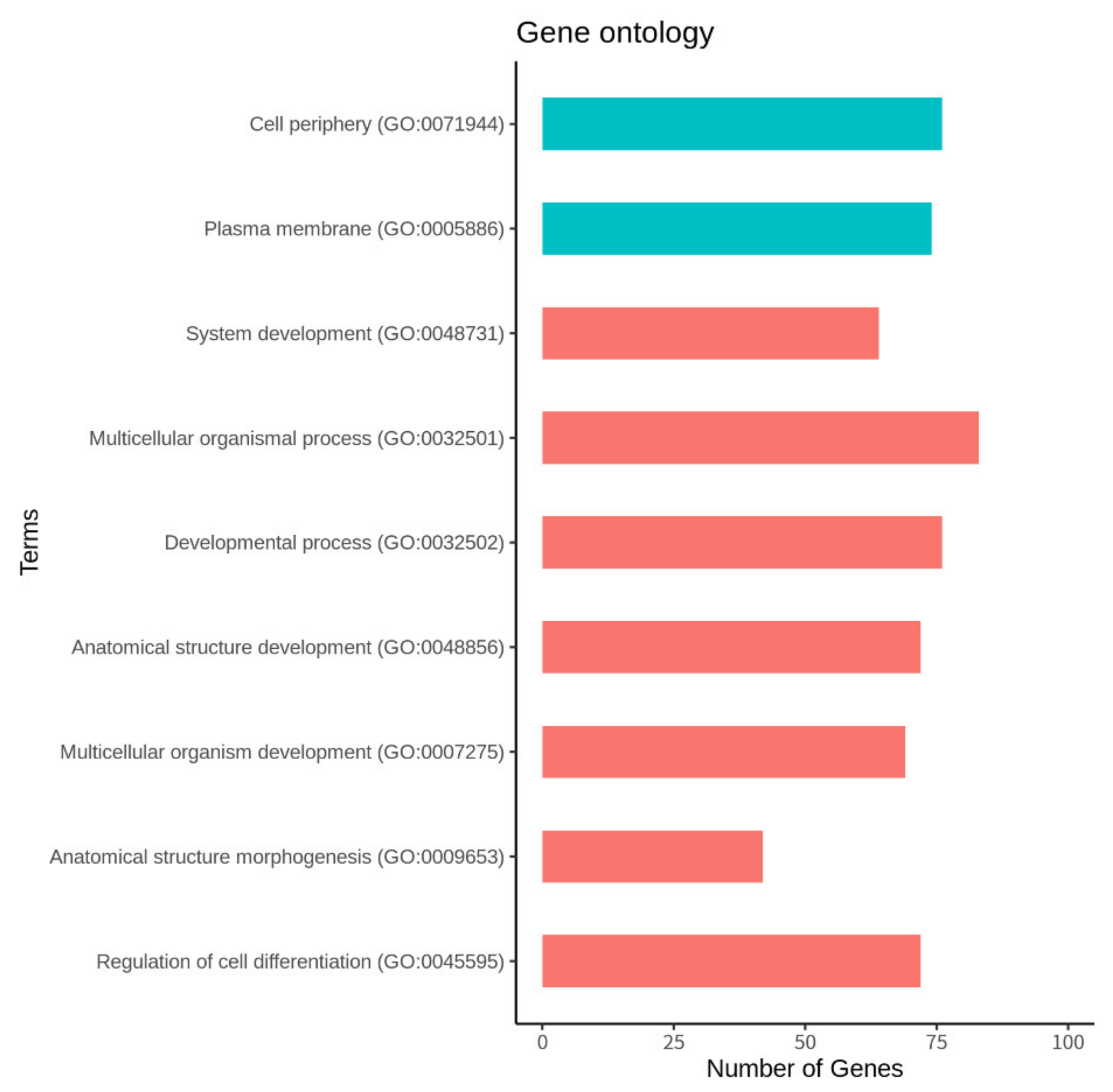
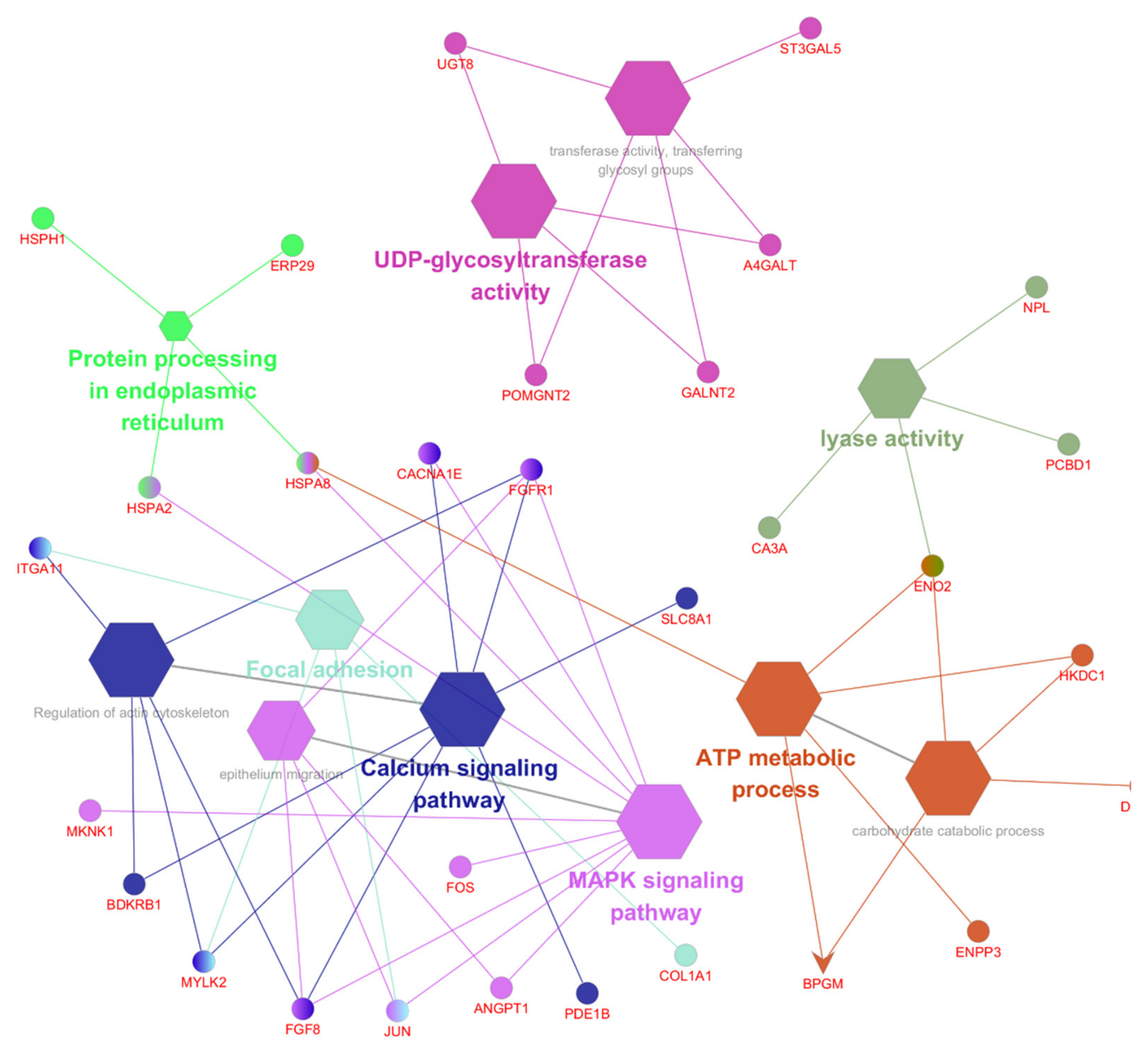
3.4. Function and Pathway Analysis of the DEGs
3.5. The Top 10 KEGG Pathways
4. Discussion
4.1. Changes in the Physiological Parameters in a Heat Stress Environment
4.2. Transcriptomic Changes in a Heat Stress Environment
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Babinszky, L.; Halas, V.; Verstegen, M.W. Impacts of Climate Change on Animal Production and Quality of Animal Food Products. In Climate Change: Socioeconomic Effects; InTech: Rijeka, Croatia, 2011; pp. 165–190. [Google Scholar] [CrossRef]
- Nawab, A.; Ibtisham, F.; Li, G.; Kieser, B.; Wu, J.; Liu, W.; Zhao, Y.; Nawab, Y.; Li, K.; Xiao, M. Heat stress in poultry production: Mitigation strategies to overcome the future challenges facing the global poultry industry. J. Therm. Biol. 2018, 78, 131–139. [Google Scholar] [CrossRef]
- Akbarian, A.; Michiels, J.; Degroote, J.; Majdeddin, M.; Golian, A.; De Smet, S. Association between heat stress and oxidative stress in poultry; mitochondrial dysfunction and dietary interventions with phytochemicals. J. Anim. Sci. Biotechnol. 2016, 7, 37. [Google Scholar] [CrossRef] [PubMed]
- Deyhim, F.; Teeter, R. Research note: Sodium and potassium chloride drinking water supplementation effects on acid-base balance and plasma corticosterone in broilers reared in thermoneutral and heat-distressed environments. Poult. Sci. 1991, 70, 2551–2553. [Google Scholar] [CrossRef]
- May, J.; Deaton, J.; Reece, F.; Branton, S. Effect of acclimation and heat stress on thyroid hormone concentration. Poult. Sci. 1986, 65, 1211–1213. [Google Scholar] [CrossRef]
- Mashaly, M.; Hendricks, G., 3rd; Kalama, M.; Gehad, A.; Abbas, A.; Patterson, P. Effect of heat stress on production parameters and immune responses of commercial laying hens. Poult. Sci. 2004, 83, 889–894. [Google Scholar] [CrossRef] [PubMed]
- Varasteh, S.; Braber, S.; Akbari, P.; Garssen, J.; Fink-Gremmels, J. Differences in susceptibility to heat stress along the chicken intestine and the protective effects of galacto-oligosaccharides. PLoS ONE 2015, 10, e0138975. [Google Scholar]
- Lin, H.; Jiao, H.; Buyse, J.; Decuypere, E. Strategies for preventing heat stress in poultry. World’s Poult. Sci. J. 2006, 62, 71–86. [Google Scholar] [CrossRef]
- St-Pierre, N.; Cobanov, B.; Schnitkey, G. Economic losses from heat stress by US livestock industries. J. Dairy Sci. 2003, 86, E52–E77. [Google Scholar] [CrossRef]
- Wasti, S.; Sah, N.; Mishra, B. Impact of heat stress on poultry health and performances, and potential mitigation strategies. Animals 2020, 10, 1266. [Google Scholar] [CrossRef] [PubMed]
- Teeter, R.G.; Belay, T. Broiler management during acute heat stress. Anim. Feed Sci. Technol. 1996, 58, 127–142. [Google Scholar] [CrossRef]
- Pawar, S.; Sajjanar, B.; Lonkar, V.; Kurade, N.; Kadam, A.; Nirmal, A.; Brahmane, M.; Bal, S. Assessing and mitigating the impact of heat stress in poultry. Adv. Anim. Vet. Sci. 2016, 4, 332–341. [Google Scholar] [CrossRef]
- Charles, D.R.; Walker, A.W. Poultry Environment Problems: A Guide to Solutions; Nottingham University Press: Nottingham, UK, 2002. [Google Scholar]
- Lara, L.J.; Rostagno, M.H. Impact of heat stress on poultry production. Animals 2013, 3, 356–369. [Google Scholar] [CrossRef] [PubMed]
- Daramola, J.O.; Abioja, M.O.; Onagbesan, O.M. Heat stress impact on livestock production. In Environmental Stress and Amelioration in Livestock Production; Springer: Berlin/Heidelberg, Germany, 2012; pp. 53–73. [Google Scholar]
- Whitley, D.; Goldberg, S.P.; Jordan, W.D. Heat shock proteins: A review of the molecular chaperones. J. Vasc. Surg. 1999, 29, 748–751. [Google Scholar] [CrossRef]
- Calderwood, S.K.; Murshid, A.; Gong, J. Heat shock proteins: Conditional mediators of inflammation in tumor immunity. Front. Immunol. 2012, 3, 75. [Google Scholar] [CrossRef]
- Ryan, J.; Hightower, L. Stress proteins as molecular biomarkers for environmental toxicology. Stress Inducible Cell. Responses 1996, 411–424. [Google Scholar] [CrossRef]
- Baler, R.; Zou, J.; Voellmy, R. Evidence for a role of Hsp 70 in the regulation of the heat shock response in mammalian cells. Cell Stress Chaperones 1996, 1, 33. [Google Scholar] [CrossRef]
- Cedraz, H.; Gromboni, J.G.G.; Garcia, A.A.P.; Farias Filho, R.V.; Souza, T.M.; Oliveira, E.R.D.; Oliveira, E.B.D.; Nascimento, C.S.D.; Meneghetti, C.; Wenceslau, A.A. Heat stress induces expression of HSP genes in genetically divergent chickens. PLoS ONE 2017, 12, e0186083. [Google Scholar] [CrossRef]
- Allen, R.; Tresini, M. Oxidative stress and gene regulation. Free Radic. Biol. Med. 2000, 28, 463–499. [Google Scholar] [CrossRef]
- Gabai, V.L.; Meriin, A.B.; Mosser, D.D.; Caron, A.; Rits, S.; Shifrin, V.I.; Sherman, M.Y. Hsp70 prevents activation of stress kinases: A novel pathway of cellular thermotolerance. J. Biol. Chem. 1997, 272, 18033–18037. [Google Scholar] [CrossRef]
- De Nadal, E.; Ammerer, G.; Posas, F. Controlling gene expression in response to stress. Nat. Rev. Genet. 2011, 12, 833–845. [Google Scholar] [CrossRef]
- Johnson, G.L.; Dohlman, H.G.; Graves, L.M. MAPK kinase kinases (MKKKs) as a target class for small-molecule inhibition to modulate signaling networks and gene expression. Curr. Opin. Chem. Biol. 2005, 9, 325–331. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Jiang, Z.; Ning, L.; Zhao, Z.; Yang, N.; Chen, L.; Ma, H.; Li, L.; Fu, Y.; Zhu, H. Protective HSP70 induction by Z-ligustilide against oxygen glucose deprivation injury via activation of the MAPK pathway but not of HSF1. Biol. Pharm. Bull. 2015, 1564–1572. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Qi, Z.; Qi, S.; Gui, L.; Shen, L.; Feng, Z. Daphnetin protects oxidative stress-induced neuronal apoptosis via regulation of MAPK signaling and HSP70 expression. Oncol. Lett. 2016, 12, 1959–1964. [Google Scholar] [CrossRef]
- Cao, M.-H.; Xu, J.; Cai, H.-D.; Lv, Z.-W.; Feng, Y.-J.; Li, K.; Chen, C.-Q.; Li, Y.-Y. p38 MAPK inhibition alleviates experimental acute pancreatitis in mice. Hepatobiliary Pancreat. Dis. Int. 2015, 14, 101–106. [Google Scholar] [CrossRef]
- Plotnikov, A.; Zehorai, E.; Procaccia, S.; Seger, R. The MAPK cascades: Signaling components, nuclear roles and mechanisms of nuclear translocation. Biochim. Biophys. Acta (BBA) Mol. Cell Res. 2011, 1813, 1619–1633. [Google Scholar] [CrossRef]
- Hao, Y.; Feng, Y.; Li, J.; Gu, X. Role of MAPKs in HSP70′s protection against heat stress-induced injury in rat small intestine. BioMed Res. Int. 2018, 2018. [Google Scholar] [CrossRef]
- Mustafi, S.B.; Chakraborty, P.K.; Dey, R.S.; Raha, S. Heat stress upregulates chaperone heat shock protein 70 and antioxidant manganese superoxide dismutase through reactive oxygen species (ROS), p38MAPK, and Akt. Cell Stress Chaperones 2009, 14, 579–589. [Google Scholar] [CrossRef] [PubMed]
- Azad, M.A.K.; Kikusato, M.; Maekawa, T.; Shirakawa, H.; Toyomizu, M. Metabolic characteristics and oxidative damage to skeletal muscle in broiler chickens exposed to chronic heat stress. Comp. Biochem. Physiol. A 2010, 155, 401–406. [Google Scholar] [CrossRef]
- Mujahid, A.; Akiba, Y.; Toyomizu, M. Olive oil-supplemented diet alleviates acute heat stress-induced mitochondrial ROS production in chicken skeletal muscle. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2009, 297, R690–R698. [Google Scholar] [CrossRef] [PubMed]
- Tao, X.; Xin, H. Temperature-humidity-velocity index for market-size broilers. In Proceedings of the 2003 ASAE Annual Meeting, Las Vegas, NV, USA, 27–30 July 2003; p. 1. [Google Scholar]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef]
- Risso, D.; Ngai, J.; Speed, T.P.; Dudoit, S. Normalization of RNA-seq data using factor analysis of control genes or samples. Nat. Biotechnol. 2014, 32, 896–902. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44. [Google Scholar]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009, 37, 1–13. [Google Scholar] [CrossRef]
- Kanehisa, M.; Furumichi, M.; Tanabe, M.; Sato, Y.; Morishima, K. KEGG: New perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017, 45, D353–D361. [Google Scholar] [CrossRef] [PubMed]
- Bindea, G.; Mlecnik, B.; Hackl, H.; Charoentong, P.; Tosolini, M.; Kirilovsky, A.; Fridman, W.-H.; Pagès, F.; Trajanoski, Z.; Galon, J. ClueGO: A Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 2009, 25, 1091–1093. [Google Scholar] [CrossRef] [PubMed]
- Garriga, C.; Hunter, R.R.; Amat, C.; Planas, J.M.; Mitchell, M.A.; Moretó, M. Heat stress increases apical glucose transport in the chicken jejunum. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2006, 290, R195–R201. [Google Scholar] [CrossRef]
- Star, L.; Decuypere, E.; Parmentier, H.; Kemp, B. Effect of single or combined climatic and hygienic stress in four layer lines: 2. Endocrine and oxidative stress responses. Poult. Sci. 2008, 87, 1031–1038. [Google Scholar] [CrossRef] [PubMed]
- Geraert, P.; Padilha, J.; Guillaumin, S. Metabolic and endocrine changes induced by chronic heatexposure in broiler chickens: Growth performance, body composition and energy retention. Br. J. Nutr. 1996, 75, 195–204. [Google Scholar] [CrossRef][Green Version]
- Belhadj Slimen, I.; Najar, T.; Ghram, A.; Abdrrabba, M. Heat stress effects on livestock: Molecular, cellular and metabolic aspects, a review. J. Anim. Physiol. Anim. Nutr. 2016, 100, 401–412. [Google Scholar] [CrossRef]
- Bolender, N.; Sickmann, A.; Wagner, R.; Meisinger, C.; Pfanner, N. Multiple pathways for sorting mitochondrial precursor proteins. EMBO Rep. 2008, 9, 42–49. [Google Scholar] [CrossRef]
- Goldenthal, M.J.; Marin-Garcia, J. Mitochondrial signaling pathways: A receiver/integrator organelle. Mol. Cell. Biochem. 2004, 262, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Plácido, A.; Pereira, C.; Duarte, A.; Candeias, E.; Correia, S.; Santos, R.; Carvalho, C.; Cardoso, S.; Oliveira, C.; Moreira, P. The role of endoplasmic reticulum in amyloid precursor protein processing and trafficking: Implications for Alzheimer’s disease. Biochim. Biophys. Acta (BBA) Mol. Basis Dis. 2014, 1842, 1444–1453. [Google Scholar] [CrossRef]
- Sitia, R.; Braakman, I. Quality control in the endoplasmic reticulum protein factory. Nature 2003, 426, 891–894. [Google Scholar] [CrossRef] [PubMed]
- Alemu, T.W.; Pandey, H.O.; Wondim, D.S.; Gebremedhn, S.; Neuhof, C.; Tholen, E.; Holker, M.; Schellander, K.; Tesfaye, D. Oxidative and endoplasmic reticulum stress defense mechanisms of bovine granulosa cells exposed to heat stress. Theriogenology 2018, 110, 130–141. [Google Scholar] [CrossRef] [PubMed]
- Mishra, D.C.; Arora, D.; Kumar, R.R.; Goswami, S.; Varshney, S.; Budhlakoti, N.; Kumar, S.; Chaturvedi, K.K.; Sharma, A.; Chinnusamy, V. Weighted gene co-expression analysis for identification of key genes regulating heat stress in wheat. Cereal Res. Commun. 2021, 49, 73–81. [Google Scholar] [CrossRef]
- Goloubinoff, P.; De Los Rios, P. The mechanism of Hsp70 chaperones:(entropic) pulling the models together. Trends Biochem. Sci. 2007, 32, 372–380. [Google Scholar] [CrossRef]
- Archana, P.; Aleena, J.; Pragna, P.; Vidya, M.; Niyas, A.; Bagath, M.; Krishnan, G.; Manimaran, A.; Beena, V.; Kurien, E. Role of heat shock proteins in livestock adaptation to heat stress. J. Dairy Vet. Anim. Res. 2017, 5, 00127. [Google Scholar]
- Sousa, R.; Lafer, E.M. The physics of entropic pulling: A novel model for the Hsp70 motor mechanism. Int. J. Mol. Sci. 2019, 20, 2334. [Google Scholar] [CrossRef]
- Shaul, Y.D.; Seger, R. The MEK/ERK cascade: From signaling specificity to diverse functions. Biochim. Biophys. Acta (BBA) Mol. Cell Res. 2007, 1773, 1213–1226. [Google Scholar] [CrossRef]
- Pimienta, G.; Pascual, J. Canonical and alternative MAPK signaling. Cell Cycle 2007, 6, 2628–2632. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.J.; Pan, W.W.; Liu, S.B.; Shen, Z.F.; Xu, Y.; Hu, L.L. ERK/MAPK signalling pathway and tumorigenesis. Exp. Ther. Med. 2020, 19, 1997–2007. [Google Scholar] [CrossRef]
- Toth, A.B.; Shum, A.K.; Prakriya, M. Regulation of neurogenesis by calcium signaling. Cell Calcium 2016, 59, 124–134. [Google Scholar] [CrossRef]
- Lipskaia, L.; Hulot, J.-S.; Lompré, A.-M. Role of sarco/endoplasmic reticulum calcium content and calcium ATPase activity in the control of cell growth and proliferation. Pflügers Arch. Eur. J. Physiol. 2009, 457, 673–685. [Google Scholar] [CrossRef] [PubMed]
- Nunes, P.; Demaurex, N. The role of calcium signaling in phagocytosis. J. Leukoc. Biol. 2010, 88, 57–68. [Google Scholar] [CrossRef]
- Wozniak, M.A.; Modzelewska, K.; Kwong, L.; Keely, P.J. Focal adhesion regulation of cell behavior. Biochim. Biophys. Acta (BBA) Mol. Cell Res. 2004, 1692, 103–119. [Google Scholar] [CrossRef] [PubMed]
- Maziveyi, M.; Alahari, S.K. Cell matrix adhesions in cancer: The proteins that form the glue. Oncotarget 2017, 8, 48471. [Google Scholar] [CrossRef]
- Wang, J.G.; Miyazu, M.; Matsushita, E.; Sokabe, M.; Naruse, K. Uniaxial cyclic stretch induces focal adhesion kinase (FAK) tyrosine phosphorylation followed by mitogen-activated protein kinase (MAPK) activation. Biochem. Biophys. Res. Commun. 2001, 288, 356–361. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Yin, P.; Liu, F.; Cheng, G.; Guo, K.; Lu, A.; Zhu, X.; Luan, W.; Xu, J. Effect of heat stress on the porcine small intestine: A morphological and gene expression study. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2010, 156, 119–128. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Liu, F.; Yin, P.; Zhao, H.; Luan, W.; Hou, X.; Zhong, Y.; Jia, D.; Zan, J.; Ma, W. Involvement of oxidative stress and mitogen-activated protein kinase signaling pathways in heat stress-induced injury in the rat small intestine. Stress 2013, 16, 99–113. [Google Scholar] [CrossRef]
- Leeb-Lundberg, L.F.; Kang, D.S.; Lamb, M.E.; Fathy, D.B. The human B1 bradykinin receptor exhibits high ligand-independent, constitutive activity: Roles of residues in the fourth intracellular and third transmembrane domains. J. Biol. Chem. 2001, 276, 8785–8792. [Google Scholar] [CrossRef] [PubMed]
- Ignjacev-Lazich, I.; Kintsurashvili, E.; Johns, C.; Vitseva, O.; Duka, A.; Shenouda, S.; Gavras, I.; Gavras, H. Angiotensin-converting enzyme regulates bradykinin receptor gene expression. Am. J. Physiol. Heart Circ. Physiol. 2005, 289, H1814–H1820. [Google Scholar] [CrossRef]
- Duka, I.; Kintsurashvili, E.; Gavras, I.; Johns, C.; Bresnahan, M.; Gavras, H. Vasoactive potential of the B1 bradykinin receptor in normotension and hypertension. Circ. Res. 2001, 88, 275–281. [Google Scholar] [CrossRef]
- Soung, Y.H.; Lee, J.W.; Kim, S.Y.; Nam, S.W.; Park, W.S.; Lee, J.Y.; Yoo, N.J.; Lee, S.H. Mutational analysis of the kinase domain of MYLK2 gene in common human cancers. Pathol. Res. Pract. 2006, 202, 137–140. [Google Scholar] [CrossRef]
- Stachowiak, E.K.; Benson, C.A.; Narla, S.T.; Dimitri, A.; Bayona Chuye, L.E.; Dhiman, S.; Harikrishnan, K.; Elahi, S.; Freedman, D.; Brennand, K.J.; et al. Cerebral organoids reveal early cortical maldevelopment in schizophrenia—Computational anatomy and genomics, role of FGFR1. Transl. Psychiatry 2017, 7, 6. [Google Scholar] [CrossRef]
- Li, C.; Xu, X.K.D.; Nelson, D.K.; Williams, T.; Kuehn, M.R.; Deng, C. FGFR1 function at the earliest stages of mouse limb development plays an indispensable role in subsequent autopod morphogenesis. Development 2005, 132, 4755–4764. [Google Scholar] [CrossRef][Green Version]
- Chow, K.; Lu, M.; Wu, M. Expression of dihydrodiol dehydrogenase plays important roles in apoptosis and drug resistance of A431 squamous cell carcinoma. J. Dermatol. 2006, 41, 205–212. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Chow, K.; Wu, Y.; Lin, T.; Li, W. Inverse expression of dihydrodiol dehydrogenase and glutathione-S-transferase in patients with esophageal squamous cell carcinoma. Int. J. Cancer 2004, 111, 246–251. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, H.; Kim, H.; Seong, P.; Arora, D.; Shin, D.; Park, W.; Park, J.-E. Transcriptomic Response under Heat Stress in Chickens Revealed the Regulation of Genes and Alteration of Metabolism to Maintain Homeostasis. Animals 2021, 11, 2241. https://doi.org/10.3390/ani11082241
Kim H, Kim H, Seong P, Arora D, Shin D, Park W, Park J-E. Transcriptomic Response under Heat Stress in Chickens Revealed the Regulation of Genes and Alteration of Metabolism to Maintain Homeostasis. Animals. 2021; 11(8):2241. https://doi.org/10.3390/ani11082241
Chicago/Turabian StyleKim, Hana, Hyeran Kim, Pilnam Seong, Devender Arora, Donghyun Shin, Woncheoul Park, and Jong-Eun Park. 2021. "Transcriptomic Response under Heat Stress in Chickens Revealed the Regulation of Genes and Alteration of Metabolism to Maintain Homeostasis" Animals 11, no. 8: 2241. https://doi.org/10.3390/ani11082241
APA StyleKim, H., Kim, H., Seong, P., Arora, D., Shin, D., Park, W., & Park, J.-E. (2021). Transcriptomic Response under Heat Stress in Chickens Revealed the Regulation of Genes and Alteration of Metabolism to Maintain Homeostasis. Animals, 11(8), 2241. https://doi.org/10.3390/ani11082241





