A Microsatellite Genotyping-Based Genetic Study of Interspecific Hybridization between the Red and Sika Deer in the Western Czech Republic
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Individuals in the Study
2.2. DNA Extraction and Genotype Determination
2.3. Statistical Analyses of the Genetic Data
2.4. Ethical Approval
3. Results
3.1. Microsatellite Genetic Diversity and Population Differentiation
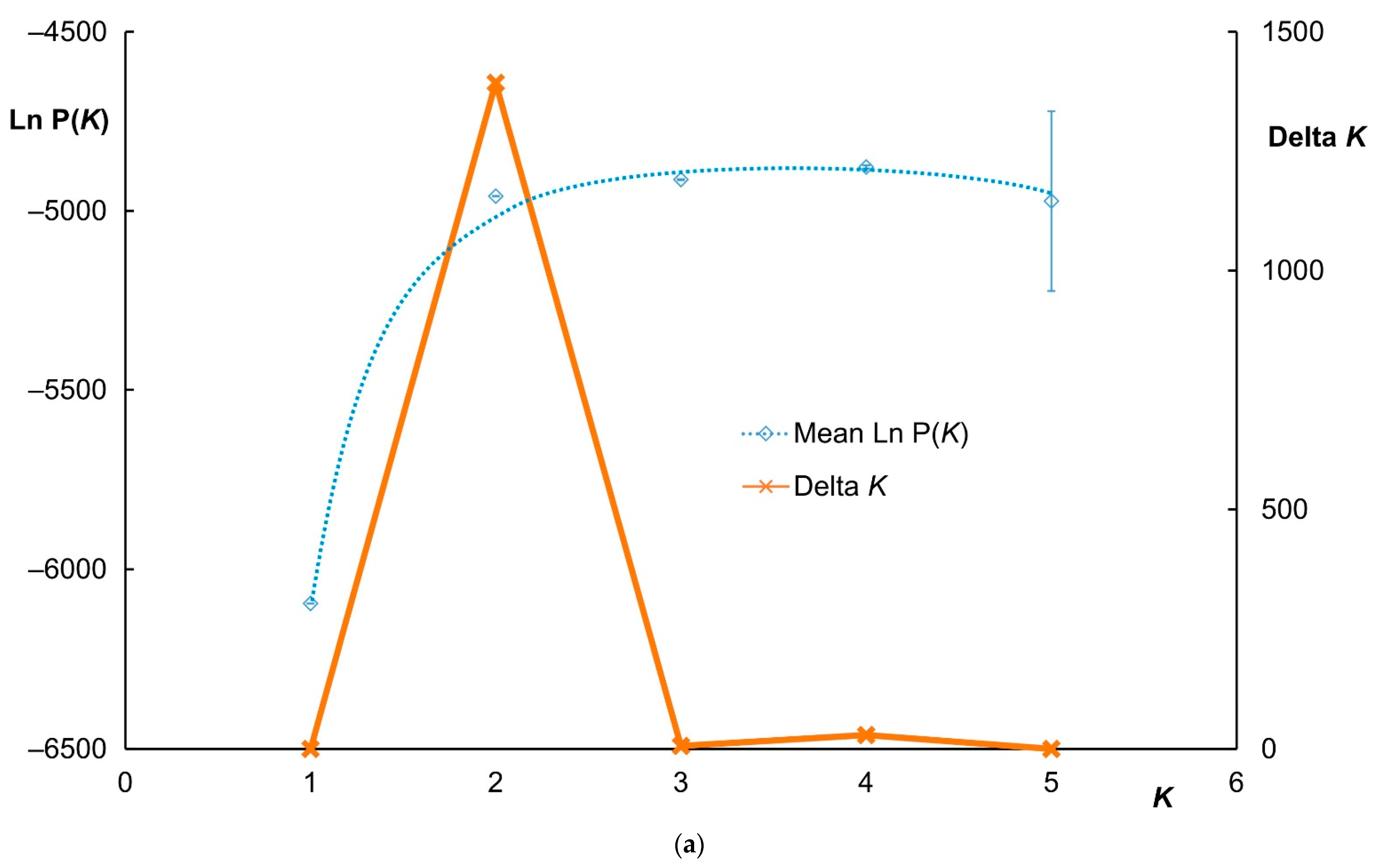
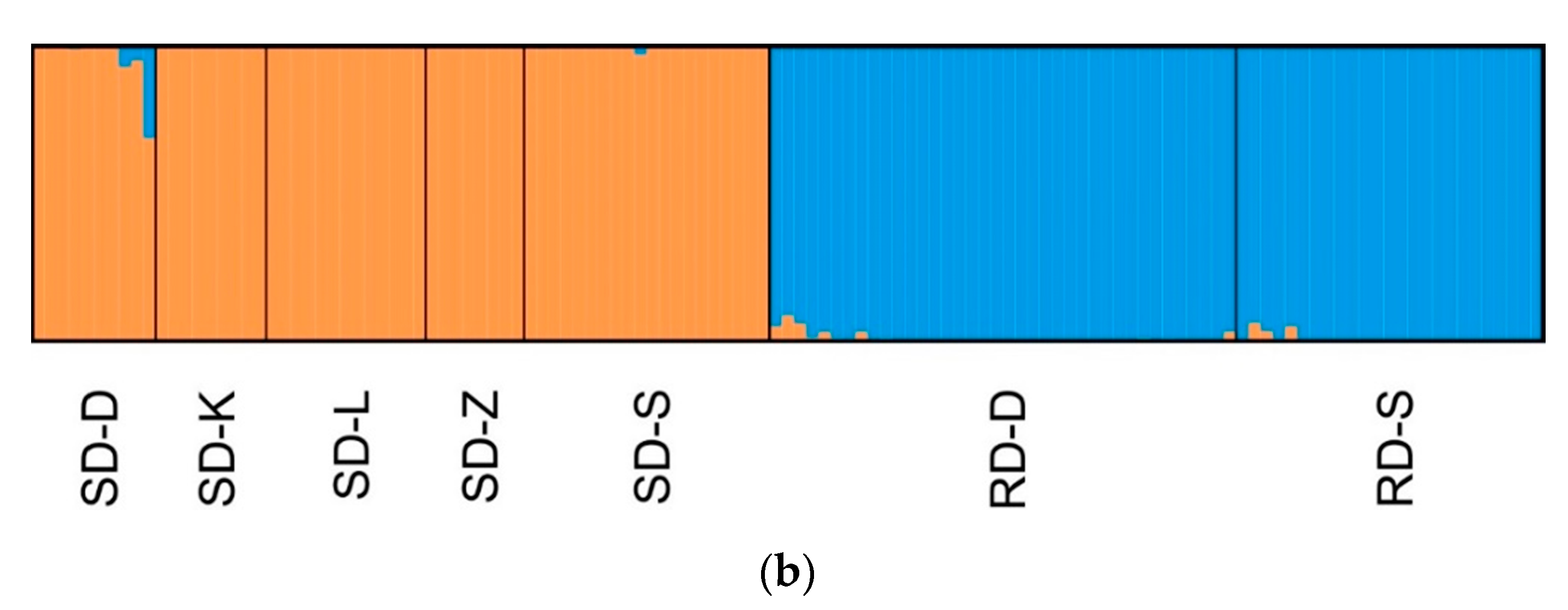
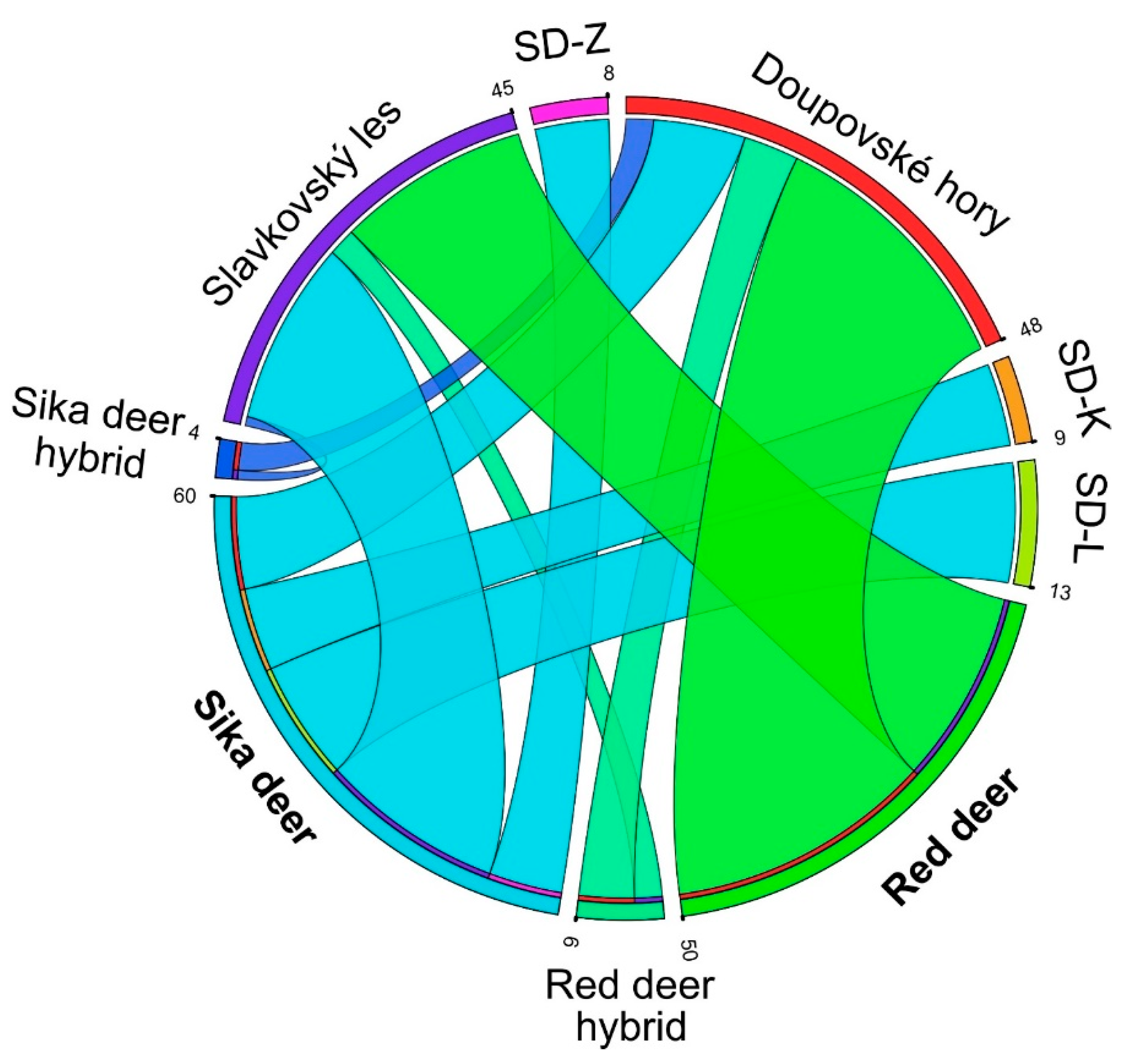
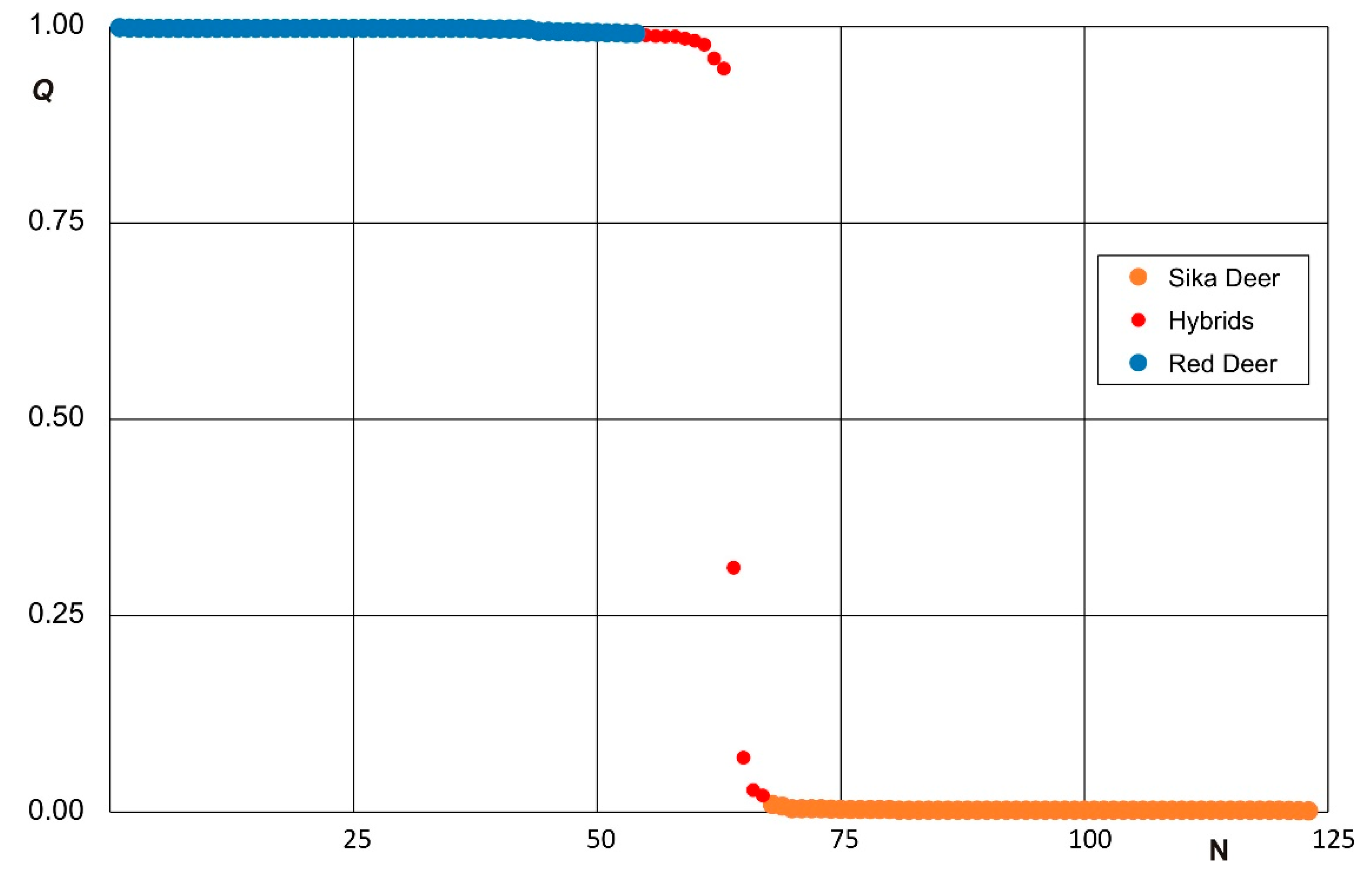
3.2. Clustering Analysis and Interspecific Hybridization
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Simberloff, D. Hybridization between native and introduced wildlife species: Importance for conservation. Wildl. Biol. 1996, 2, 143–150. [Google Scholar] [CrossRef]
- Huxel, G.R. Rapid displacement of native species by invasive species: Effects of hybridization. Biol. Cons. 1999, 89, 143–152. [Google Scholar] [CrossRef]
- Allendorf, F.W.; Leary, R.F.; Spruell, P.; Wenburg, J.K. The problems with hybrids: Setting conservation guidelines. Trends Ecol. Evol. 2001, 16, 613–622. [Google Scholar] [CrossRef]
- Harrington, R. Hybridization among deer and its implications for conservation. Ir. For. 1973, 30, 64–78. [Google Scholar]
- Fitzpatrick, B.M.; Ryan, M.E.; Johnson, J.R.; Corush, J. Hybridization and the species problem in conservation. Curr. Zool. 2015, 61, 206–216. [Google Scholar] [CrossRef]
- Santostasi, N.L.; Ciucci, P.; Bearzi, G.S.; Bonizzoni, S.; Gimenez, O. Assessing the dynamics of hybridization through a matrix modelling approach. Ecol. Modell. 2020, 431, 1–11. [Google Scholar] [CrossRef]
- Keller, R.P.; Geist, J.; Jeschke, J.M.; Kühn, I. Invasive species in Europe: Ecology, status, and policy. Environ. Sci. Eur. 2011, 23, 1–17. [Google Scholar] [CrossRef]
- Goodman, S.J.; Barton, N.H.; Swanson, G.; Abernethy, K.; Pemberton, J.K. Introgression through rare hybridization: A genetic study of a hybrid zone between red and sika deer (genus Cervus) in Argyll, Scotland. Genetics 1999, 152, 355–371. [Google Scholar] [CrossRef] [PubMed]
- Abbott, R.; Albach, D.; Ansell, S.; Arntzen, J.W.; Baird, J.E.; Bierne, N.; Boughman, J.; Brelsford, A.; Buerkle, C.A.; Buggs, R.; et al. Hybridization and speciation. J. Evol. Biol. 2013, 26, 229–246. [Google Scholar] [CrossRef]
- Zhang, L.; Hua, Y.; Wei, S. High Genetic Diversity of an Invasive Alien Species: Comparison between Fur-Farmed and Feral American Mink (Neovison vison) in China. Animals 2021, 11, 472. [Google Scholar] [CrossRef] [PubMed]
- Wolf, R. Jelen sika. In Husák, Wolf, Lochman. Daněk/Sika/Jelenec; SZN: Praha, Czech Republic, 1986; pp. 150–227. [Google Scholar]
- Bartoš, L. Sika deer in continental Europe. In Sika Deer: Biology and Management of Native and Introduced Populations; McCullough, D.R., Takatsuki, S., Kaji, K., Eds.; Springer: Tokyo, Japan, 2009; pp. 573–594. [Google Scholar] [CrossRef]
- Herzog, S.; Harrington, R. The role of hybridisaton in the karyotype evolution of deer (Cervidae, Artiodactyla, Mammalia). Theor. Appl. Genet. 1991, 82, 425–429. [Google Scholar] [CrossRef]
- Bartoš, L.; Vítek, M. Cluster analysis of red and sika deer phenotypes. In Deer of China: Biology and Management; Ohtaishi, N., Sheng, H.-I., Eds.; Elsevier: Amsterdam, The Netherlands, 1993; pp. 15–21. [Google Scholar]
- Senn, H.V.; Pemberton, J.M. Variable extent of hybridization between invasive sika (Cervus nippon) and native red deer (C. elaphus) in a small geographical area. Mol. Ecol. 2009, 18, 862–876. [Google Scholar] [CrossRef] [PubMed]
- Smith, S.L.; Senn, H.V.; Pérez-Espona, S.; Wyman, M.T.; Heap, E.; Pemberton, J.M. Introgression of exotic Cervus (nippon and canadensis) into red deer (Cervus elaphus) populations in Scotland and the English Lake District. Ecol. Evol. 2018, 8, 2122–2134. [Google Scholar] [CrossRef] [PubMed]
- McFarlane, S.E.; Hunter, D.C.; Senn, H.V.; Smith, S.L.; Holland, R.; Huisman, J.; Pemberton, J.M. Increased genetic marker density reveals high levels of admixture between red deer and introduced Japanese sika in Kintyre, Scotland. Evol. Appl. 2020, 13, 432–441. [Google Scholar] [CrossRef] [PubMed]
- Bartoš, L.; Žirovnický, J. Hybridization between red and sika deer, II. Phenotype analysis. Zool. Anz. 1981, 207, 271–287. [Google Scholar]
- Krojerová, J.; Barančeková, M.; Koubek, P. Jelen sika, jelen evropský nebo kříženec? Svět Mysliv. 2013, 1, 8–11. [Google Scholar]
- McDevitt, A.D.; Edwards, C.J.; O’Toole, P.; O’Sullivan, P.; O’Reilly, C.; Carden, R.F. Genetic structure of, and hybridisation between, red (Cervus elaphus) and sika (Cervus nippon) deer in Ireland. Mamm. Biol. 2009, 74, 263–273. [Google Scholar] [CrossRef]
- Biedrzycka, A.; Solarz, W.; Okarma, H. Hybridization between native and introduced species of deer in Eastern Europe. J. Mammal. 2012, 93, 1331–1341. [Google Scholar] [CrossRef]
- Krojerová-Prokešová, J.; Barančeková, M.; Koubek, P. Application of genetic methods for the detection of deer origin and existence of interspecific hybridization. In Proceedings of the Jelen Sika—Přehled Celorepublikových Poznatků, Dubina, Czech Republic, 16 May 2014; Česká Lesnická Společnost: Praha, Czech Republic, 2014; pp. 9–14. [Google Scholar]
- Smith, S.L.; Carden, R.F.; Coad, B.; Birkitt, T.; Pemberton, J.M. A survey of the hybridisation status of Cervus deer species on the island of Ireland. Conserv. Genet. 2014, 15, 823–835. [Google Scholar] [CrossRef][Green Version]
- Ražanskė, I.; Gibiežaitė, J.M.; Paulauskas, A. Genetic Analysis of Red Deer Cervus elaphus and Sika Deer Cervus nippon to Evaluate Possible Hybridisation in Lithuania. Balt. For. 2017, 23, 683–690. [Google Scholar]
- McFarlane, S.E.; Pemberton, J.M. Detecting the true extent of introgression during anthropogenic hybridization. Trends Ecol. Evol. 2019, 34, 315–326. [Google Scholar] [CrossRef] [PubMed]
- Browett, S.S.; O’Meara, D.B.; McDevitt, A.D. Genetic tools in the management of invasive mammals: Recent trends and future perspectives. Mamm. Rev. 2020, 50, 200–210. [Google Scholar] [CrossRef]
- Frank, K.; Bana, N.Á.; Bleier, N.; Sugár, L.; Nagy, J.; Wilhelm, J.; Kálmán, Z.; Barta, E.; Orosz, L.; Horn, P.; et al. Mining the red deer genome (CerEla1.0) to develop X-and Y-chromosome-linked STR markers. PLoS ONE 2020, 15, e0242506. [Google Scholar] [CrossRef] [PubMed]
- Herzog, S.; Gehle, T. Hybridization in the Genus Cervus: Evidence for Hybridization between Red and Sika Deer in Germany. Acad. J. Appl. Sci. Res. 2016, 1, 7–10. [Google Scholar]
- Šustr, P.; Lamka, J.; Rąpała, R.; Zendulková, D.; Tesa, K.; Ernst, M.; Robovský, J.; Svobodová, K.; Pohlová, L.; Široký, Z.; et al. Jeleni v Krkonoších/Jelenie w Karkonoszach; Správa KRNAP: Vrchlabí, Czech Republic; Dyrekcja KPN: Jelenia Góra, Poland, 2015; 200p. [Google Scholar]
- Cervus Elaphus (Red Deer)—Distribution Map|BioLib.cz. Available online: https://www.biolib.cz/en/taxonmap/id40/ (accessed on 17 March 2021).
- Basic Data on Hunting Grounds, Game Stock and Hunting—From 1 April 2019 to 31 March 2020|CZSO. Available online: https://www.czso.cz/csu/czso/basic-data-on-hunting-grounds-game-stock-and-hunting-from-1-april-2019-to-31-march-2020 (accessed on 17 March 2021).
- Kokeš, O. Asijští jeleni na území Československa. Ochr. Fauny 1970, 4, 158–161. [Google Scholar]
- Vavruněk, J.; Wolf, R. Chov jelení zvěře v Západočeském kraji. In Sborník Vědeckého Lesnického Ústavu VŠZ v Praze; VŠZ: Praha, Czech Republic, 1977; Volume 20. [Google Scholar]
- Babička, C.; Drábek, M.; Štika, J.; Ženožička, J. Poznatky z chovu jelena siky. Myslivost 1977, 11, 270–271. [Google Scholar]
- Křivánek, J. Jelen sika japonský—Plíživé nebezpečí genofondu jelena evropského. Myslivost 2010, 8, 12–15. [Google Scholar]
- Dvořák, J.; Čermák, P. Jelen sika a škody působené na lesních porostech ve vybraných honitbách Plzeňska. Lesn. Práce 2008, 87, 12–14. [Google Scholar]
- Ježek, M.; Holá, M.; Kušta, T.; Hart, V.; Červený, J. Reprodukční Charakteristiky Samic Jelena Siky: Výzkumné Projekty Grantové Služby LČR; Lesy České republiky: Praha, Czech Republic, 2016; p. 43. ISBN 978-80-86945-31-6. [Google Scholar]
- Wolf, R.; Vavruněk, J. Sika východní—Cervus nippon Temm. v západních Čechách. In Sborník Vědeckého Lesnického Ústavu VŠZ v Praze; VŠZ: Praha, Czech Republic, 1975; Volume 18–19, pp. 185–199. [Google Scholar]
- Červený, J.; Anděra, M. Vývoj populací spárkaté zvěře v ČR (II.)—Jelen lesní a sika. Svět Mysliv. 2012, 13, 8–11. [Google Scholar]
- Dvořák, J.; Palyzová, L. Analysis of the Development and Spatial Distribution of Sika Deer (Cervus Nippon) Populations on the Territory of the Czech Republic. Acta Univ. Agric. Silvic. Mendel. Brun. 2016, 64, 1507–1515. [Google Scholar] [CrossRef]
- Ježek, M.; Kušta, T.; Trombik, J. Soubor Map: Odstřel Jelena Siky (Cervus Nippon) v Letech 2006 a 2016 na Území České Republiky; Czech University of Life Sciences Prague: Prague, Czech Republic, 2017; p. 10. ISBN 978-80-213-2824-2. [Google Scholar]
- Barančeková, M.; Krojerová-Prokešová, J.; Voloshina, I.V.; Myslenkov, A.I.; Kawata, Y.; Oshida, T.; Lamka, J.; Koubek, P. The origin and genetic variability of the Czech sika deer population. Ecol. Res. 2012, 27, 991–1003. [Google Scholar] [CrossRef]
- Cervus Nippon (Sika Deer)—Distribution Map|BioLib.cz. Available online: https://www.biolib.cz/en/taxonmap/id41/ (accessed on 17 March 2021).
- Dvořák, J.; Kamler, J. Jelen Sika—Problematika Chovu: Komplexní Řešení Problematiky Chovu Jelena Siky Včetně Škod Působených Touto Zvěří v Plzeňském Regionu a Navazující Části Karlovarského Regionu; Lesy České Republiky: Hradec Králové, Czech Republic, 2009; p. 65. ISBN 978-80-86945-08-8. [Google Scholar]
- Cukor, J.; Vacek, Z.; Linda, R.; Vacek, S.; Marada, P.; Šimůnek, V.; Havránek, F. Effects of Bark Stripping on Timber Production and Structure of Norway Spruce Forests in Relation to Climatic Factors. Forests 2019, 10, 320. [Google Scholar] [CrossRef]
- Bartoš, L.; Hyánek, J.; Žirovnický, J. Hybridization between red and sika deer (Cervus nippon) in Czechoslovakia. Folia Zool. Brno 1981, 31, 195–208. [Google Scholar]
- Bartoš, L.; Žirovnický, J. Hybridization between red and sika deer, III. Interspecific behaviour. Zool. Anz. 1982, 208, 30–36. [Google Scholar]
- Powerscourt, V. On the acclimatization of the Japanese deer at Powerscourt. In Proceedings of the Zoological Society of London, London, UK, 1 March 1884; pp. 207–209. [Google Scholar] [CrossRef]
- Bishop, M.D.; Kappes, S.M.; Keele, J.W.; Stone, R.T.; Sunden, S.L.; Hawkins, G.A.; Toldo, S.S.; Fries, R.; Grosz, M.D.; Yoo, J.; et al. A genetic linkage map for cattle. Genetics 1994, 136, 619–639. [Google Scholar] [CrossRef] [PubMed]
- Barendse, W.; Armitage, S.M.; Kossarek, L.M.; Shalom, A.; Kirkpatrick, B.W.; Ryan, A.M.; Clayton, D.; Li, L.; Neibergs, H.L.; Zhang, N.; et al. A genetic linkage map of the bovine genome. Nat. Genet. 1994, 6, 227–235. [Google Scholar] [CrossRef]
- Steffen, P.; Eggen, A.; Dietz, A.B.; Womack, J.E.; Stranzinger, G.; Fies, R. Isolation and mapping of polymorphic microsatellites in cattle. Anim. Genet. 1993, 24, 121–124. [Google Scholar] [CrossRef] [PubMed]
- Buchanan, F.C.; Galloway, S.M.; Crawford, A.M. Ovine microsatellites at the OarFCB5, OarFCB19, OarFCB20, OarFCB48, OarFCB129 and OarFCB226 loci. Anim. Genet. 1994, 25, 60. [Google Scholar] [CrossRef]
- Wilson, G.A.; Strobeck, C.; Wu, L.; Coffin, J.W. Characterization of microsatellite loci in caribou Rangifer tarandus, and their use in other artiodactyls. Mol. Ecol. 1997, 6, 697–699. [Google Scholar] [CrossRef]
- Jones, K.C.; Levine, K.F.; Banks, J.D. Characterization of 11 polymorphic tetranucleotide microsatellites for forensic applications in California elk (Cervus elaphus canadensis). Mol. Ecol. Resour. 2002, 2, 425–427. [Google Scholar] [CrossRef]
- Liu, K.; Muse, S.V. PowerMarker: Integrated analysis environment for genetic marker data. Bioinformatics 2005, 21, 2128–2129. [Google Scholar] [CrossRef]
- FSTAT. Available online: http://www2.unil.ch/popgen/softwares/fstat.htm (accessed on 17 March 2021).
- Rousset, F. Genepop’007: A complete reimplementation of the Genepop software for Windows and Linux. Mol. Ecol. Resour. 2008, 8, 103–106. [Google Scholar] [CrossRef]
- Nei, M.; Tajima, F.; Tateno, Y. Accuracy of estimated phylogenetic trees from molecular data. J. Mol. Evol. 1983, 19, 153–170. [Google Scholar] [CrossRef]
- Reynolds, J.; Weir, B.S.; Cockerham, C.C. Estimation of the Coancestry coefficient: Basic for a short-term genetic distance. Genetics 1983, 105, 767–779. [Google Scholar] [CrossRef]
- Huson, D.H.; Bryant, D. Application of Phylogenetic Networks in Evolutionary Studies. Mol. Biol. Evol. 2006, 23, 254–267. [Google Scholar] [CrossRef] [PubMed]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef]
- Earl, D.A.; von Holdt, B.M. STRUCTURE HARVESTER: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 2012, 4, 359–361. [Google Scholar] [CrossRef]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef] [PubMed]
- Jakobsson, M.; Rosenberg, N.A. CLUMPP: A cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 2007, 23, 1801–1806. [Google Scholar] [CrossRef]
- Rosenberg, N.A. Distruct: A program for the graphical display of population structure. Mol. Ecol. Notes 2004, 4, 137–138. [Google Scholar] [CrossRef]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef] [PubMed]
- Slate, J.; Coltman, D.W.; Goodman, S.J.; MacLean, I.; Pemberton, J.M.; Williams, J.L. Bovine microsatellite loci are highly conserved in red deer (Cervus elaphus), sika deer (Cervus nippon) and Soay sheep (Ovis aries). Anim. Genet. 1998, 29, 307–315. [Google Scholar] [CrossRef] [PubMed]
- Krojerová-Prokešová, J.; Barančeková, M.; Kawata, Y.; Oshida, T.; Igota, H.; Koubek, P. Genetic differentiation between introduced Central European sika and source populations in Japan: Effects of isolation and demographic events. Biol. Invasions 2017, 19, 2125–2141. [Google Scholar] [CrossRef]
- Ziegrosser, P. Sika—Nepůvodní—Invazivní—Druh. Myslivost 2017, 12, 18. [Google Scholar]
- Pipek, P. Seek Sika. Invasion of Sika Deer in the Czech Republic and Europe. Živa 2018, 5, 280–281. [Google Scholar]
- Beysard, M.; Heckel, G. Structure and dynamics of hybrid zones at different stages of speciation in the common vole (Microtus arvalis). Mol. Ecol. 2014, 23, 673–687. [Google Scholar] [CrossRef] [PubMed]
- Macháček, Z.; Dvořák, S.; Ježek, M.; Zahradník, D. Impact of interspecific relations between native red deer (Cervus elaphus) and introduced sika deer (Cervus nippon) on their rutting season in the Doupovské hory Mts. J. For. Sci. 2014, 60, 272–280. [Google Scholar] [CrossRef]
- Wyman, M.; Locatelli, Y.; Charlton, B.; Reby, D. Female Sexual Preferences Toward Conspecific and Hybrid Male Mating Calls in Two Species of Polygynous Deer, Cervus elaphus and C. nippon. Evol. Biol. 2016, 43, 227–241. [Google Scholar] [CrossRef]
- Li, Z.; Wright, A.G.; Si, H.; Wang, X.; Qian, W.; Zhang, Z.; Li, G. Changes in the rumen microbiome and metabolites reveal the effect of host genetics on hybrid crosses. Environ. Microbiol. Rep. 2016, 8, 1016–1023. [Google Scholar] [CrossRef] [PubMed]
- Vähä, J.P.; Primmer, C.R. Efficiency of model-based Bayesian methods for detecting hybrid individuals under different hybridization scenarios and with different numbers of loci. Mol. Ecol. 2006, 15, 63–72. [Google Scholar] [CrossRef] [PubMed]


| Marker | MAF | Genotype No. | No. of Observations | Allele No. | Availability | HE | HO | PIC | F |
|---|---|---|---|---|---|---|---|---|---|
| OarFCB5 | 0.4024 | 34 | 123 | 13 | 1.0000 | 0.7781 | 0.6992 | 0.7561 | 0.1055 |
| T156 | 0.2125 | 55 | 120 | 17 | 0.9756 | 0.8996 | 0.8167 | 0.8920 | 0.0964 |
| T26 | 0.1595 | 48 | 116 | 16 | 0.9431 | 0.9052 | 0.7586 | 0.8975 | 0.1661 |
| BM888 | 0.2500 | 50 | 120 | 22 | 0.9756 | 0.8739 | 0.6917 | 0.8628 | 0.2125 |
| RM188 | 0.2479 | 31 | 117 | 17 | 0.9512 | 0.8650 | 0.5812 | 0.8521 | 0.3319 |
| RT1 | 0.2764 | 31 | 123 | 13 | 1.0000 | 0.8211 | 0.6423 | 0.7998 | 0.2217 |
| T501 | 0.3696 | 34 | 115 | 14 | 0.9350 | 0.8050 | 0.6000 | 0.7868 | 0.2587 |
| RT13 | 0.2059 | 43 | 119 | 17 | 0.9675 | 0.8737 | 0.5966 | 0.8612 | 0.3209 |
| T193 | 0.2292 | 49 | 120 | 22 | 0.9756 | 0.9034 | 0.8417 | 0.8971 | 0.0725 |
| BM1818 | 0.3824 | 30 | 119 | 10 | 0.9675 | 0.7943 | 0.6471 | 0.7745 | 0.1894 |
| ETH225 | 0.2967 | 39 | 123 | 15 | 1.0000 | 0.8162 | 0.6585 | 0.7941 | 0.1971 |
| Mean/Overall | 0.2757 | 40.3636 | 119.5455 | 16 | 0.9719 | 0.8487 | 0.6849 | 0.8340 | 0.1971 |
| Population | Code | MAF | Genotype No. | n | No. of Observations | Allele No. | Availability | HE | HO | PIC | F |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Red deer Doupovské hory | RD-D | 0.2897 | 22.2727 | 38 | 36.8182 | 10.9091 | 0.9689 | 0.8276 | 0.7454 | 0.8096 | 0.1129 |
| Red deer Slavkovský les | RD-S | 0.2866 | 16.9091 | 25 | 24.1818 | 10.3636 | 0.9673 | 0.8242 | 0.7078 | 0.8057 | 0.1619 |
| Sika deer Doupovské hory | SD-D | 0.4742 | 6.3636 | 10 | 9.7273 | 5.7273 | 0.9727 | 0.6621 | 0.6364 | 0.6240 | 0.0930 |
| Sika deer Slavkovský les | SD-S | 0.5323 | 7.3636 | 20 | 19.2727 | 5.2727 | 0.9636 | 0.6088 | 0.5564 | 0.5612 | 0.1126 |
| Sika deer Kladská | SD-K | 0.4380 | 5.1818 | 9 | 8.8182 | 4.3636 | 0.9798 | 0.6806 | 0.8398 | 0.6254 | −0.1766 |
| Sika deer Lány | SD-L | 0.5584 | 5.7273 | 13 | 12.7273 | 4.2727 | 0.9790 | 0.5850 | 0.5757 | 0.5345 | 0.0574 |
| Sika deer Žlutice | SD-Z | 0.4659 | 4.9091 | 8 | 8.0000 | 4.2727 | 1.0000 | 0.6584 | 0.7159 | 0.6052 | −0.0208 |
| Marker | AR | HE | HO | PIC | FNull | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Red Deer | Sika Deer | Red Deer | Sika Deer | Red Deer | Sika Deer | Red Deer | Sika Deer | Red Deer | Sika Deer | |
| OarFCB5 | 11.690 | 6.982 | 0.8644 | 0.5986 | 0.8254 | 0.5667 | 0.8416 | 0.5658 | 0.0300 | 0.0357 |
| T156 | 12.484 | 9.931 | 0.8846 | 0.8352 | 0.8065 | 0.8276 | 0.8650 | 0.8107 | 0.0493 | 0.0000 |
| T26 | 12.789 | 10.724 | 0.8921 | 0.8505 | 0.8103 | 0.7069 | 0.8732 | 0.8240 | 0.0412 | 0.0832 |
| BM888 | 16.653 | 6.991 | 0.8417 | 0.6915 | 0.7333 | 0.6500 | 0.8182 | 0.6514 | 0.0767 | 0.0120 |
| RM188 | 9.675 | 8.000 | 0.7873 | 0.6731 | 0.5079 | 0.6667 | 0.7508 | 0.6397 | 0.1624 | 0.0179 |
| RT1 | 11.390 | 5.899 | 0.6989 | 0.6423 | 0.6984 | 0.5833 | 0.6668 | 0.5980 | 0.0266 | 0.0474 |
| T501 | 10.000 | 9.492 | 0.8111 | 0.5363 | 0.6607 | 0.5424 | 0.7859 | 0.5038 | 0.1126 | 0.0000 |
| RT13 | 15.721 | 6.931 | 0.8824 | 0.7652 | 0.5410 | 0.6552 | 0.8637 | 0.7242 | 0.1917 | 0.0630 |
| T193 | 14.771 | 6.900 | 0.9090 | 0.7182 | 0.9000 | 0.7833 | 0.8929 | 0.6767 | 0.0000 | 0.0000 |
| BM1818 | 8.000 | 4.000 | 0.8291 | 0.6628 | 0.7742 | 0.5088 | 0.8037 | 0.5933 | 0.0086 | 0.1740 |
| ETH225 | 13.810 | 4.791 | 0.8555 | 0.5167 | 0.7778 | 0.5333 | 0.8337 | 0.4120 | 0.0626 | 0.0000 |
| Mean/Overall | 12.453 | 7.331 | 0.8415 | 0.6809 | 0.7305 | 0.6386 | 0.8178 | 0.6363 | 0.0692 | 0.0394 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Štohlová Putnová, L.; Štohl, R.; Ernst, M.; Svobodová, K. A Microsatellite Genotyping-Based Genetic Study of Interspecific Hybridization between the Red and Sika Deer in the Western Czech Republic. Animals 2021, 11, 1701. https://doi.org/10.3390/ani11061701
Štohlová Putnová L, Štohl R, Ernst M, Svobodová K. A Microsatellite Genotyping-Based Genetic Study of Interspecific Hybridization between the Red and Sika Deer in the Western Czech Republic. Animals. 2021; 11(6):1701. https://doi.org/10.3390/ani11061701
Chicago/Turabian StyleŠtohlová Putnová, Lenka, Radek Štohl, Martin Ernst, and Kateřina Svobodová. 2021. "A Microsatellite Genotyping-Based Genetic Study of Interspecific Hybridization between the Red and Sika Deer in the Western Czech Republic" Animals 11, no. 6: 1701. https://doi.org/10.3390/ani11061701
APA StyleŠtohlová Putnová, L., Štohl, R., Ernst, M., & Svobodová, K. (2021). A Microsatellite Genotyping-Based Genetic Study of Interspecific Hybridization between the Red and Sika Deer in the Western Czech Republic. Animals, 11(6), 1701. https://doi.org/10.3390/ani11061701






