Fertility-Associated Polymorphism within Bovine ITGβ5 and Its Significant Correlations with Ovarian and Luteal Traits
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Collection of Ovarian Tissue Samples from Cows
2.2. Extraction of Total DNA
2.3. Selection of Indel Sites and Design of Primers
2.4. Identification of Indel Loci, Genotyping and DNA Sequencing
2.5. Statistical Analyses
3. Results
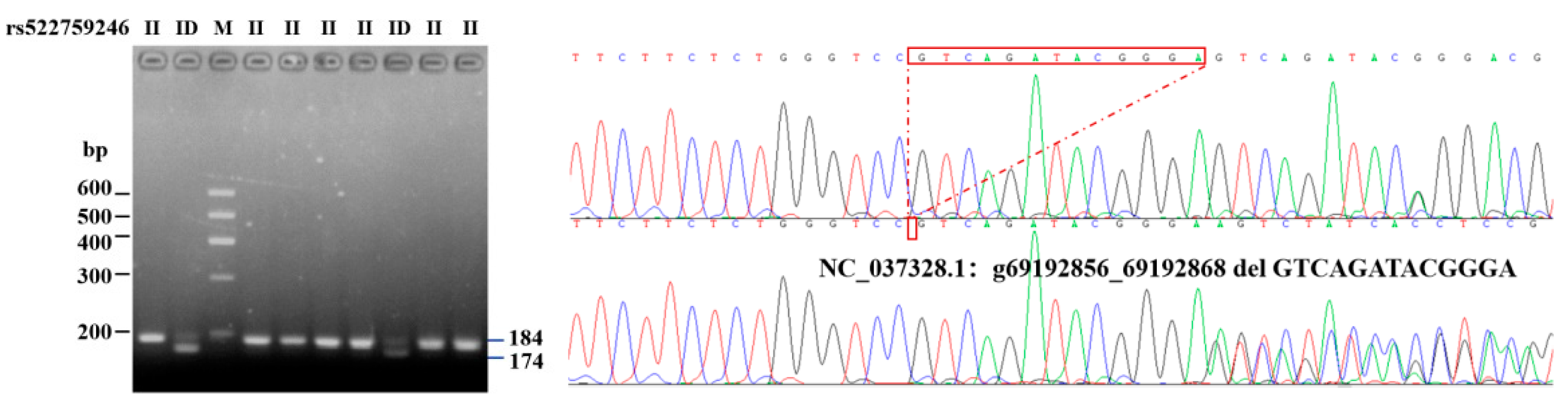
3.1. Identification of 13-bp Indel Polymorphism of the Bovine ITGβ5 Gene
3.2. Genetic Diversity of the Novel 13-bp Indel Locus within ITGβ5 in the Population
3.3. Association Analysis between 13-bp Indel of ITGβ5 and Phenotypic Characteristics of the Bovine Ovary
3.4. Relevance Analysis of the 13-bp Indel of ITGβ5 with Corpus Luteum Traits, Mature Follicle Follicles Traits and Corpus Albican Traits of the Bovine Ovary
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cai, Z.; Guldbrandtsen, B.; Lund, M.S.; Sahana, G. Prioritizing candidate genes for fertility in dairy cows using gene-based analysis, functional annotation and differential gene expression. BMC Genom. 2019, 20, 255. [Google Scholar] [CrossRef] [PubMed]
- Lucy, M.C. Reproductive loss in high-producing dairy cattle: Where will it end? J. Dairy Sci. 2001, 84, 1277–1293. [Google Scholar] [CrossRef]
- Royal, M.D.; Darwash, A.O.; Flint, A.P.E.; Webb, R.; Woolliams, J.A.; Lamming, G.E. Declining fertility in dairy cattle, changes in traditional and endocrine parameters of fertility. Anim. Sci. 2000, 70, 487–501. [Google Scholar] [CrossRef]
- Snelling, W.M.; Cushman, R.A.; Keele, J.W.; Maltecca, C.; Thomas, M.G.; Fortes, M.R.; Reverter, A. Breeding and Genetics Symposium, networks and pathways to guide genomic selection. J. Anim. Sci. 2013, 91, 537–552. [Google Scholar] [CrossRef] [PubMed]
- Lande, R.; Thompson, R. Efficiency of marker-assisted selection in the improvement of quantitative traits. Genetics 1990, 124, 743–756. [Google Scholar] [CrossRef] [PubMed]
- Schulman, N.F.; Sahana, G.; Lund, M.S.; Viitala, S.M.; Vilkki, J.H. Quantitative trait loci for fertility traits in Finnish Ayrshire cattle. Genet. Sel. Evol. 2008, 40, 195–214. [Google Scholar]
- Guo, X.; Pei, J.; Wu, X.; Bao, P.; Ding, X.; Xiong, L.; Chu, M.; Lan, X.; Yan, P. Detection of InDel and CNV of SPAG17 gene and their associations with bovine growth traits. Anim. Biotechnol. 2020, 21, 1–8. [Google Scholar] [CrossRef]
- Olsen, H.G.; Hayes, B.J.; Kent, M.P.; Nome, T.; Svendsen, M.; Larsgard, A.G.; Lien, S. Genome-wide association mapping in Norwegian Red cattle identifies quantitative trait loci for fertility and milk production on BTA12. Anim. Genet. 2011, 42, 466–474. [Google Scholar] [CrossRef]
- Höglund, J.K.; Buitenhuis, B.; Guldbrandtsen, B.; Lund, M.S.; Sahana, G. Genome-wide association study for female fertility in Nordic Red cattle. BMC Genet. 2015, 16, 110. [Google Scholar] [CrossRef] [PubMed]
- Moore, S.G.; Pryce, J.E.; Hayes, B.J.; Chamberlain, A.J.; Kemper, K.E.; Berry, D.P.; McCabe, M.; Cormican, P.; Lonergan, P.; Fair, T.; et al. Differentially expressed genes in endometrium and corpus luteum of holstein cows selected for high and low fertility are enriched for sequence variants associated with fertility. Biol. Reprod. 2016, 94, 19. [Google Scholar] [CrossRef]
- Akanno, E.C.; Plastow, G.; Fitzsimmons, C.; Miller, S.P.; Baron, V.; Ominski, K.; Basarab, J.A. Genome-wide association for heifer reproduction and calf performance traits in beef cattle. Genome 2015, 58, 549–557. [Google Scholar] [CrossRef] [PubMed]
- Bi, Y.; Zhang, S.; Li, J.; He, L.; Kang, Y.; Chen, H.; Lan, X.; Pan, C. The mRNA expression profile of the goat prion protein testis-specific (PRNT) gene and its associations with litter size. Theriogenology 2021, 165, 69–75. [Google Scholar] [CrossRef] [PubMed]
- Shaw, L.M. Integrin function in breast carcinoma progression. J. Mammary Gland Biol. Neoplasia 1999, 4, 367–376. [Google Scholar] [CrossRef] [PubMed]
- Taddei, I.; Faraldo, M.M.; Teulière, J.; Deugnier, M.A.; Thiery, J.P.; Glukhova, M.A. Integrins in mammary gland development and differentiation of mammary epithelium. J. Mammary Gland Biol. Neoplasia 2003, 8, 383–394. [Google Scholar] [CrossRef] [PubMed]
- Herrera, L.; Ottolenghi, C.; Garcia-Ortiz, J.E.; Pellegrini, M.; Manini, F.; Ko, M.S.; Nagaraja, R.; Forabosco, A.; Schlessinger, D. Mouse ovary developmental RNA and protein markers from gene expression profiling. Dev. Biol. 2005, 279, 271–290. [Google Scholar] [CrossRef]
- Liu, Z.; Youngquist, R.S.; Garverick, H.A.; Antoniou, E. Molecular mechanisms regulating bovine ovarian follicular selection. Mol. Reprod. Dev. 2009, 76, 351–366. [Google Scholar] [CrossRef] [PubMed]
- Hatzirodos, N.; Irving-Rodgers, H.F.; Hummitzsch, K.; Harland, M.L.; Morris, S.E.; Rodgers, R.J. Transcriptome profiling of granulosa cells of bovine ovarian follicles during growth from small to large antral sizes. BMC Genom. 2014, 15, 24. [Google Scholar] [CrossRef]
- Honkatukia, M.; Tuiskula-Haavisto, M.; Arango, J.; Tabell, J.; Schmutz, M.; Preisinger, R.; Vilkki, J. QTL mapping of egg albumen quality in egg layers. Genet. Sel. Evol. 2013, 45, 31. [Google Scholar] [CrossRef]
- Sun, Y.; Wu, Q.; Pan, J.; Li, T.; Liu, L.; Chen, D.; Zhang, X.; Chen, H.; Li, Y.; Lin, R. Identification of differentially expressed genes and signalling pathways in the ovary of higher and lower laying ducks. Br. Poult. Sci. 2020, 61, 609–614. [Google Scholar] [CrossRef]
- Rimon-Dahari, N.; Yerushalmi-Heinemann, L.; Alyagor, L.; Dekel, N. Ovarian Folliculogenesis. Results Probl. Cell Differ. 2016, 58, 167–190. [Google Scholar]
- Edson, M.A.; Nagaraja, A.K.; Matzuk, M.M. The mammalian ovary from genesis to revelation. Endocr. Rev. 2009, 30, 624–712. [Google Scholar] [CrossRef]
- Stocco, C.; Telleria, C.; Gibori, G. The molecular control of corpus luteum formation, function, and regression. Endocr. Rev. 2007, 28, 117–149. [Google Scholar] [CrossRef]
- Li, J.; Shen, C.; Zhang, K.; Niu, Z.; Liu, Z.; Zhang, S.; Wang, Y.; Lan, X. Polymorphic variants of bovine ADCY5 gene identified in GWAS analysis were significantly associated with ovarian morphological related traits. Gene 2021, 766, 145158. [Google Scholar] [CrossRef]
- Li, J.; Zhang, S.; Shen, C.; Niu, Z.; Yang, H.; Zhang, K.; Liu, Z.; Wang, Y.; Lan, X. Indel mutations within the bovine HSD17B3 gene are significantly associated with ovary morphological traits and mature follicle number. J. Steroid Biochem. Mol. Biol. 2021, 209, 105833. [Google Scholar] [CrossRef] [PubMed]
- Xia, X.; Qu, K.; Zhang, G.; Jia, Y.; Ma, Z.; Zhao, X.; Huang, Y.; Chen, H.; Huang, B.; Lei, C. Comprehensive analysis of the mitochondrial DNA diversity in Chinese cattle. Anim. Genet. 2019, 50, 70–73. [Google Scholar] [CrossRef]
- Aljanabi, S.M.; Martinez, I. Universal and rapid salt-extraction of high quality genomic DNA for PCR-based techniques. Nucleic Acids Res. 1997, 25, 4692–4693. [Google Scholar] [CrossRef] [PubMed]
- Botstein, D.; White, R.L.; Skolnick, M.; Davis, R.W. Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am. J. Hum. Genet. 1980, 32, 314–331. [Google Scholar] [PubMed]
- Kouamo, J.; Dawaye, S.M.; Zoli, A.P.; Bah, G.S. Evaluation of bovine (Bos indicus) ovarian potential for in vitro embryo production in the Adamawa plateau (Cameroon). Open Vet. J. 2014, 4, 128–136. [Google Scholar] [PubMed]
- Strobel, T.; Cannistra, S.A. Beta1-integrins partly mediate binding of ovarian cancer cells to peritoneal mesothelium in vitro. Gynecol. Oncol. 1999, 73, 362–367. [Google Scholar] [CrossRef] [PubMed]
- Cannistra, S.A.; Kansas, G.S.; Niloff, J.; DeFranzo, B.; Kim, Y.; Ottensmeier, C. Binding of ovarian cancer cells to peritoneal mesothelium in vitro is partly mediated by CD44H. Cancer Res. 1993, 53, 3830–3838. [Google Scholar]
- Gillan, L.; Matei, D.; Fishman, D.A.; Gerbin, C.S.; Karlan, B.Y.; Chang, D.D. Periostin secreted by epithelial ovarian carcinoma is a ligand for alpha(V)beta(3) and alpha(V)beta(5) integrins and promotes cell motility. Cancer Res. 2002, 62, 5358–5364. [Google Scholar] [PubMed]
- Velho, A.; Wang, H.; Koenig, L.; Grant, K.E.; Menezes, E.S.; Kaya, A.; Moura, A.; Memili, E. Expression dynamics of Integrin Subunit Beta 5 in bovine gametes and embryos imply functions in male fertility and early embryonic development. Andrologia 2019, 51, e13305. [Google Scholar] [CrossRef] [PubMed]
- Sueoka, K.; Shiokawa, S.; Miyazaki, T.; Kuji, N.; Tanaka, M.; Yoshimura, Y. Integrins and reproductive physiology, expression and modulation in fertilization, embryogenesis, and implantation. Fertil. Steril. 1997, 67, 799–811. [Google Scholar] [CrossRef]
- Brendle, A.; Lei, H.; Brandt, A.; Johansson, R.; Enquist, K.; Henriksson, R.; Hemminki, K.; Lenner, P.; Försti, A. Polymorphisms in predicted microRNA-binding sites in integrin genes and breast cancer: ITGB4 as prognostic marker. Carcinogenesis 2008, 29, 1394–1399. [Google Scholar] [CrossRef]
- Dalmay, T.; Edwards, D.R. MicroRNAs and the hallmarks of cancer. Oncogene 2006, 25, 6170–6175. [Google Scholar] [CrossRef]
- Brendle, A.; Slack, F.J. Oncomirs-microRNAs with a role in cancer. Nat. Rev. Cancer 2006, 6, 259–269. [Google Scholar]
| Locus | Rs Number | Primer Sequences (5′–3′) | Product Size (bp) | Tm (°C) | Region |
|---|---|---|---|---|---|
| P1-D13-bp | rs522759246 | F1: GTTCCTGCTCAAGTCTCGGG R1: CTTCCACTCTCACCCCCAAC | 187/174 | 60.39 | Downstream 4000 bp |
| P2-D12-bp | rs450642714 | F2: TGACTGCCTGCCAAATGTCC R2: TCTGCTCTCCCTCCTCCTTT | 247/235 | 60.90 | Downstream 4000 bp |
| P3-D10-bp | rs135754430 | F3: CTGCTCAAGTCTCGGGGATT R3: TCTTCCACTCTCACCCCCAA | 184/174 | 60.10 | Downstream 4000 bp |
| P4-D10-bp | rs797103610 | F4: CTGTCTCCCCTTCCACACAC R4: TCTCCTAGAAGGTCACCGCA | 151/141 | 59.96 | Downstream 2000 bp |
| P5-D9-bp | rs468306533 | F5: TTTCTCCCGTGCGTGTATGT R5: CCCCTCTGATCTCCCCATCT | 247/238 | 59.81 | Downstream 1000 bp |
| P6-D9-bp | rs136097870 | F6: CAGCACACAGAGGCAACAAC R6: AATCACCGCCAGCTTTGAAC | 239/230 | 59.97 | Downstream 1000 bp |
| Sizes | Genotypic Frequencies | Allelic Frequencies | HWE p Values | Population Parameters | |||||
| II | ID | DD | I | D | 1.2 × 10−23 | He | Ne | PIC | |
| 696 | 0.768 | 0.160 | 0.072 | 0.848 | 0.152 | 0.257 | 1.346 | 0.224 | |
| Sizes | Traits (Units) | Observed Genotypes (Mean ± SE) | p Values | ||
|---|---|---|---|---|---|
| II (n) | ID (n) | DD (n) | |||
| 611 | Ovarian length (mm) | 42.25 ± 0.41 (462) | 41.78 ± 0.93 (106) | 40.40 ± 1.47 (43) | 0.409 |
| 611 | Ovarian width (mm) | 23.49 b ± 0.35 (462) | 25.75 a ± 0.65 (106) | 24.60 ab ± 1.13 (43) | 0.015 |
| 610 | Ovarian height (mm) | 25.44 ± 0.35 (461) | 24.95 ± 0.68 (106) | 26.84 ± 1.05 (43) | 0.368 |
| 611 | Ovarian weight (g) | 11.49 ± 0.24 (463) | 11.06 ± 0.52 (105) | 10.45 ± 0.73 (43) | 0.371 |
| Sizes | Traits (Units) | Observed Genotypes (Mean ± SE) | p Values | ||
|---|---|---|---|---|---|
| II (n) | ID (n) | DD (n) | |||
| 528 | Corpus luteum diameter (mm) | 13.82 a ± 0.44 (400) | 10.60 b ± 0.78 (95) | 13.33 ab ± 1.55 (33) | 0.005 |
| 531 | Number of corpora lutea | 1.55 ± 0.48 (402) | 1.81 ± 0.13 (95) | 1.68 ± 0.20 (34) | 0.075 |
| 157 | Number of mature follicles | 1.26 ± 0.05 (105) | 1.35 ± 0.11 (37) | 1.27 ± 0.12 (15) | 0.672 |
| 156 | Mature follicle diameter (mm) | 12.27 ± 0.44 (105) | 12.43 ± 0.79 (36) | 13.90 ± 1.07 (15) | 0.426 |
| 123 | Number of corpora albicantia | 1.55 ± 0.12 (92) | 1.22 ± 0.88 (23) | 1.75 ± 0.49 (8) | 0.315 |
| 122 | Corpus albican diameter (mm) | 4.76 ± 0.36 (91) | 4.70 ± 0.68 (23) | 5.91 ± 1.71 (8) | 0.653 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, J.; Li, J.; Jiang, F.; Song, E.; Lan, X.; Zhao, H. Fertility-Associated Polymorphism within Bovine ITGβ5 and Its Significant Correlations with Ovarian and Luteal Traits. Animals 2021, 11, 1579. https://doi.org/10.3390/ani11061579
Zhao J, Li J, Jiang F, Song E, Lan X, Zhao H. Fertility-Associated Polymorphism within Bovine ITGβ5 and Its Significant Correlations with Ovarian and Luteal Traits. Animals. 2021; 11(6):1579. https://doi.org/10.3390/ani11061579
Chicago/Turabian StyleZhao, Jianing, Jie Li, Fugui Jiang, Enliang Song, Xianyong Lan, and Haiyu Zhao. 2021. "Fertility-Associated Polymorphism within Bovine ITGβ5 and Its Significant Correlations with Ovarian and Luteal Traits" Animals 11, no. 6: 1579. https://doi.org/10.3390/ani11061579
APA StyleZhao, J., Li, J., Jiang, F., Song, E., Lan, X., & Zhao, H. (2021). Fertility-Associated Polymorphism within Bovine ITGβ5 and Its Significant Correlations with Ovarian and Luteal Traits. Animals, 11(6), 1579. https://doi.org/10.3390/ani11061579






