The Kidney-Associated Microbiome of Wild-Caught Artibeus spp. in Grenada, West Indies
Abstract
Simple Summary
Abstract
1. Introduction
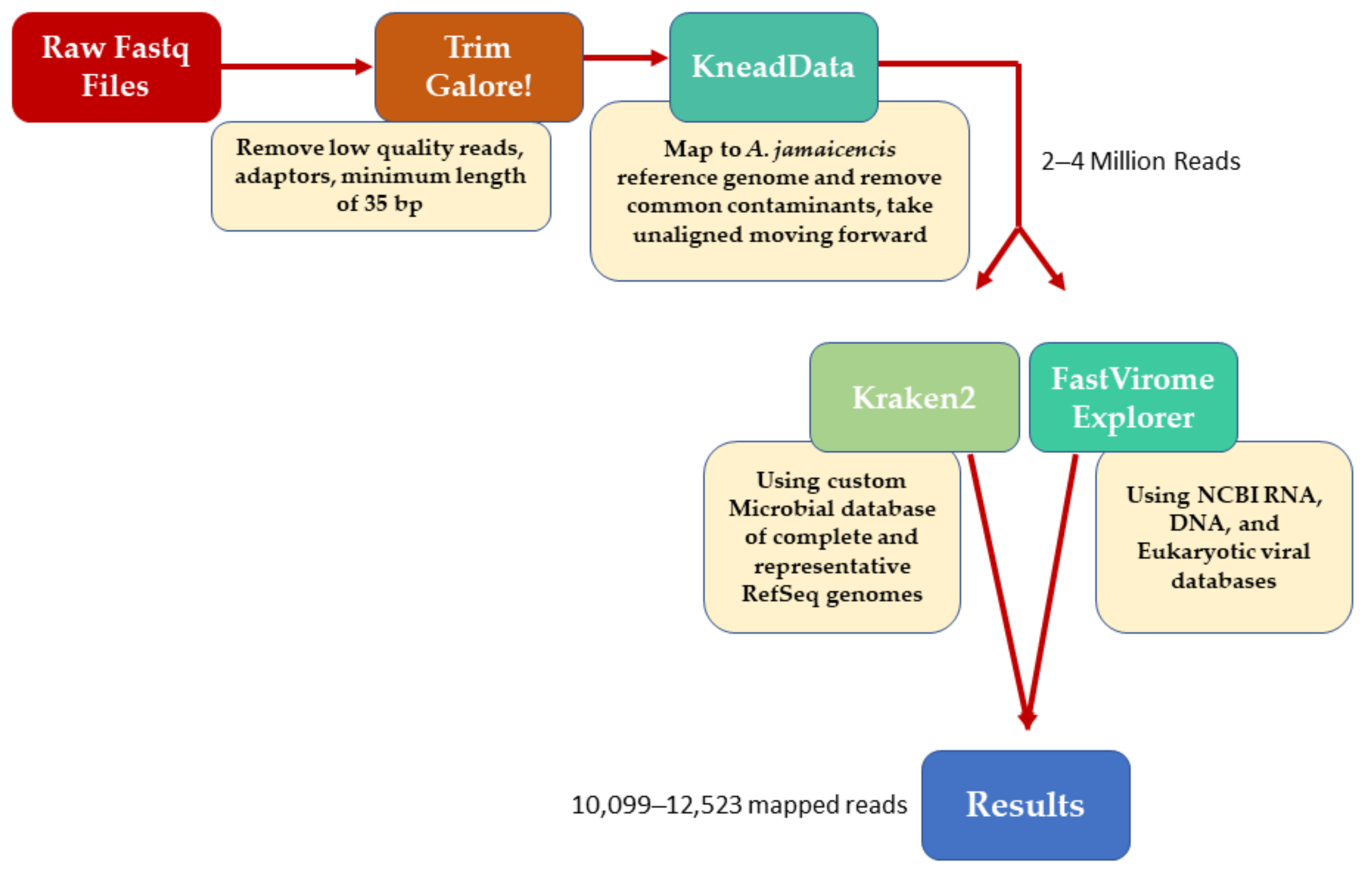
2. Materials and Methods
2.1. Bat Processing
2.2. Histopathology
2.3. Total RNA Extraction and RNA-Seq
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chung, H.; Pamp, S.J.; Hill, J.A.; Surana, N.K.; Edelman, S.M.; Troy, E.B.; Reading, N.C.; Villablanca, E.J.; Wang, S.; Mora, J.R.; et al. Gut immune maturation depends on colonization with a host-specific microbiota. Cell 2012, 149, 1578–1593. [Google Scholar] [CrossRef]
- Mazmanian, S.K.; Liu, C.H.; Tzianabos, A.O.; Kasper, D.L. An Immunomodulatory Molecule of Symbiotic Bacteria Directs Maturation of the Host Immune System. Cell 2005, 122, 107–118. [Google Scholar] [CrossRef] [PubMed]
- Shekhar, S.; Petersen, F.C. The dark side of antibiotics: Adverse effects on the infant immune defense against infection. Front. Pediatr. 2020, 8, 544460. [Google Scholar] [CrossRef] [PubMed]
- Turnbaugh, P.J.; Ley, R.E.; Mahowald, M.A.; Magrini, V.; Mardis, E.R.; Gordon, J.I. An obesity-associated gut microbiome with increased capacity for energy harvest. Nat. Cell Biol. 2006, 444, 1027–1031. [Google Scholar] [CrossRef] [PubMed]
- Bäckhed, F.; Ley, R.E.; Sonnenburg, J.L.; Peterson, D.A.; Gordon, J.I. Host-bacterial mutualism in the human intestine. Science 2005, 307, 1915–1920. [Google Scholar] [CrossRef] [PubMed]
- Dinan, T.G.; Stilling, R.M.; Stanton, C.; Cryan, J.F. Collective unconscious: How gut microbes shape human behavior. J. Psychiatr. Res. 2015, 63, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Sharon, G.; Segal, D.; Ringo, J.M.; Hefetz, A.; Zilber-Rosenberg, I.; Rosenberg, E. Commensal bacteria play a role in mating preference of Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 2010, 107, 20051–20056. [Google Scholar] [CrossRef]
- Sharon, G.; Segal, D.; Zilber-Rosenberg, I.; Rosenberg, E. Symbiotic bacteria are responsible for diet-induced mating preference in Drosophila melanogaster, providing support for the hologenome concept of evolution. Gut Microbes 2011, 2, 190–192. [Google Scholar] [CrossRef]
- Brucker, R.M.; Bordenstein, S.R. The hologenomic basis of speciation: Gut bacteria cause hybrid lethality in the genus Nasonia. Science 2013, 341, 667–669. [Google Scholar] [CrossRef]
- Hird, S.M. Evolutionary biology needs wild microbiomes. Front. Microbiol. 2017, 8, 725. [Google Scholar] [CrossRef]
- Solari, S.; Baker, R.J. Mammal species of the world: A taxonomic and geographic reference. J. Mammal. 2007, 88, 824–830. [Google Scholar] [CrossRef]
- Calisher, C.H.; Childs, J.E.; Field, H.E.; Holmes, K.V.; Schountz, T. Bats: Important reservoir hosts of emerging viruses. Clin. Microbiol. Rev. 2006, 19, 531–545. [Google Scholar] [CrossRef]
- He, B.; Li, Z.; Yang, F.; Zheng, J.; Feng, Y.; Guo, H.; Li, Y.; Wang, Y.; Su, N.; Zhang, F.; et al. Virome profiling of bats from myanmar by metagenomic analysis of tissue samples reveals more novel mammalian viruses. PLoS ONE 2013, 8, e61950. [Google Scholar] [CrossRef]
- Serra-Cobo, J.; López-Roig, M. Bats and emerging infections: An ecological and virological puzzle. In Advances in Experimental Medicine and Biology; Springer: New York, NY, USA, 2017; pp. 35–48. Available online: https://pubmed.ncbi.nlm.nih.gov/27726073 (accessed on 21 May 2021).
- Banerjee, A.; Baker, M.L.; Kulcsar, K.; Misra, V.; Plowright, R.; Mossman, K. Novel insights into immune systems of bats. Front. Immunol. 2020, 11, 26. [Google Scholar] [CrossRef]
- Subudhi, S.; Rapin, N.; Misra, V. Immune system modulation and viral persistence in bats: Understanding viral spillover. Viruses 2019, 11, 192. [Google Scholar] [CrossRef]
- Omatsu, T.; Watanabe, S.; Akashi, H.; Yoshikawa, Y. Biological characters of bats in relation to natural reservoir of emerging viruses. Comp. Immunol. Microbiol. Infect. Dis. 2007, 30, 357–374. [Google Scholar] [CrossRef] [PubMed]
- Pourrut, X.; Souris, M.; Towner, J.S.; Rollin, P.E.; Nichol, S.T.; Gonzalez, J.-P.; Leroy, E. Large serological survey showing cocirculation of Ebola and Marburg viruses in Gabonese bat populations, and a high seroprevalence of both viruses in Rousettus aegyptiacus. BMC Infect. Dis. 2009, 9, 159. [Google Scholar] [CrossRef]
- Li, W.; Shi, Z.; Yu, M.; Ren, W.; Smith, C.; Epstein, J.H.; Wang, H.; Crameri, G.; Hu, Z.; Zhang, H.; et al. Bats are natural reservoirs of SARS-like coronaviruses. Science 2005, 310, 676–679. [Google Scholar] [CrossRef]
- Wang, L.F.; Shi, Z.; Zhang, S.; Field, H.; Daszak, P.; Eaton, B.T. Review of bats and SARS. Emerg. Infect. Dis. 2006, 12, 1834–1840. [Google Scholar] [CrossRef]
- Allocati, N.; Petrucci, A.G.; Di Giovanni, P.; Masulli, M.; Di Ilio, C.; De Laurenzi, V. Bat–man disease transmission: Zoonotic pathogens from wildlife reservoirs to human populations. Cell Death Discov. 2016, 2, 16048. [Google Scholar] [CrossRef]
- Geldenhuys, M.; Weyer, J.; Nel, L.H.; Markotter, W. Coronaviruses in South African bats. Vector-Borne Zoonotic Dis. 2013, 13, 516–519. [Google Scholar] [CrossRef] [PubMed]
- Kasso, M.; Balakrishnan, M. Ecological and economic importance of bats (order chiroptera). ISRN Biodivers. 2013, 2013, 1–9. [Google Scholar] [CrossRef]
- Boyles, J.G.; Cryan, P.M.; McCracken, G.F.; Kunz, T.H. Economic importance of bats in agriculture. Science 2011, 332, 41–42. [Google Scholar] [CrossRef] [PubMed]
- Ducummon, S.L. Ecological and Economic Importance of Bats; Bat Conservation International, Inc.: Austin, TX, USA, 2001. [Google Scholar]
- Marí Saéz, A.; Weiss, S.; Nowak, K.; Lapeyre, V.; Zimmermann, F.; Düx, A.; Kühl, H.S.; Kaba, M.; Regnaut, S.; Merkel, K.; et al. Investigating the zoonotic origin of the West African Ebola epidemic. EMBO Mol. Med. 2015, 7, 17–23. [Google Scholar] [CrossRef]
- Donaldson, E.F.; Haskew, A.N.; Gates, J.E.; Huynh, J.; Moore, C.J.; Frieman, M.B. Metagenomic analysis of the viromes of three North American bat species: Viral diversity among different bat species that share a common habitat. J. Virol. 2010, 84, 13004–13018. [Google Scholar] [CrossRef]
- Li, L.; Victoria, J.G.; Wang, C.; Jones, M.; Fellers, G.M.; Kunz, T.H.; Delwart, E. Bat guano virome: Predominance of dietary viruses from insects and plants plus novel mammalian viruses. J. Virol. 2010, 84, 6955–6965. [Google Scholar] [CrossRef]
- Zheng, X.Y.; Qiu, M.; Guan, W.J.; Li, J.M.; Chen, S.W.; Cheng, M.J.; Huo, S.T.; Chen, Z.; Wu, Y.; Jiang, L.N.; et al. Viral metagenomics of six bat species in close contact with humans in southern China. Arch. Virol. 2018, 163, 73–88. [Google Scholar] [CrossRef]
- Yang, F.; Wang, Y.; Zheng, W.; He, B.; Jiang, T.; Li, Y.; Xia, L.; Feng, Y.; Fan, Q.; Tu, C. Metagenomic analysis of bat virome in several Chinese regions. Shengwu Gongcheng Xuebao/Chin. J. Biotechnol. 2013, 29, 586–600. [Google Scholar]
- Dacheux, L.; Cervantes-Gonzalez, M.; Guigon, G.; Thiberge, J.M.; Vandenbogaert, M.; Maufrais, C.; Caro, V.; Bourhy, H. A preliminary study of viral metagenomics of french bat species in contact with humans: Identification of new mammalian viruses. PLoS ONE 2014, 9, e87194. [Google Scholar] [CrossRef]
- Avena, C.V.; Parfrey, L.W.; Leff, J.W.; Archer, H.M.; Frick, W.F.; Langwig, K.E.; Kilpatrick, A.M.; Powers, K.E.; Foster, J.T.; McKenzie, V.J. Deconstructing the bat skin microbiome: Influences of the host and the environment. Front. Microbiol. 2016, 7, 1753. [Google Scholar] [CrossRef]
- Vengust, M.; Knapic, T.; Weese, J.S. The fecal bacterial microbiota of bats; Slovenia. PLoS ONE 2018, 13, e0196728. [Google Scholar] [CrossRef] [PubMed]
- Dietrich, M.; Kearney, T.; Seamark, E.C.J.; Paweska, J.T.; Markotter, W. Synchronized shift of oral, faecal and urinary microbiotas in bats and natural infection dynamics during seasonal reproduction. R. Soc. Open Sci. 2018, 5, 180041. [Google Scholar] [CrossRef] [PubMed]
- Fofanov, V.Y.; Furstenau, T.N.; Sanchez, D.; Hepp, C.M.; Cocking, J.; Sobek, C.; Pagel, N.; Walker, F.; Chambers, C.L. Guano exposed: Impact of aerobic conditions on bat fecal microbiota. Ecol. Evol. 2018, 8, 5563–5574. [Google Scholar] [CrossRef] [PubMed]
- Carrillo-Araujo, M.; Tas, N.; Alcántara-Hernández, R.J.; Gaona, O.; Schondube, J.E.; Medellín, R.A.; Jansson, J.K.; Falkón, L.I. Phyllostomid bat microbiome composition is associated to host phylogeny and feeding strategies. Front. Microbiol. 2015. [Google Scholar] [CrossRef] [PubMed]
- Sikes, R.S.; Gannon, W.L. Guidelines of the American Society of mammalogists for the use of wild mammals in research. J. Mammal. 2011, 97, 663–688. [Google Scholar] [CrossRef] [PubMed]
- Lim, B.K. Book reviews: Bats of Trinidad and Tobago: A field guide and natural History. Acta Chiropterologica 2015, 17, 199–200. [Google Scholar] [CrossRef]
- Larsen, P.A.; Hoofer, S.R.; Bozeman, M.C.; Pedersen, S.C.; Genoways, H.H.; Phillips, C.J.; Pumo, D.E.; Baker, R.J. Phylogenetics and phylogeography of the Artibeus jamaicensis complex based on cytochrome-b DNA sequences. J. Mammal. 2007, 88, 712–727. [Google Scholar] [CrossRef]
- Bevans, A.I.; Fitzpatrick, D.M.; Stone, D.M.; Butler, B.P.; Smith, M.P.; Cheetham, S. Phylogenetic relationships and diversity of bat-associated leptospira and the histopathological evaluation of these infections in bats from Grenada, West Indies. PLoS Negl. Trop Dis. 2020, 14, e0007940. [Google Scholar] [CrossRef]
- Ramos-Nino, M.E.; Fitzpatrick, D.M.; Tighe, S.; Eckstrom, K.M.; Hattaway, L.M.; Hsueh, A.N.; Stone, D.M.; Dragon, J.; Cheetham, S. High prevalence of Phasi Charoen-like virus from wild-caught Aedes aegypti in Grenada, W.I. As revealed by metagenomic analysis. PLoS ONE 2020, 15, e0227998. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Nowak, K.; Fahr, J.; Weber, N.; Lübke-Becker, A.; Semmler, T.; Weiss, S.; Mombouli, J.-V.; Wieler, L.H.; Guenther, S.; Leendertz, F.H.; et al. Highly diverse and antimicrobial susceptible Escherichia coli display a naïve bacterial population in fruit bats from the Republic of Congo. PLoS ONE 2017, 12, e0178146. [Google Scholar] [CrossRef] [PubMed]
- Tulsiani, S.M.; Graham, G.C.; Dohnt, M.F.; Burns, M.; Craig, S.B. Maximizing the chances of detecting pathogenic leptospires in mammals: The evaluation of field samples and a multi-sample-per-mammal, multi-test approach. Ann. Trop. Med. Parasitol. 2011, 105, 145–162. [Google Scholar] [CrossRef] [PubMed]
- Chander, A.M.; Nair, R.G.; Kaur, G.; Kochhar, R.; Dhawan, D.K.; Bhadada, S.K.; Mayilraj, S. Genome Insight and Comparative Pathogenomic Analysis of Nesterenkonia jeotgali Strain CD08_7 Isolated from Duodenal Mucosa of Celiac Disease Patient. Front. Microbiol. 2017, 8, 129. [Google Scholar] [CrossRef] [PubMed]
- Vaziri, N.D.; Wong, J.; Pahl, M.; Piceno, Y.M.; Yuan, J.; DeSantis, T.Z.; Ni, Z.; Nguyen, T.-H.; Andersen, G.L. Chronic kidney disease alters intestinal microbial flora. Kidney Int. 2013, 83, 308–315. [Google Scholar] [CrossRef] [PubMed]
- Koerner, R.J.; Goodfellow, M.; Jones, A.L. The genusDietzia: A new home for some known and emerging opportunist pathogens. FEMS Immunol. Med. Microbiol. 2009, 55, 296–305. [Google Scholar] [CrossRef]
- Long, S.S.; Pickering, L.K.; Prober, C.G. Principles and Practice of Pediatric Infectious Diseases, 4th ed.; Elsevier: Amsterdam, The Netherlands, 2012. [Google Scholar]
- Gessler, C.; Pertot, I.; Perazzolli, M. Plasmopara viticola: A review of knowledge on downy mildew of grapevine and effective disease management. Phytopathol. Mediterr. 2011, 50, 3–44. [Google Scholar]
- Furtado, B.G.; Savi, G.D.; Angioletto, E.; Carvalho, F. Filamentous fungi occurrence on Molossus molossus (Pallas, 1766) (Chiroptera: Molossidae) present in an Atlantic Forest remnant in Southern Brazil. Braz. J. Biol. 2020, 81, 1073–1080. [Google Scholar] [CrossRef]
- Escalera-Zamudio, M.; Mendoza, M.L.Z.; Heeger, F.; Loza-Rubio, E.; Rojas-Anaya, E.; Méndez-Ojeda, M.L.; Taboada, B.; Mazzoni, C.J.; Arias, C.F.; Greenwood, A.D. A novel endogenous betaretrovirus in the common vampire bat (desmodus rotundus) suggests multiple independent infection and cross-species transmission events. J. Virol. 2015, 89, 5180–5184. [Google Scholar] [CrossRef][Green Version]
- Filho, L.C.F.; Barata, R.R.; Cardoso, J.F.; de Vasconcelos Massafra, J.M.; da Silva Lemos, P.; Casseb, L.M.N.; Cruz, A.C.R.; Nunes, M.R.T. Complete endogenous retrovirus genome sequence from a Brazilian vampire bat (desmodus rotundus). Microbiol. Resour. Announc. 2019, 8, e01497–e01518. [Google Scholar] [CrossRef]
- Hayward, J.A.; Tachedjian, M.; Cui, J.; Field, H.; Holmes, E.C.; Wang, L.F.; Tachedjian, G. Identification of diverse full-length endogenous betaretroviruses in megabats and microbats. Retrovirology 2013, 10, 35. [Google Scholar] [CrossRef]
| Organism | Sample #1 | Sample #2 |
|---|---|---|
| Bacteria | 8174 | 6675 |
| Fungi | 4100 | 3083 |
| Viruses | 249 | 241 |
| Organism | Sample #1 | Sample #2 | ||||
|---|---|---|---|---|---|---|
| Phylum | Class | Family | Genus | |||
| Bacteria | 65.27 | 66.09 | ||||
| Proteobacteria | Gammaproteo-bacteria | Enterobacteriaceae | Escherichia | 51.22 | 49.01 | |
| Spirochaetes | Spirochaetia | Leptospiraceae | Leptospira | 12.82 | 15.24 | |
| Actinobacteria | Actinobacteria | Micrococcaceae | Nesterenkonia | 0.11 | 0.11 | |
| Actinobacteria | Actinobacteria | Dietziaceae | Dietzia | 0.10 | 0.11 | |
| Proteobacteria | Betaproteobacteria | Comamonadaceae | Acidovorax | 0.09 | 0.08 | |
| Firmicutes | Clostridia | Clostridiaceae | Clostridium | 0.01 | 0.2 | |
| Fungi | 32.74 | 30.53 | ||||
| Oomycetes | Peronosporaceae | Plasmopara | 31.36 | 30.00 | ||
| Ascomycota | Eurotiomycetes | Aspergillaceae | Aspergillus | 0.34 | 0.42 | |
| Viruses | Retroviridae | Betaretrovirus * | 1.99 | 3.38 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ramos-Nino, M.E.; Fitzpatrick, D.M.; Eckstrom, K.M.; Tighe, S.; Dragon, J.A.; Cheetham, S. The Kidney-Associated Microbiome of Wild-Caught Artibeus spp. in Grenada, West Indies. Animals 2021, 11, 1571. https://doi.org/10.3390/ani11061571
Ramos-Nino ME, Fitzpatrick DM, Eckstrom KM, Tighe S, Dragon JA, Cheetham S. The Kidney-Associated Microbiome of Wild-Caught Artibeus spp. in Grenada, West Indies. Animals. 2021; 11(6):1571. https://doi.org/10.3390/ani11061571
Chicago/Turabian StyleRamos-Nino, Maria E., Daniel M. Fitzpatrick, Korin M. Eckstrom, Scott Tighe, Julie A. Dragon, and Sonia Cheetham. 2021. "The Kidney-Associated Microbiome of Wild-Caught Artibeus spp. in Grenada, West Indies" Animals 11, no. 6: 1571. https://doi.org/10.3390/ani11061571
APA StyleRamos-Nino, M. E., Fitzpatrick, D. M., Eckstrom, K. M., Tighe, S., Dragon, J. A., & Cheetham, S. (2021). The Kidney-Associated Microbiome of Wild-Caught Artibeus spp. in Grenada, West Indies. Animals, 11(6), 1571. https://doi.org/10.3390/ani11061571






