Aspergillus oryzae and Aspergillus niger Co-Cultivation Extract Affects In Vitro Degradation, Fermentation Characteristics, and Bacterial Composition in a Diet-Specific Manner
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Experiment Design and In Vitro Batch Culture
2.3. Sample Collection and Measurement
2.4. DNA Extraction and Sequencing
2.5. Sequencing Data Processing
2.6. Statistical Analyses
3. Results
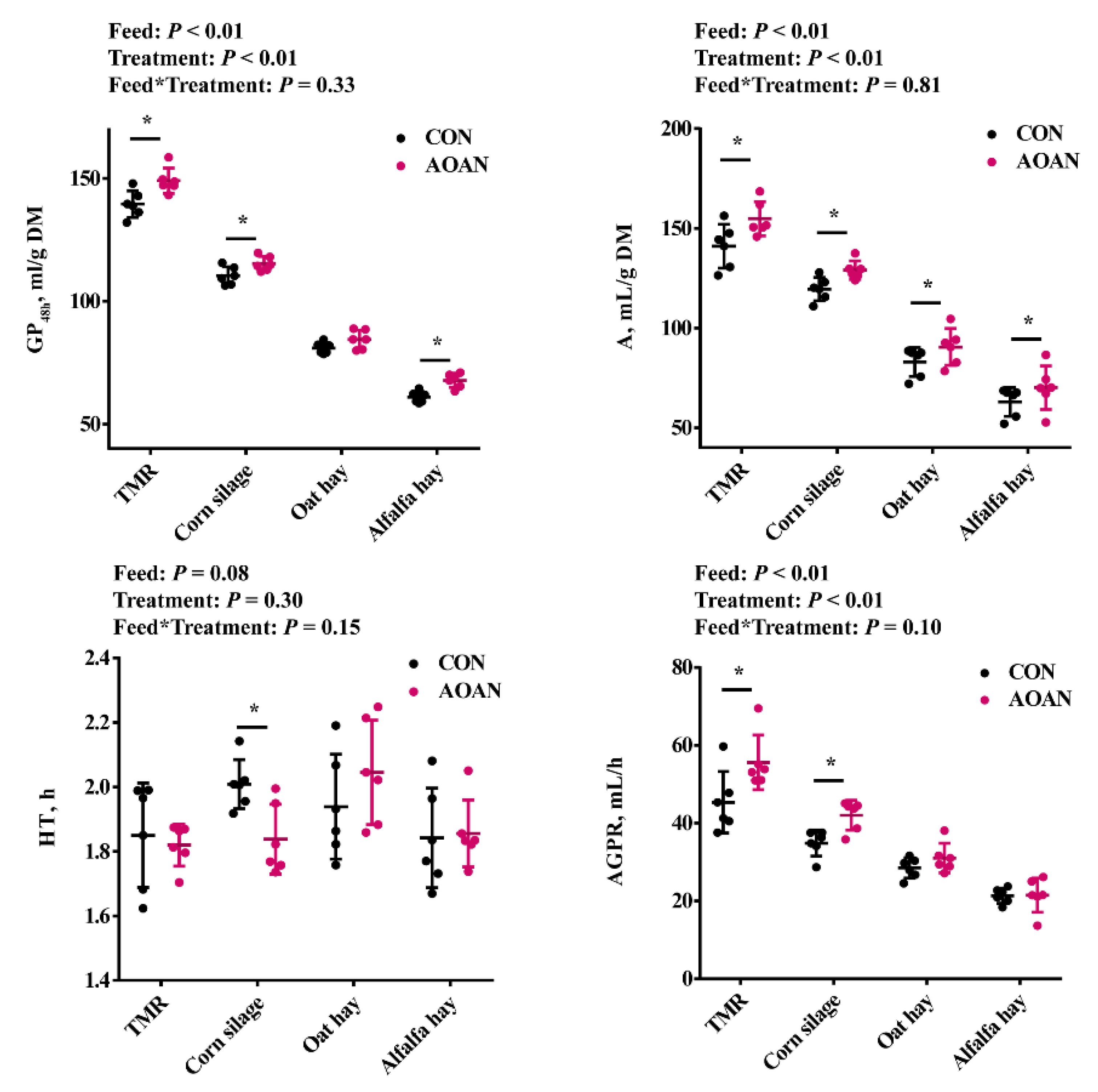
3.1. Gas Production Kinetics Parameters
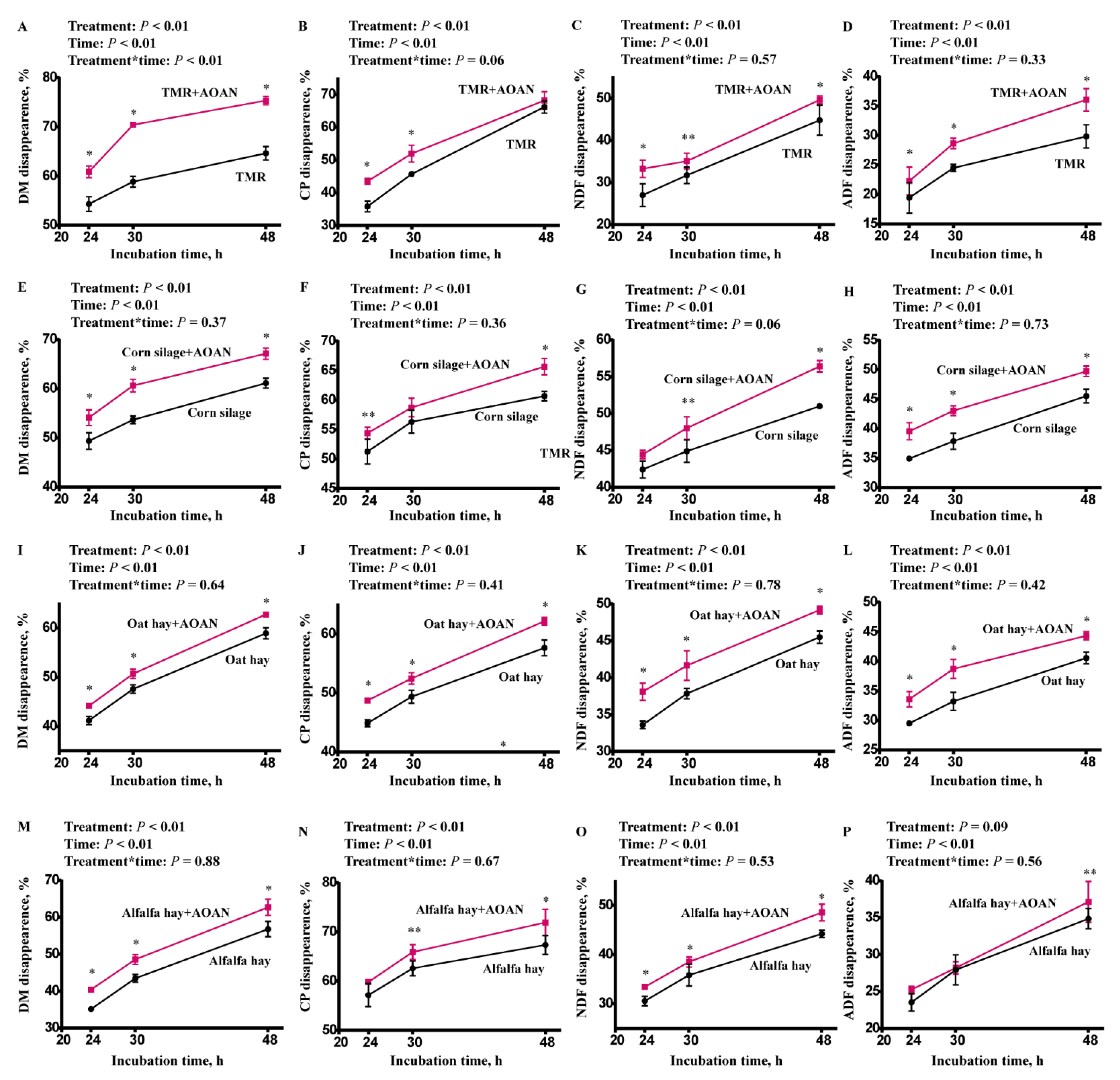
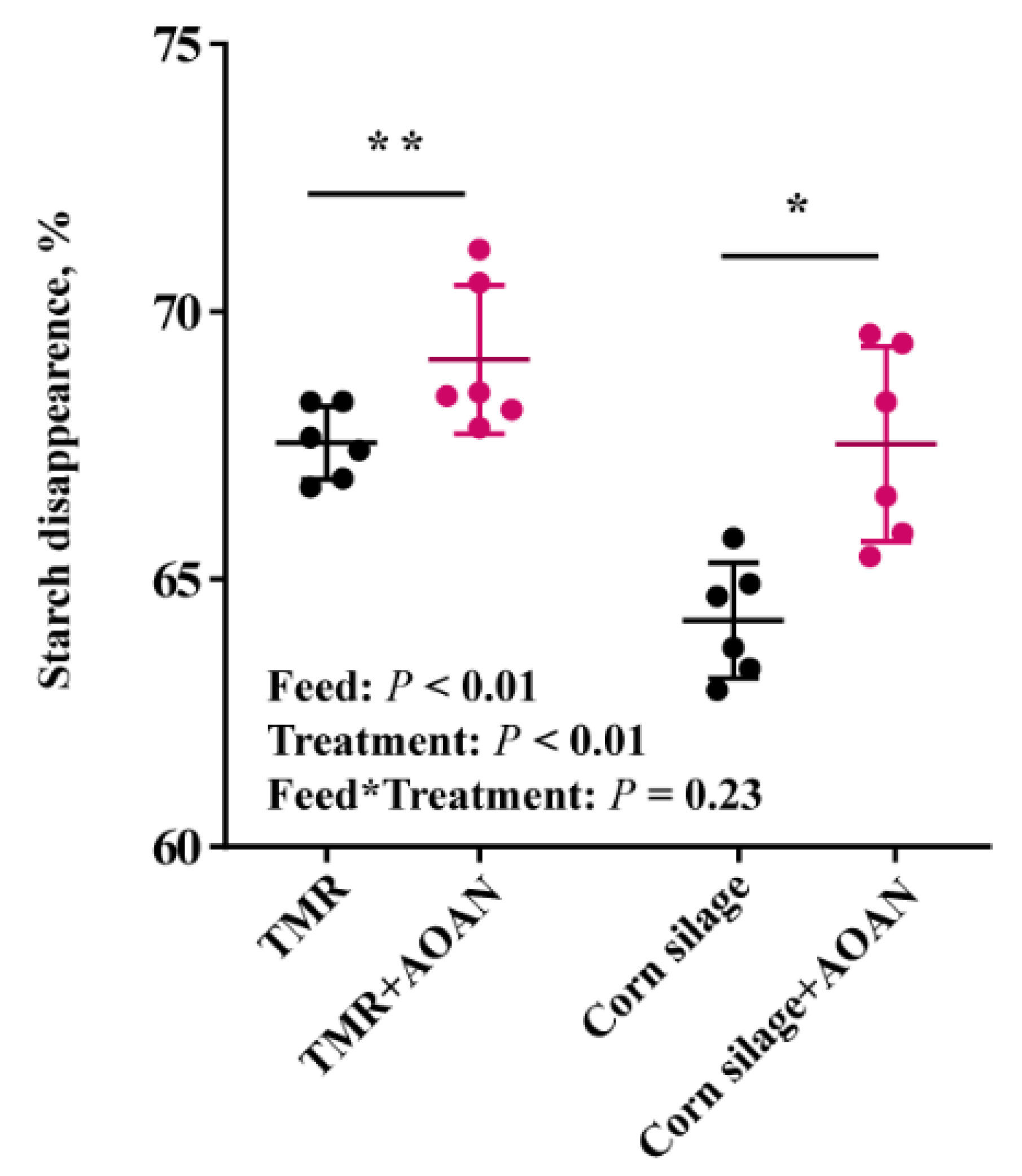
3.2. Nutrient Digestibility
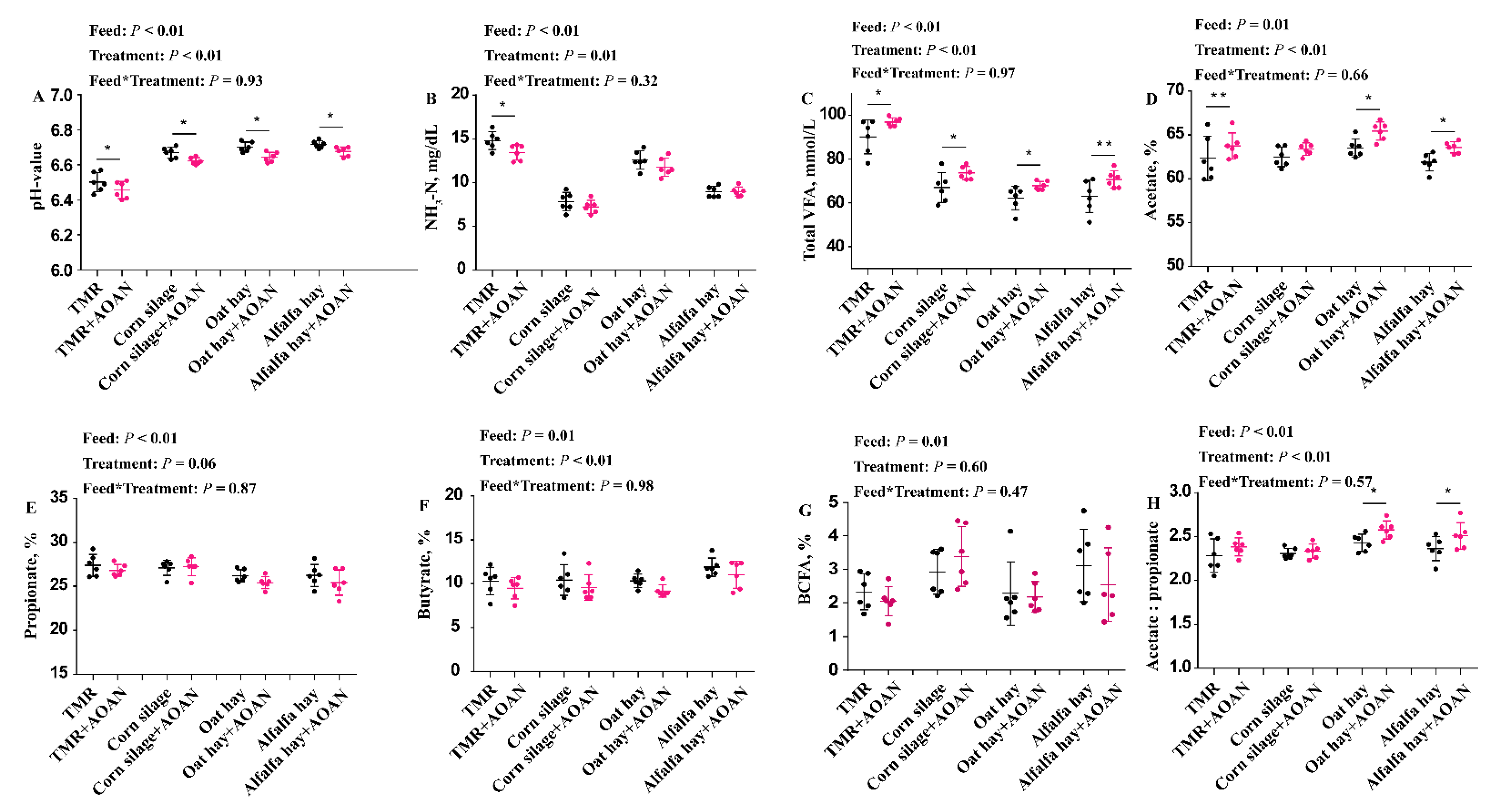
3.3. Fermentation Parameters
3.4. Bacterial Community
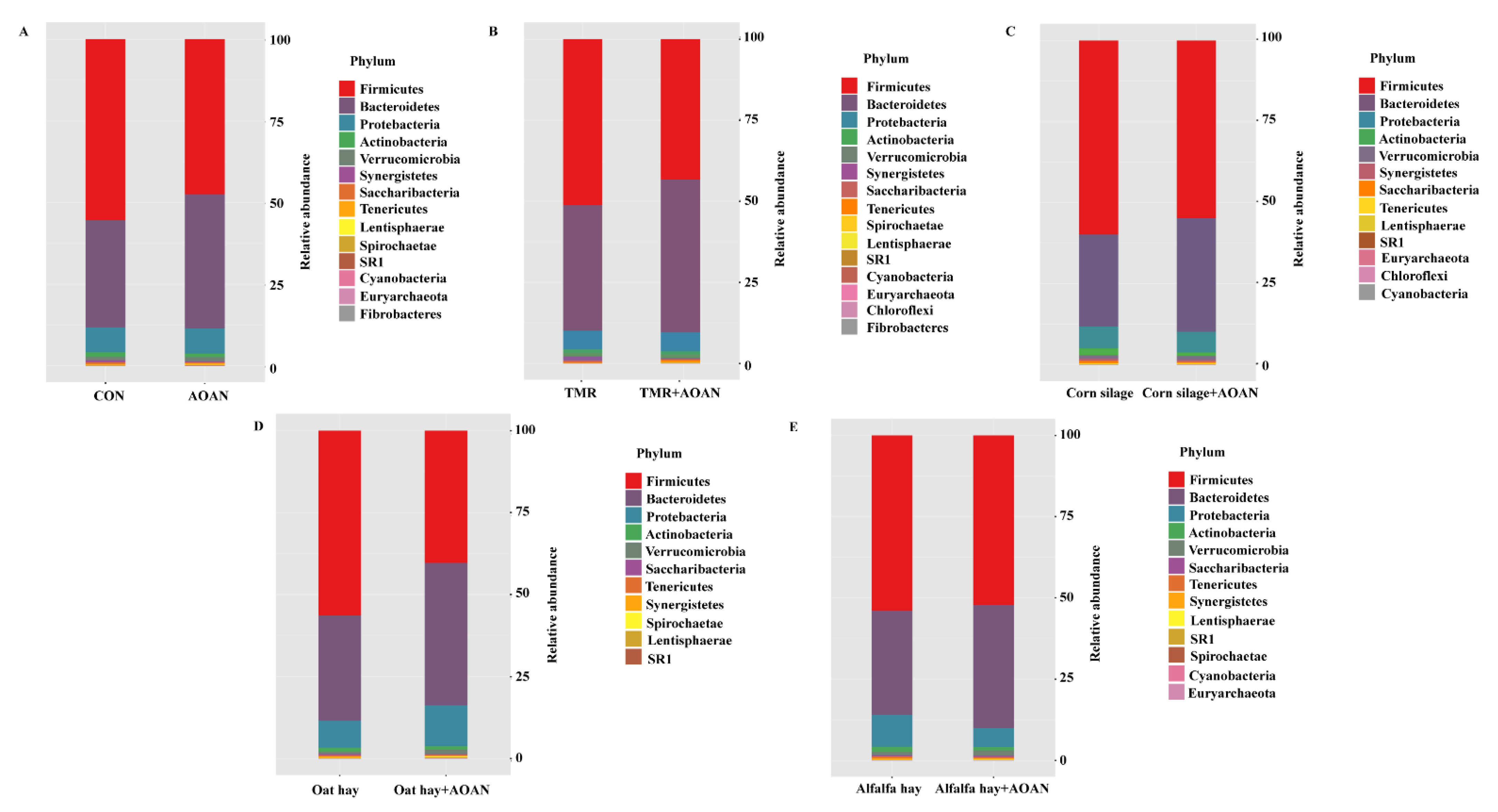
3.4.1. Sequence Depth, Diversity, and Composition
3.4.2. The β Diversity and Bacterial Composition
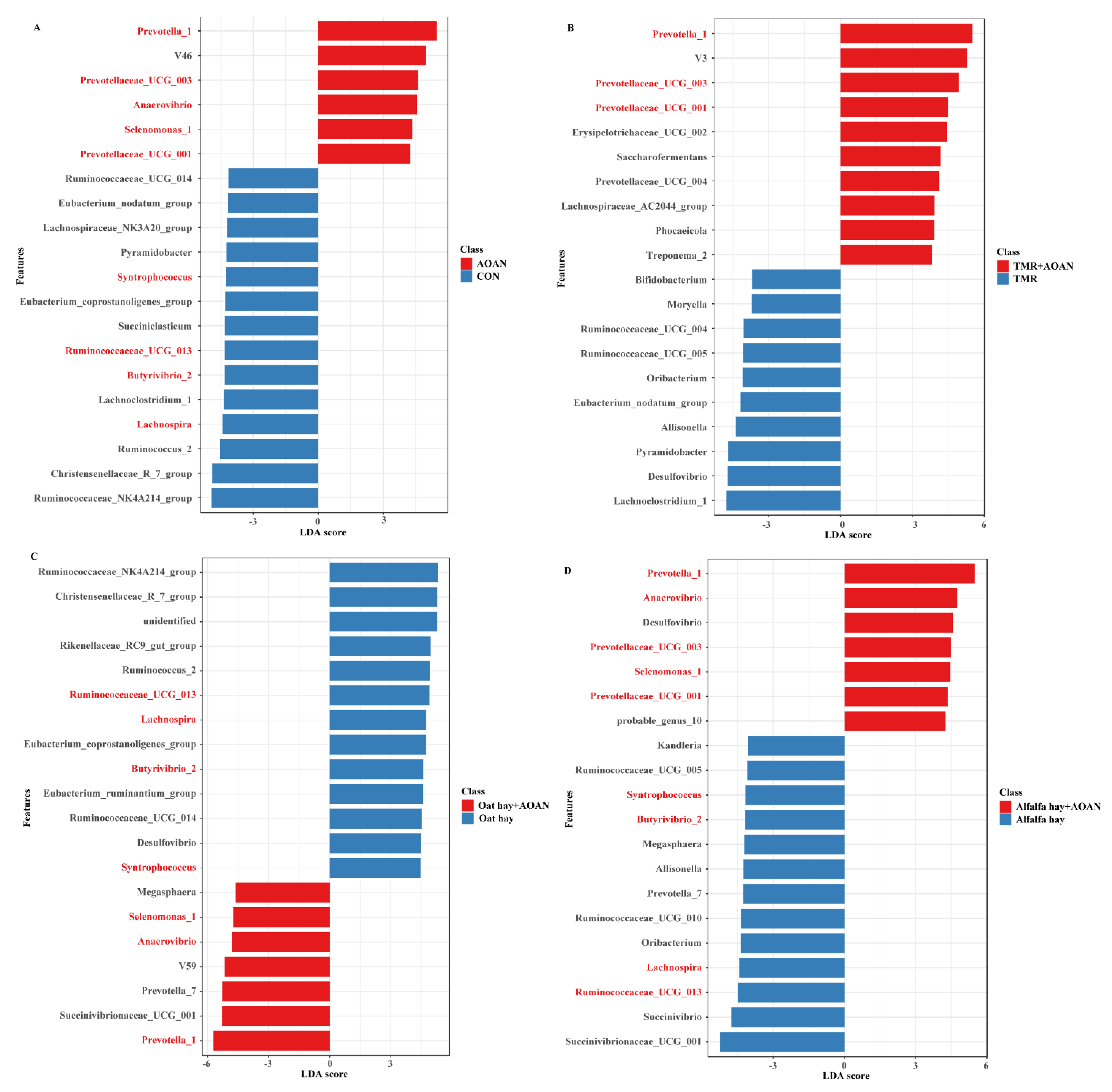
3.4.3. Differential Genera
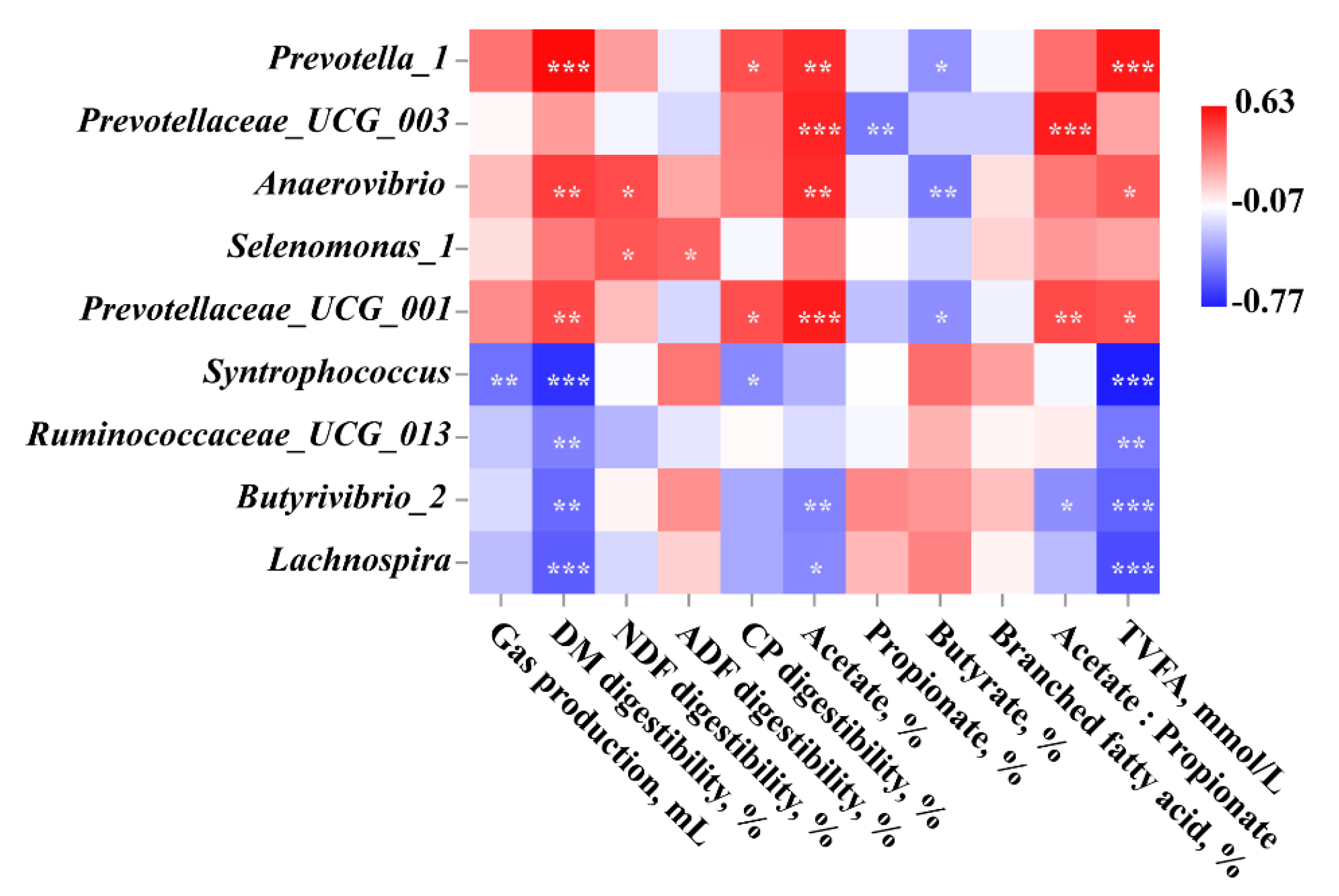
3.4.4. Spearman Correlation Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sucu, E.; Moore, C.; VanBaale, M.J.; Jensen, H.; Sanz-Fernandez, M.V.; Baumgard, L.H. Effects of feeding aspergillus oryzae fermentation product to transition holstein cows on performance and health. Can. J. Anim. Sci. 2019, 99, 237–243. [Google Scholar] [CrossRef]
- Martarello, R.D.A.; Cunha, L.; Cardoso, S.L.; de Freitas, M.M.; Silveira, D.; Fonseca-Bazzo, Y.M.; Homem-de-Mello, M.; Ferreira Filho, E.X.; Magalhães, P.O. Optimization and partial purification of beta-galactosidase production by Aspergillus niger isolated from Brazilian soils using soybean residue. AMB Express 2019, 9, 81. [Google Scholar] [CrossRef]
- Cong, S.; Tian, K.; Zhang, X.; Lu, F.; Singh, S.; Prior, B.; Wang, Z.-X. Synthesis of flavor esters by a novel lipase from Aspergillus niger in a soybean-solvent system. 3 Biotech 2019, 9, 244. [Google Scholar] [CrossRef] [PubMed]
- Hyeon, H.; Min, C.W.; Moon, K.; Cha, J.; Gupta, R.; Park, S.U.; Kim, S.T.; Kim, J.K. Metabolic profiling-based evaluation of the fermentative behavior of Aspergillus oryzae and Bacillus subtilis for soybean residues treated at different temperatures. Foods 2020, 9, 117. [Google Scholar] [CrossRef] [PubMed]
- Sadh, P.K.; Chawla, P.; Bhandari, L.; Kaushik, R.; Duhan, J.S. In vitro assessment of bio-augmented minerals from peanut oil cakes fermented by Aspergillus oryzae through Caco-2 cells. J. Food Sci. Tech. 2017, 54, 3640–3649. [Google Scholar] [CrossRef] [PubMed]
- De Vries, R.P.; Visser, J. Aspergillus enzymes involved in degradation of plant cell wall polysaccharides. Microbiol. Mol. Biol. Rev. 2001, 65, 497–522. [Google Scholar] [CrossRef]
- Fu, G.; Wang, Y.; Wang, D.; Zhou, C. Cloning, expression, and characterization of an GHF 11 Xylanase from aspergillus niger XZ-3S. Indian J. Microbiol. 2012, 52, 682–688. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Moreira, L.R.d.S.; Ferreira, G.V.; Santos, S.S.T.; Ribeiro, A.P.S.; Siqueira, F.G.; Ferreira Filho, E.X. The hydrolysis of agro-industrial residues by holocellulose-degrading enzymes. Braz. J. Microbiol. 2012, 43, 498–505. [Google Scholar] [CrossRef]
- Li, L.-h.; King, K. Fractionation of β-glucosidases and related extracellular enzymes from Aspergillus niger. Appl. Microbiol. 1963, 11, 320–325. [Google Scholar] [CrossRef]
- Altop, A.; Güngör, E.; Erener, G. Aspergillus niger may improve nutritional quality of grape seed and its usability in animal nutrition through solid-state fermentation. Int. Adv. Res. Eng. J. 2018, 2, 273–277. [Google Scholar]
- Ong, L.; Abd-Aziz, S.; Noraini, S.; Karim, M.; Hassan, M. Enzyme production and profile by Aspergillus niger during solid substrate fermentation using palm kernel cake as substrate. Appl. Biochem. Biotech. 2004, 118, 73–79. [Google Scholar] [CrossRef]
- Wang, J.; Cao, F.; Su, E.; Zhao, L.; Qin, W. Improvement of animal feed additives of Ginkgo leaves through solid-state fermentation using Aspergillus niger. Int. J. Biol. Sci. 2018, 14, 736–747. [Google Scholar] [CrossRef]
- Sun, H.; Wu, Y.; Wang, Y.; Liu, J.; Myung, K. Effects of Aspergillus oryzae culture and 2-hydroxy-4-(methylthio)-butanoic acid on in vitro rumen fermentation and microbial populations between different roughage sources. Asian Austral. J. Anim. 2014, 27, 1285–1292. [Google Scholar] [CrossRef]
- Hong, K.-J.; Lee, C.-H.; Kim, S.W. Aspergillus oryzae GB-107 fermentation improves nutritional quality of food soybeans and feed soybean meals. J. Med. Food 2004, 7, 430–435. [Google Scholar] [CrossRef]
- Sun, H.; Wu, Y.; Wang, Y.; Wang, C.; Liu, J. Effects of addition of Aspergillus oryzae culture and 2-hydroxyl-4-(methylthio) butanoic acid on milk performance and rumen fermentation of dairy cows. Anim. Sci. J. 2017, 88, 602–609. [Google Scholar] [CrossRef] [PubMed]
- Ominski, K.; Wittenberg, K.; Kennedy, A.; Moshtaghi-Nia, S. Physiological and production responses when feeding Aspergillus oryzae to dairy cows during short-term, moderate heat stress. Anim. Sci. 2003, 77, 485–490. [Google Scholar] [CrossRef]
- Sievert, S.; Shaver, R. Carbohydrate and Aspergillus oryzae effects on intake, digestion, and milk production by dairy cows. J. Dairy Sci. 1993, 76, 245–254. [Google Scholar] [CrossRef]
- Hu, H.; Van den Brink, J.; Gruben, B.; Wösten, H.; Gu, J.-D.; De Vries, R. Improved enzyme production by co-cultivation of Aspergillus niger and Aspergillus oryzae and with other fungi. Int. Biodeter. Biodegr. 2011, 65, 248–252. [Google Scholar] [CrossRef]
- Xue, M.-Y.; Sun, H.-Z.; Wu, X.-H.; Liu, J.-X.; Guan, L.L. Multi-omics reveals that the rumen microbiome and its metabolome together with the host metabolome contribute to individualized dairy cow performance. Microbiome 2020, 8, 1–19. [Google Scholar] [CrossRef]
- Zhou, M.; Peng, Y.-J.; Chen, Y.; Klinger, C.M.; Oba, M.; Liu, J.-X.; Guan, L.L. Assessment of microbiome changes after rumen transfaunation: Implications on improving feed efficiency in beef cattle. Microbiome 2018, 6, 62. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Lv, M.; Wang, J.; Tian, Z.; Yu, B.; Wang, B.; Liu, J.; Liu, H. Dandelion (Taraxacum mongolicum Hand.-Mazz.) Supplementation-enhanced rumen fermentation through the interaction between ruminal microbiome and metabolome. Microorganisms 2021, 9, 83. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Guo, J.; Li, W.; Wu, Z.; Yu, Z. Effects of ferulic acid esterase-producing lactic acid bacteria and storage temperature on the fermentation quality, in vitro digestibility and phenolic acid extraction yields of sorghum (sorghum bicolor L.) Silage. Microorganisms 2021, 9, 114. [Google Scholar] [CrossRef]
- Ma, T.; Wu, W.; Tu, Y.; Zhang, N.; Diao, Q. Resveratrol affects in vitro rumen fermentation, methane production and prokaryotic community composition in a time-and diet-specific manner. Microb. Biotechnol. 2020, 13, 1118–1131. [Google Scholar] [CrossRef]
- Zhang, D.F.; Yang, H.J. Combination effects of nitrocompounds, pyromellitic diimide, and 2-bromoethanesulfonate on in vitro ruminal methane production and fermentation of a grain-rich feed. J. Agric. Food Chem. 2012, 60, 364–371. [Google Scholar] [CrossRef]
- Yáñez-Ruiz, D.R.; Bannink, A.; Dijkstra, J.; Kebreab, E.; Morgavi, D.P.; O’Kiely, P.; Reynolds, C.K.; Schwarm, A.; Shingfield, K.J.; Yu, Z.; et al. Design, implementation and interpretation of in vitro batch culture experiments to assess enteric methane mitigation in ruminants—A review. Anim. Feed Sci. Tech. 2016, 216, 1–18. [Google Scholar] [CrossRef]
- Menke, K.H. Estimation of the energetic feed value obtained from chemical analysis and in vitro gas production using rumen fluid. Anim. Res. Dev. 1988, 28, 7–55. [Google Scholar]
- Kong, F.; Gao, Y.; Tang, M.; Fu, T.; Diao, Q.; Bi, Y.; Tu, Y. Effects of dietary rumen-protected Lys levels on rumen fermentation and bacterial community composition in Holstein heifers. Appl. Microbiol. Biotechnol. 2020, 104, 6623–6634. [Google Scholar] [CrossRef] [PubMed]
- Bi, Y.; Zeng, S.; Zhang, R.; Diao, Q.; Tu, Y. Effects of dietary energy levels on rumen bacterial community composition in Holstein heifers under the same forage to concentrate ratio condition. BMC Microbiol. 2018, 18, 69. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Pena, A.G.; Goodrich, J.K.; Gordon, J.I. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef]
- Haas, B.J.; Gevers, D.; Earl, A.M.; Feldgarden, M.; Ward, D.V.; Giannoukos, G.; Ciulla, D.; Tabbaa, D.; Highlander, S.K.; Sodergren, E.; et al. Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome. Res. 2011, 21, 494–504. [Google Scholar] [CrossRef]
- Pruesse, E.; Quast, C.; Knittel, K.; Fuchs, B.M.; Ludwig, W.; Peplies, J.; Glöckner, F.O. SILVA: A comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 2007, 35, 7188–7196. [Google Scholar] [CrossRef]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef]
- Chong, J.; Liu, P.; Zhou, G.; Xia, J. Using MicrobiomeAnalyst for comprehensive statistical, functional, and meta-analysis of microbiome data. Nat. Protoc. 2020, 15, 799–821. [Google Scholar] [CrossRef] [PubMed]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome. Biol. 2011, 12, R60. [Google Scholar] [CrossRef] [PubMed]
- France, J.; Dijkstra, J.; Dhanoa, M.; Lopez, S.; Bannink, A. Estimating the extent of degradation of ruminant feeds from a description of their gas production profiles observed in vitro: Derivation of models and other mathematical considerations. Br. J. Nutr. 2000, 83, 143–150. [Google Scholar] [CrossRef]
- García-Martínez, R.; Ranilla, M.; Tejido, M.; Carro, M. Effects of disodium fumarate on in vitro rumen microbial growth, methane production and fermentation of diets differing in their forage: Concentrate ratio. Br. J. Nutr. 2005, 94, 71–77. [Google Scholar] [CrossRef]
- Hristov, A.; Harper, M.; Roth, G.; Canale, C.; Huhtanen, P.; Richard, T.; DiMarco, K. Effects of ensiling time on corn silage neutral detergent fiber degradability and relationship between laboratory fiber analyses and in vivo digestibility. J. Dairy Sci. 2020, 103, 2333–2346. [Google Scholar] [CrossRef]
- Richards, C.; Pedersen, J.F.; Britton, R.; Stock, R.; Krehbiel, C. In vitro starch disappearance procedure modifications. Anim. Deed. Sci. Tech. 1995, 55, 35–45. [Google Scholar] [CrossRef]
- Varel, V.H.; Kreikemeier, K.K.; Jung, H.-J.G.; Hatfield, R.D. In vitro stimulation of forage fiber degradation by ruminal microorganisms with Aspergillus oryzae fermentation extract. Appl. Environ. Microbiol. 1993, 59, 3171–3176. [Google Scholar] [CrossRef] [PubMed]
- Miranda, R.; Mendoza, M.; Bárcena-Gama, J.; González, M.; Ferrara, R.; Ortega, C.; Cobos, P. Effect of Saccharomyces cerevisiae or Aspergillus oryzae cultures and NDF level on parameters of ruminal fermentation. Anim. Deed. Sci. Tech. 1996, 63, 289–296. [Google Scholar] [CrossRef]
- Newbold, C.; Brock, R.; Wallace, R. Influence of autoclaved or irradiated Aspergillus oryzae fermentation extract on fermentation in the rumen simulation technique (Rusitec). J. Agr. Sci. 1991, 116, 159–162. [Google Scholar] [CrossRef]
- Wiedmeier, R.; Arambel, M.; Walters, J. Effect of yeast culture and Aspergillus oryzae fermentation extract on ruminal characteristics and nutrient digestibility. J. Dairy Sci. 1987, 70, 2063–2068. [Google Scholar] [CrossRef]
- Gomez-Alarcon, R.; Dudas, C.; Huber, J. Influence of cultures of Aspergillus oryzae on rumen and total tract digestibility of dietary components. J. Dairy Sci. 1990, 73, 703–710. [Google Scholar] [CrossRef]
- Raffrenato, E.; Fievisohn, R.; Cotanch, K.W.; Grant, R.J.; Chase, L.E.; Van Amburgh, M.E. Effect of lignin linkages with other plant cell wall components on in vitro and in vivo neutral detergent fiber digestibility and rate of digestion of grass forages. J. Dairy Sci. 2017, 100, 8119–8131. [Google Scholar] [CrossRef]
- Khazaal, K.; Dentinho, M.T.; Ribeiro, J.M.; Ørskov, E.R. Prediction of apparent digestibility and voluntary intake of hays fed to sheep: Comparison between using fiber components, in vitro digestibility or characteristics of gas production or nylon bag degradation. Anim. Sci. 1995, 61, 527–538. [Google Scholar] [CrossRef]
- Zicarelli, F.; Addi, L.; Tudisco, R.; Calabrò, S.; Lombardi, P.; Cutrignelli, M.I.; Moniello, G.; Grossi, M.; Tozzi, B.; Musco, N.; et al. The influence of diet supplementation with Saccharomyces cerevisiae or Saccharomyces cerevisiae plus Aspergillus oryzae on milk yield of Cilentana grazing dairy goats. Small Rumin. Res. 2016, 135, 90–94. [Google Scholar] [CrossRef]
- Campanile, G.; Zicarelli, F.; Vecchio, D.; Pacelli, C.; Neglia, G.; Balestrieri, A.; Di Palo, R.; Infascelli, F. Effects of Saccharomyces cerevisiae on in vivo organic matter digestibility and milk yield in buffalo cows. Livest. Sci. 2008, 114, 358–361. [Google Scholar] [CrossRef]
- Higginbotham, G.E.; Santos, J.E.; Juchem, S.O.; DePeters, E.J. Effect of feeding Aspergillus oryzae extract on milk production and rumen parameters. Livest. Prod. Sci. 2004, 86, 55–59. [Google Scholar] [CrossRef]
- Chiquette, J. Saccharomyces cerevisiae and Aspergillus oryzae, used alone or in combination, as a feed supplement for beef and dairy cattle. Can. J. Anim. Sci. 1995, 75, 405–415. [Google Scholar] [CrossRef]
- Chuang, W.Y.; Hsieh, Y.C.; Lee, T.-T. The effects of fungal feed additives in animals: A review. Animals 2020, 10, 805. [Google Scholar] [CrossRef]
- Martin, S.; Nisbet, D. Effects of Aspergillus oryzae fermentation extract on fermentation of amino acids, bermudagrass and starch by mixed ruminal microorganisms in vitro. J. Anim. Sci. 1990, 68, 2142–2149. [Google Scholar] [CrossRef]
- Takiya, C.S.; Calomeni, G.D.; Silva, T.H.; Vendramini, T.H.A.; Silva, G.G.; Consentini, C.E.C.; Bertoni, J.C.; Zilio, E.M.C.; Rennó, F.P. Increasing dietary doses of an Aspergillus oryzae extract with alpha-amylase activity on nutrient digestibility and ruminal fermentation of lactating dairy cows. Anim. Feed Sci. Tech. 2017, 228, 159–167. [Google Scholar] [CrossRef]
- Sato, Y.; Tominaga, K.; Aoki, H.; Murayama, M.; Oishi, K.; Hirooka, H.; Yoshida, T.; Kumagai, H. Calcium salts of long-chain fatty acids from linseed oil decrease methane production by altering the rumen microbiome in vitro. PLoS ONE 2020, 15, e0242158. [Google Scholar] [CrossRef]
- Shen, J.; Liu, Z.; Yu, Z.; Zhu, W. Monensin and nisin affect rumen fermentation and microbiota differently in vitro. Front. Microbiol. 2017, 8, 1111. [Google Scholar] [CrossRef] [PubMed]
- Yoon, I.; Stern, M.D. Effects of Saccharomyces cerevisiae and Aspergillus oryzae cultures on ruminal fermentation in dairy cows. J. Dairy Sci. 1996, 79, 411–417. [Google Scholar] [CrossRef]
- Beharka, A.; Nagaraja, T. Effect of Aspergillus oryzae extract alone or in combination with antimicrobial compounds on ruminal bacteria. J. Dairy Sci. 1998, 81, 1591–1598. [Google Scholar] [CrossRef]
- Janssen, P.H. Identifying the dominant soil bacterial taxa in libraries of 16S rRNA and 16S rRNA genes. Appl. Environ. Microbiol. 2006, 72, 1719–1728. [Google Scholar] [CrossRef]
- Jose, V.L.; Appoothy, T.; More, R.P.; Arun, A.S. Metagenomic insights into the rumen microbial fibrolytic enzymes in Indian crossbred cattle fed finger millet straw. AMB Express 2017, 7, 13. [Google Scholar] [CrossRef]
- Zeitz, J.O.; Guertler, P.; Pfaffl, M.W.; Eisenreich, R.; Wiedemann, S.; Schwarz, F.J. Effect of non-starch-polysaccharide-degrading enzymes as feed additive on the rumen bacterial population in non-lactating cows quantified by real-time PCR. J. Anim. Physiol. Anim. Nutr. 2013, 97, 1104–1113. [Google Scholar] [CrossRef] [PubMed]
- Bora, L.; Bora, M. Optimization of extracellular thermophilic highly alkaline lipase from thermophilic bacillus sp isolated from hotspring of Arunachal Pradesh, India. Braz. J. Microbiol. 2012, 43, 30–42. [Google Scholar] [CrossRef] [PubMed]
- Watabe, Y.; Suzuki, Y.; Koike, S.; Shimamoto, S.; Kobayashi, Y. Cellulose acetate, a new candidate feed supplement for ruminant animals: In vitro evaluations. J. Dairy Sci. 2018, 101, 10929–10938. [Google Scholar] [CrossRef] [PubMed]


| Ingredient | Level (%) |
|---|---|
| Corn | 4.95 |
| Soybean hull | 2.78 |
| Soybean meal | 7.61 |
| Molasses beet | 2.23 |
| Whole cottonseed | 2.88 |
| NaHCO3 | 0.28 |
| Yeast powder | 0.06 |
| Steam flaked corn | 9.74 |
| Alfalfa hay | 7.93 |
| Whole corn silage | 50.07 |
| Rumen-pass fatty acid | 0.42 |
| Corn bran | 2.70 |
| Distillers dried grains and soluble | 4.17 |
| Oat hay | 2.31 |
| Mycotoxin remover agent | 0.06 |
| Premix 1 | 1.81 |
| Item 1 | TMR | Corn Silage | Oat Hay | Alfalfa Hay |
|---|---|---|---|---|
| DM, % | 94.6 ± 0.3 | 93.5 ± 0.2 | 94.1 ± 0.2 | 93.4 ± 0.3 |
| CP, % | 16.0 ± 0.3 | 8.6 ± 0.1 | 6.2 ± 0.2 | 21.4 ± 0.5 |
| NDF, % | 38.2 ± 0.6 | 54.0 ± 0.9 | 56.9 ± 1.1 | 41.1 ± 0.6 |
| ADF, % | 25.3 ± 0.4 | 34.3 ± 0.6 | 33.9 ± 0.4 | 26.6 ± 0.3 |
| Starch, % | 26.8 ± 0.3 | 28.6 ± 0.2 | - | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kong, F.; Lu, N.; Liu, Y.; Zhang, S.; Jiang, H.; Wang, H.; Wang, W.; Li, S. Aspergillus oryzae and Aspergillus niger Co-Cultivation Extract Affects In Vitro Degradation, Fermentation Characteristics, and Bacterial Composition in a Diet-Specific Manner. Animals 2021, 11, 1248. https://doi.org/10.3390/ani11051248
Kong F, Lu N, Liu Y, Zhang S, Jiang H, Wang H, Wang W, Li S. Aspergillus oryzae and Aspergillus niger Co-Cultivation Extract Affects In Vitro Degradation, Fermentation Characteristics, and Bacterial Composition in a Diet-Specific Manner. Animals. 2021; 11(5):1248. https://doi.org/10.3390/ani11051248
Chicago/Turabian StyleKong, Fanlin, Na Lu, Yanfang Liu, Shu Zhang, Hongqin Jiang, Haomin Wang, Wei Wang, and Shengli Li. 2021. "Aspergillus oryzae and Aspergillus niger Co-Cultivation Extract Affects In Vitro Degradation, Fermentation Characteristics, and Bacterial Composition in a Diet-Specific Manner" Animals 11, no. 5: 1248. https://doi.org/10.3390/ani11051248
APA StyleKong, F., Lu, N., Liu, Y., Zhang, S., Jiang, H., Wang, H., Wang, W., & Li, S. (2021). Aspergillus oryzae and Aspergillus niger Co-Cultivation Extract Affects In Vitro Degradation, Fermentation Characteristics, and Bacterial Composition in a Diet-Specific Manner. Animals, 11(5), 1248. https://doi.org/10.3390/ani11051248





