The Influence of Diet Change and Oral Metformin on Blood Glucose Regulation and the Fecal Microbiota of Healthy Horses
Abstract
Simple Summary
Abstract
1. Introduction
2. Methods
2.1. Horses and Experimental Design
2.2. Metformin Compounding and Administration
2.3. Clinical Pathology Testing
2.3.1. Plasma ACTH Concentration
2.3.2. Blood Glucose Concentration
2.3.3. Serum Insulin Concentration
2.3.4. Oral Sugar Test
2.4. Fecal Sample Collection and DNA Extraction
2.5. 16S rRNA Amplicon Library Preparation and Sequencing
2.6. Bioinformatics
2.7. Data Availability
2.8. Statistics
3. Results
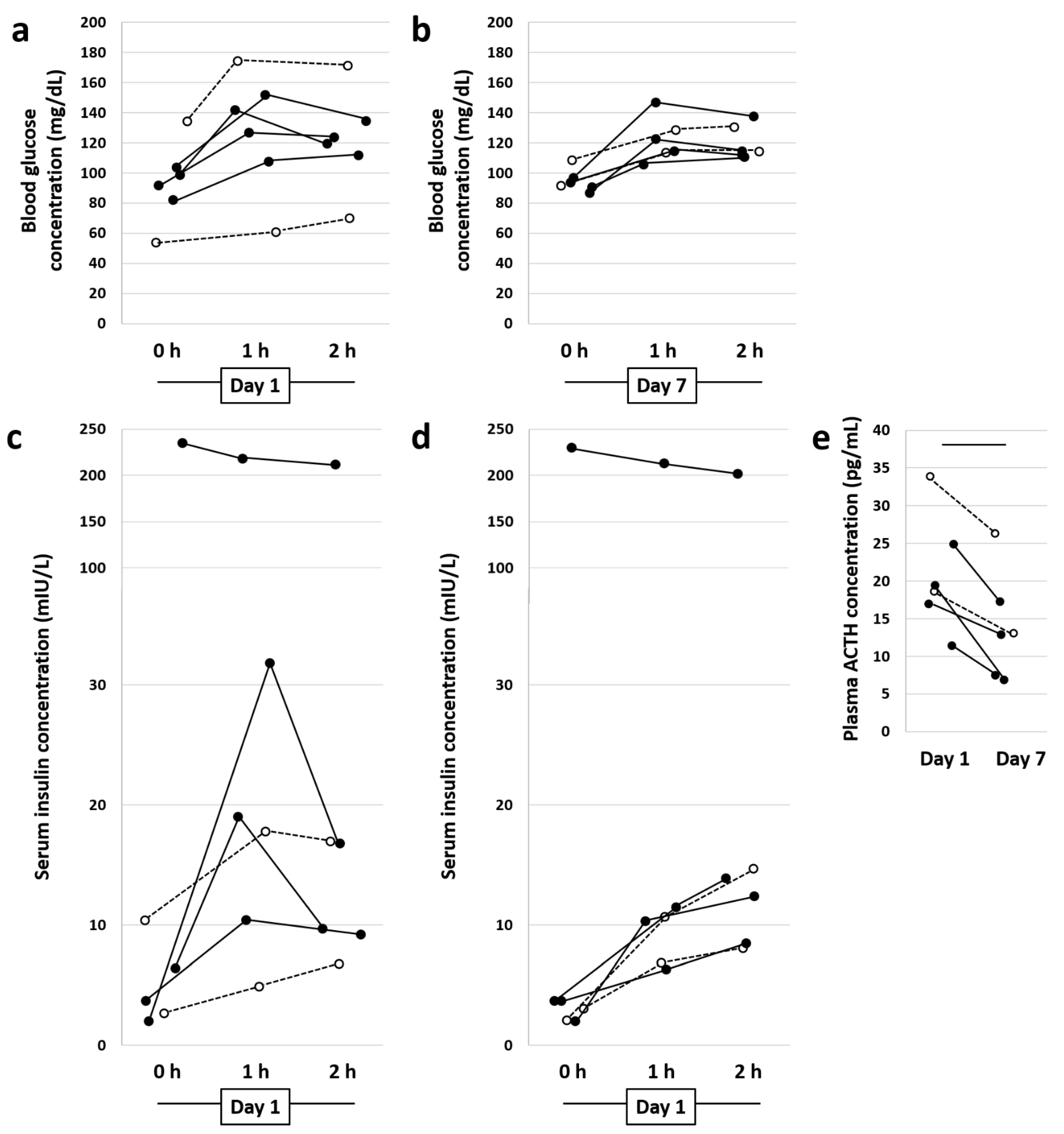
3.1. Diet Change and Metformin Result in Normalization of Blood Glucose and Plasma Serum Insulin
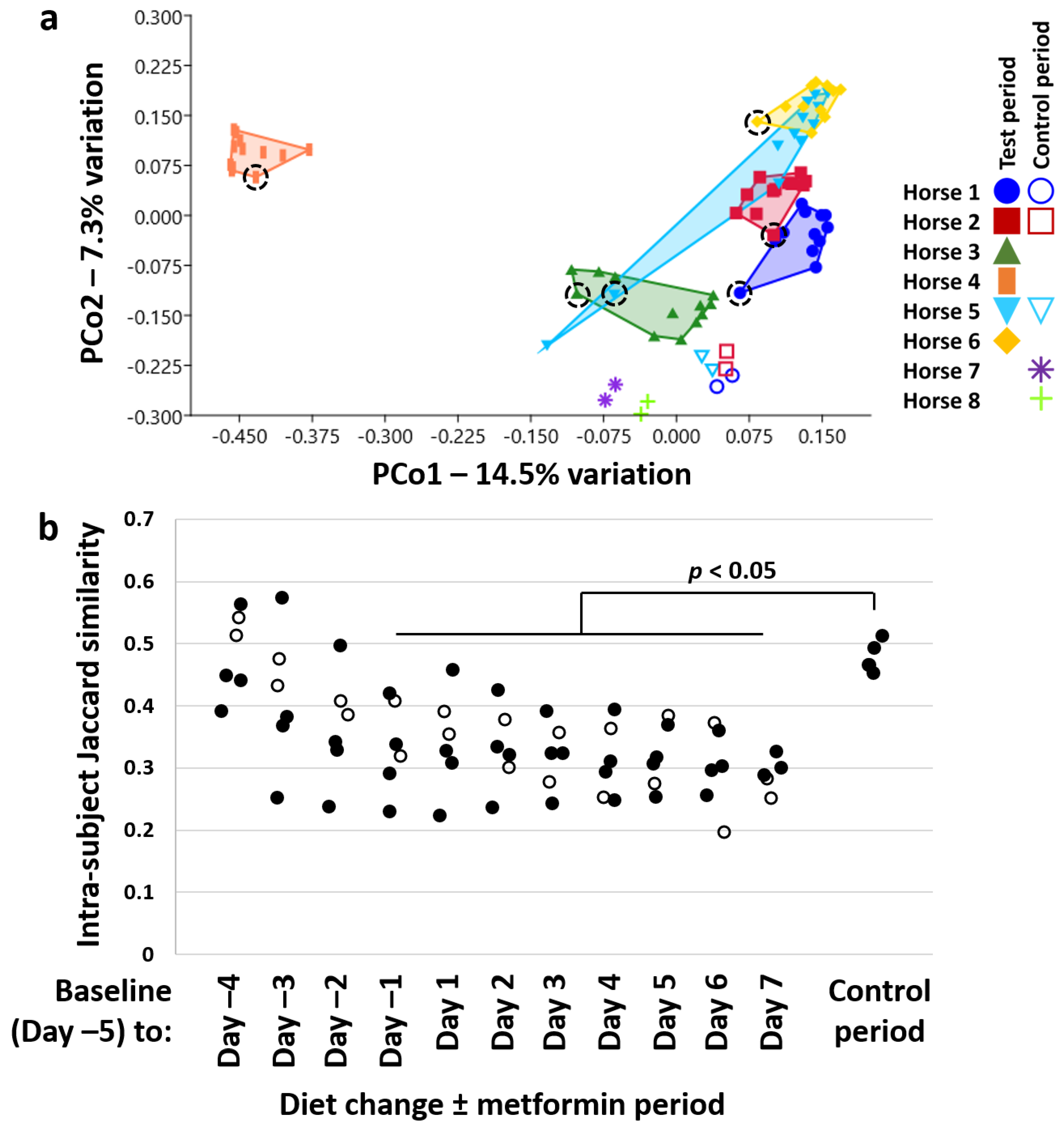
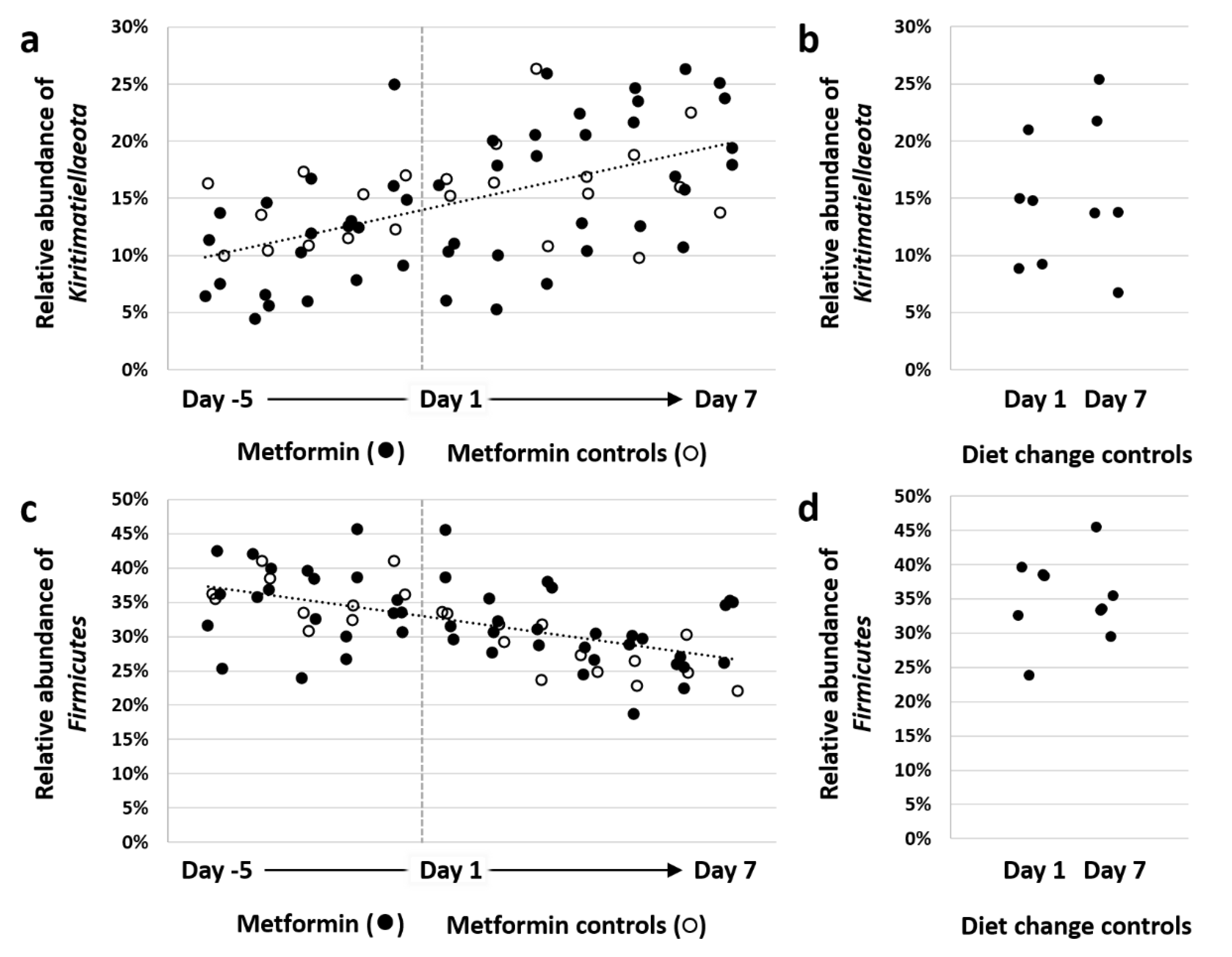
3.2. Diet Change and Metformin Result in Shift in β-Diversity and Proliferation of the Phylum Kiritimatiellaeota
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Frank, N.; Geor, R.J.; Bailey, S.R.; Durham, A.E.; Johnson, P.J.; American College of Veterinary Internal, M. Equine metabolic syndrome. J. Vet. Intern. Med. 2010, 24, 467–475. [Google Scholar] [CrossRef] [PubMed]
- Johnson, P.J. The equine metabolic syndrome peripheral Cushing’s syndrome. Vet. Clin. North. Am. Equine Pract. 2002, 18, 271–293. [Google Scholar] [CrossRef]
- Muno, J.; Gallatin, L.; Geor, R.J.; Anderson, G.A.; Hinchcliff, K.W. Prevalence and risk factors for hyperinsulinemia in clinically normal horses in central Ohio. J. Vet. Intern. Med. 2009, 23, 721. [Google Scholar]
- Pleasant, R.S.; Suagee, J.K.; Thatcher, C.D.; Elvinger, F.; Geor, R.J. Adiposity, plasma insulin, leptin, lipids, and oxidative stress in mature light breed horses. J. Vet. Intern. Med. 2013, 27, 576–582. [Google Scholar] [CrossRef] [PubMed]
- Morgan, R.A.; McGowan, T.W.; McGowan, C.M. Prevalence and risk factors for hyperinsulinaemia in ponies in Queensland, Australia. Aust. Vet. J. 2014, 92, 101–106. [Google Scholar] [CrossRef] [PubMed]
- Sessions-Bresnahan, D.R.; Carnevale, E.M. The effect of equine metabolic syndrome on the ovarian follicular environment. J. Anim. Sci. 2014, 92, 1485–1494. [Google Scholar] [CrossRef] [PubMed]
- Durham, A.E.; Frank, N.; McGowan, C.M.; Menzies-Gow, N.J.; Roelfsema, E.; Vervuert, I.; Feige, K.; Fey, K. ECEIM consensus statement on equine metabolic syndrome. J. Vet. Intern. Med. 2019, 33, 335–349. [Google Scholar] [CrossRef] [PubMed]
- Shu, Y.; Sheardown, S.A.; Brown, C.; Owen, R.P.; Zhang, S.; Castro, R.A.; Ianculescu, A.G.; Yue, L.; Lo, J.C.; Burchard, E.G.; et al. Effect of genetic variation in the organic cation transporter 1 (OCT1) on metformin action. J. Clin. Investig. 2007, 117, 1422–1431. [Google Scholar] [CrossRef]
- Wang, D.S.; Jonker, J.W.; Kato, Y.; Kusuhara, H.; Schinkel, A.H.; Sugiyama, Y. Involvement of organic cation transporter 1 in hepatic and intestinal distribution of metformin. J. Pharmacol. Exp. Ther. 2002, 302, 510–515. [Google Scholar] [CrossRef] [PubMed]
- de la Cuesta-Zuluaga, J.; Mueller, N.T.; Corrales-Agudelo, V.; Velasquez-Mejia, E.P.; Carmona, J.A.; Abad, J.M.; Escobar, J.S. Metformin Is Associated with Higher Relative Abundance of Mucin-Degrading Akkermansia muciniphila and Several Short-Chain Fatty Acid-Producing Microbiota in the Gut. Diabetes Care 2017, 40, 54–62. [Google Scholar] [CrossRef]
- Shin, N.R.; Lee, J.C.; Lee, H.Y.; Kim, M.S.; Whon, T.W.; Lee, M.S.; Bae, J.W. An increase in the Akkermansia spp. population induced by metformin treatment improves glucose homeostasis in diet-induced obese mice. Gut 2014, 63, 727–735. [Google Scholar] [CrossRef]
- Sun, L.; Xie, C.; Wang, G.; Wu, Y.; Wu, Q.; Wang, X.; Liu, J.; Deng, Y.; Xia, J.; Chen, B.; et al. Gut microbiota and intestinal FXR mediate the clinical benefits of metformin. Nat. Med. 2018, 24, 1919–1929. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Esteve, E.; Tremaroli, V.; Khan, M.T.; Caesar, R.; Manneras-Holm, L.; Stahlman, M.; Olsson, L.M.; Serino, M.; Planas-Felix, M.; et al. Metformin alters the gut microbiome of individuals with treatment-naive type 2 diabetes, contributing to the therapeutic effects of the drug. Nat. Med. 2017, 23, 850–858. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Lee, Y.; Kim, J.; An, J.; Lee, S.; Kong, H.; Song, Y.; Lee, C.K.; Kim, K. Modulation of the gut microbiota by metformin improves metabolic profiles in aged obese mice. Gut Microbes 2018, 9, 155–165. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhao, Y.; Xu, J.; Xue, Z.; Zhang, M.; Pang, X.; Zhang, X.; Zhao, L. Modulation of gut microbiota by berberine and metformin during the treatment of high-fat diet-induced obesity in rats. Sci. Rep. 2015, 5, 14405. [Google Scholar] [CrossRef] [PubMed]
- Ericsson, A.C.; Johnson, P.J.; Lopes, M.A.; Perry, S.C.; Lanter, H.R. A Microbiological Map of the Healthy Equine Gastrointestinal Tract. PLoS ONE 2016, 11, e0166523. [Google Scholar] [CrossRef] [PubMed]
- Elzinga, S.E.; Weese, J.S.; Adams, A.A. Comparison of the fecal microbiota in horses with equine metabolic syndrome and metabolically normal controls fed a similar all-forage diet. J. Equine Vet. Sci 2016, 44, 9–16. [Google Scholar] [CrossRef]
- Tinworth, K.D.; Boston, R.C.; Harris, P.A.; Sillence, M.N.; Raidal, S.L.; Noble, G.K. The effect of oral metformin on insulin sensitivity in insulin-resistant ponies. Vet. J. 2012, 191, 79–84. [Google Scholar] [CrossRef]
- Vick, M.M.; Sessions, D.R.; Murphy, B.A.; Kennedy, E.L.; Reedy, S.E.; Fitzgerald, B.P. Obesity is associated with altered metabolic and reproductive activity in the mare: Effects of metformin on insulin sensitivity and reproductive cyclicity. Reprod. Fertil. Dev. 2006, 18, 609–617. [Google Scholar] [CrossRef] [PubMed]
- Durham, A.E.; Rendle, D.I.; Newton, J.E. The effect of metformin on measurements of insulin sensitivity and beta cell response in 18 horses and ponies with insulin resistance. Equine Vet. J. 2008, 40, 493–500. [Google Scholar] [CrossRef]
- Rendle, D.I.; Rutledge, F.; Hughes, K.J.; Heller, J.; Durham, A.E. Effects of metformin hydrochloride on blood glucose and insulin responses to oral dextrose in horses. Equine Vet. J. 2013, 45, 751–754. [Google Scholar] [CrossRef] [PubMed]
- Tinworth, K.D.; Edwards, S.; Noble, G.K.; Harris, P.A.; Sillence, M.N.; Hackett, L.P. Pharmacokinetics of metformin after enteral administration in insulin-resistant ponies. Am. J. Vet. Res. 2010, 71, 1201–1206. [Google Scholar] [CrossRef]
- Hustace, J.L.; Firshman, A.M.; Mata, J.E. Pharmacokinetics and bioavailability of metformin in horses. Am. J. Vet. Res. 2009, 70, 665–668. [Google Scholar] [CrossRef]
- Smith, S.; Harris, P.A.; Menzies-Gow, N.J. Comparison of the in-feed glucose test and the oral sugar test. Equine Vet. J. 2016, 48, 224–227. [Google Scholar] [CrossRef] [PubMed]
- Walters, W.A.; Caporaso, J.G.; Lauber, C.L.; Berg-Lyons, D.; Fierer, N.; Knight, R. PrimerProspector: De novo design and taxonomic analysis of barcoded polymerase chain reaction primers. Bioinformatics 2011, 27, 1159–1161. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Lauber, C.L.; Walters, W.A.; Berg-Lyons, D.; Lozupone, C.A.; Turnbaugh, P.J.; Fierer, N.; Knight, R. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. USA 2011, 108, 4516–4522. [Google Scholar] [CrossRef]
- Loy, A.; Maixner, F.; Wagner, M.; Horn, M. probeBase—An online resource for rRNA-targeted oligonucleotide probes: New features 2007. Nucleic Acids Res. 2007, 35, D800–D804. [Google Scholar] [CrossRef] [PubMed]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glockner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- Xia, J.; Psychogios, N.; Young, N.; Wishart, D.S. MetaboAnalyst: A web server for metabolomic data analysis and interpretation. Nucleic Acids Res. 2009, 37, W652–W660. [Google Scholar] [CrossRef] [PubMed]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Hammer, O.; Harper, D.A.T. PAST: Paleontological statistics software package for education and data analysis. Palaeontol. Electron. 2011, 4, 1–9. [Google Scholar]
- Spring, S.; Bunk, B.; Sproer, C.; Schumann, P.; Rohde, M.; Tindall, B.J.; Klenk, H.P. Characterization of the first cultured representative of Verrucomicrobia subdivision 5 indicates the proposal of a novel phylum. ISME J. 2016, 10, 2801–2816. [Google Scholar] [CrossRef] [PubMed]
- Hugenholtz, P.; Goebel, B.M.; Pace, N.R. Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity. J. Bacteriol. 1998, 180, 4765–4774. [Google Scholar] [CrossRef]
- Shepherd, M.L.; Swecker, W.S., Jr.; Jensen, R.V.; Ponder, M.A. Characterization of the fecal bacteria communities of forage-fed horses by pyrosequencing of 16S rRNA V4 gene amplicons. FEMS Microbiol. Lett. 2012, 326, 62–68. [Google Scholar] [CrossRef]
- Kuczynski, J.; Stombaugh, J.; Walters, W.A.; Gonzalez, A.; Caporaso, J.G.; Knight, R. Using QIIME to analyze 16S rRNA gene sequences from microbial communities. Curr. Protoc. Bioinform. 2011, 10, 10–17. [Google Scholar] [CrossRef]
- Bokulich, N.A.; Kaehler, B.D.; Rideout, J.R.; Dillon, M.; Bolyen, E.; Knight, R.; Huttley, G.A.; Gregory Caporaso, J. Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2′s q2-feature-classifier plugin. Microbiome 2018, 6, 90. [Google Scholar] [CrossRef] [PubMed]
- Willing, B.; Voros, A.; Roos, S.; Jones, C.; Jansson, A.; Lindberg, J.E. Changes in faecal bacteria associated with concentrate and forage-only diets fed to horses in training. Equine Vet. J. 2009, 41, 908–914. [Google Scholar] [CrossRef] [PubMed]
- Daly, K.; Proudman, C.J.; Duncan, S.H.; Flint, H.J.; Dyer, J.; Shirazi-Beechey, S.P. Alterations in microbiota and fermentation products in equine large intestine in response to dietary variation and intestinal disease. Br. J. Nutr. 2012, 107, 989–995. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, K.A.; Kittelmann, S.; Rogers, C.W.; Gee, E.K.; Bolwell, C.F.; Bermingham, E.N.; Thomas, D.G. Faecal microbiota of forage-fed horses in New Zealand and the population dynamics of microbial communities following dietary change. PLoS ONE 2014, 9, e112846. [Google Scholar] [CrossRef] [PubMed]
- Salem, S.E.; Maddox, T.W.; Berg, A.; Antczak, P.; Ketley, J.M.; Williams, N.J.; Archer, D.C. Variation in faecal microbiota in a group of horses managed at pasture over a 12-month period. Sci. Rep. 2018, 8, 8510. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ericsson, A.C.; Johnson, P.J.; Gieche, L.M.; Zobrist, C.; Bucy, K.; Townsend, K.S.; Martin, L.M.; LaCarrubba, A.M. The Influence of Diet Change and Oral Metformin on Blood Glucose Regulation and the Fecal Microbiota of Healthy Horses. Animals 2021, 11, 976. https://doi.org/10.3390/ani11040976
Ericsson AC, Johnson PJ, Gieche LM, Zobrist C, Bucy K, Townsend KS, Martin LM, LaCarrubba AM. The Influence of Diet Change and Oral Metformin on Blood Glucose Regulation and the Fecal Microbiota of Healthy Horses. Animals. 2021; 11(4):976. https://doi.org/10.3390/ani11040976
Chicago/Turabian StyleEricsson, Aaron C., Philip J. Johnson, Lyndsy M. Gieche, Chelsea Zobrist, Katie Bucy, Kile S. Townsend, Lynn M. Martin, and Alison M. LaCarrubba. 2021. "The Influence of Diet Change and Oral Metformin on Blood Glucose Regulation and the Fecal Microbiota of Healthy Horses" Animals 11, no. 4: 976. https://doi.org/10.3390/ani11040976
APA StyleEricsson, A. C., Johnson, P. J., Gieche, L. M., Zobrist, C., Bucy, K., Townsend, K. S., Martin, L. M., & LaCarrubba, A. M. (2021). The Influence of Diet Change and Oral Metformin on Blood Glucose Regulation and the Fecal Microbiota of Healthy Horses. Animals, 11(4), 976. https://doi.org/10.3390/ani11040976





