Direct Phenotyping and Principal Component Analysis of Type Traits Implicate Novel QTL in Bovine Mastitis through Genome-Wide Association
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Phenotyping
2.2. Principal Component Analysis
2.3. Genotyping and Quality Control
2.4. Genome-Wide Association
3. Results
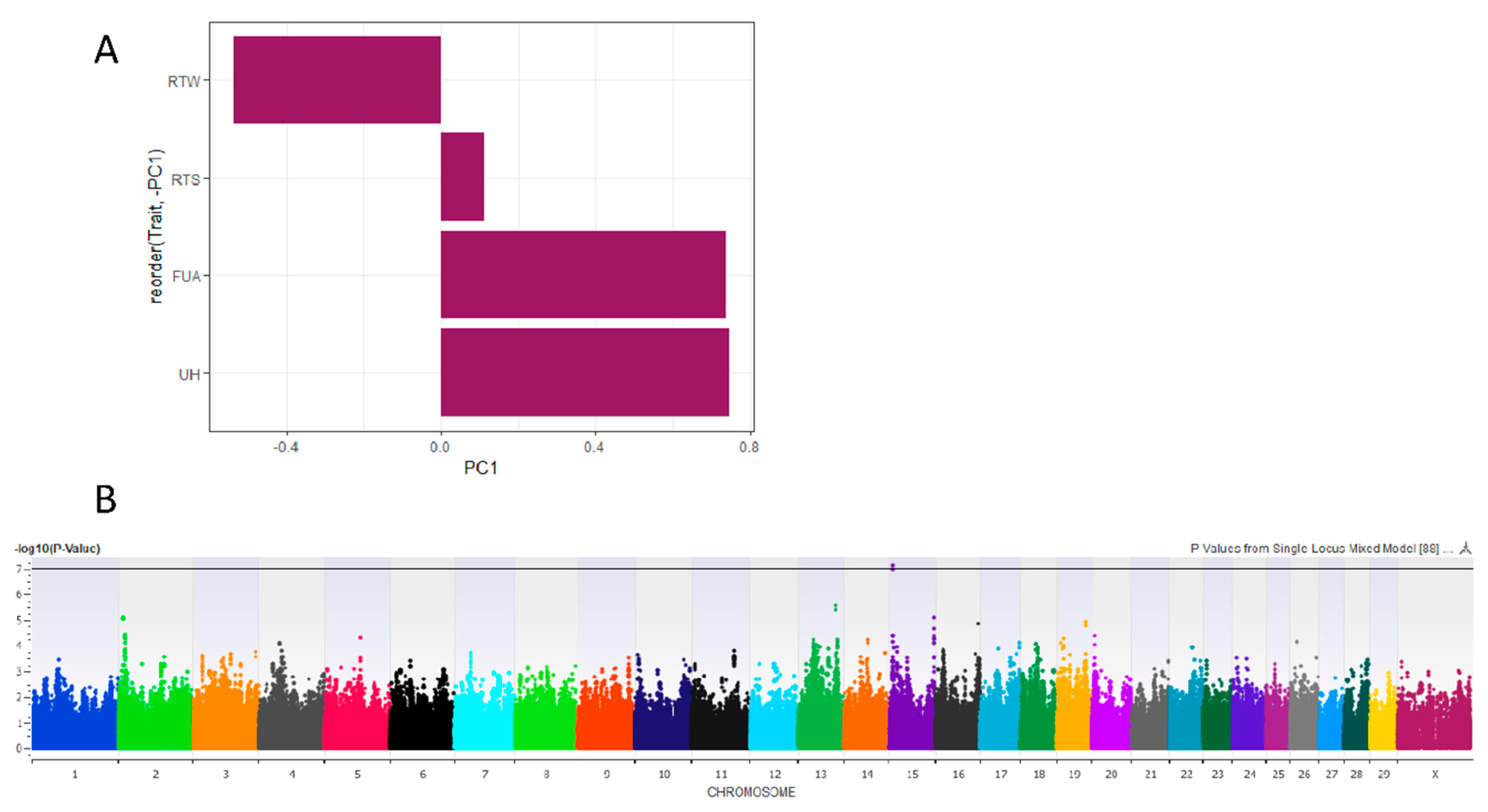
3.1. Principal Component Analysis
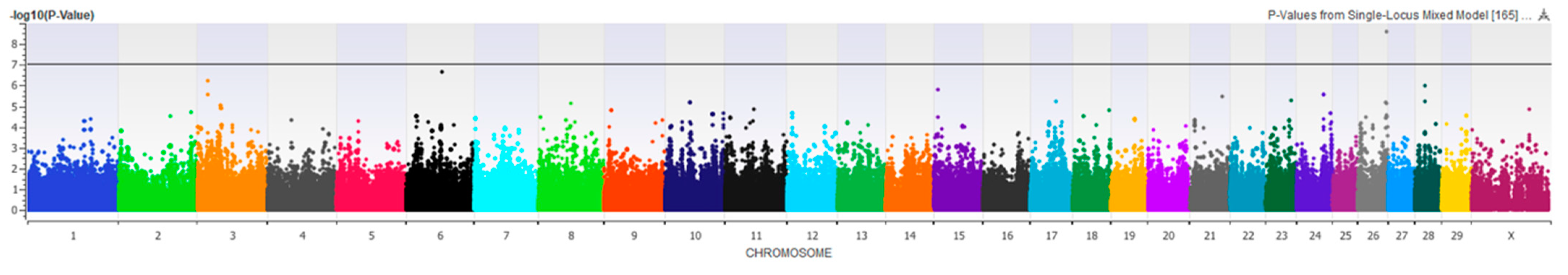
3.2. Genome-Wide Association
3.2.1. Risk PC1
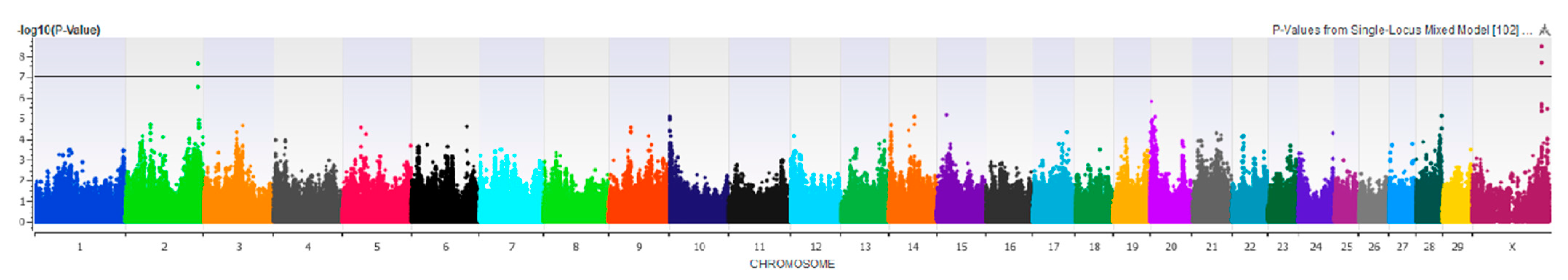
3.2.2. Fore Udder Attachment
3.2.3. Udder Depth
3.2.4. Rear Udder Height
3.2.5. Udder Width
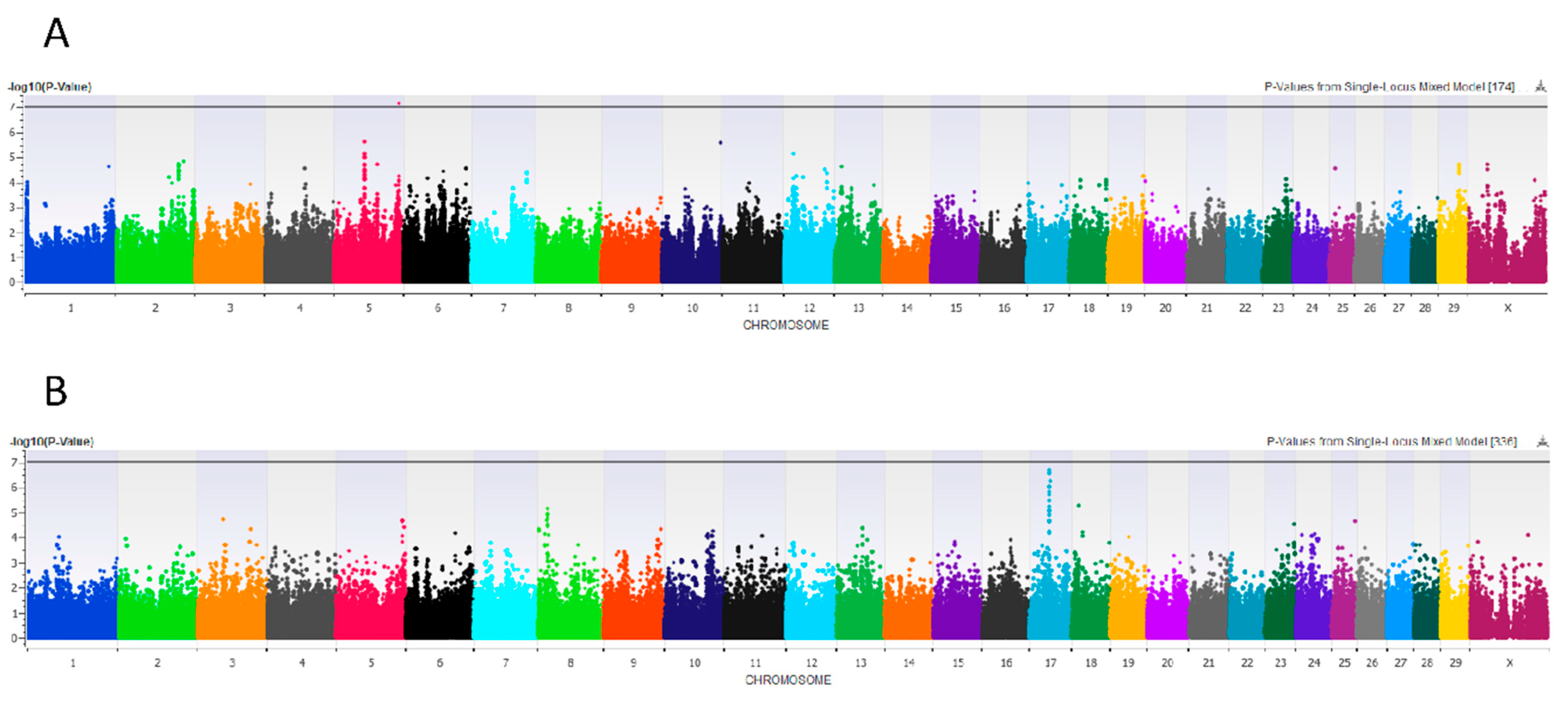
3.2.6. Front Teat Length and Width
3.2.7. Rear Teat Length, Width, and End Shape
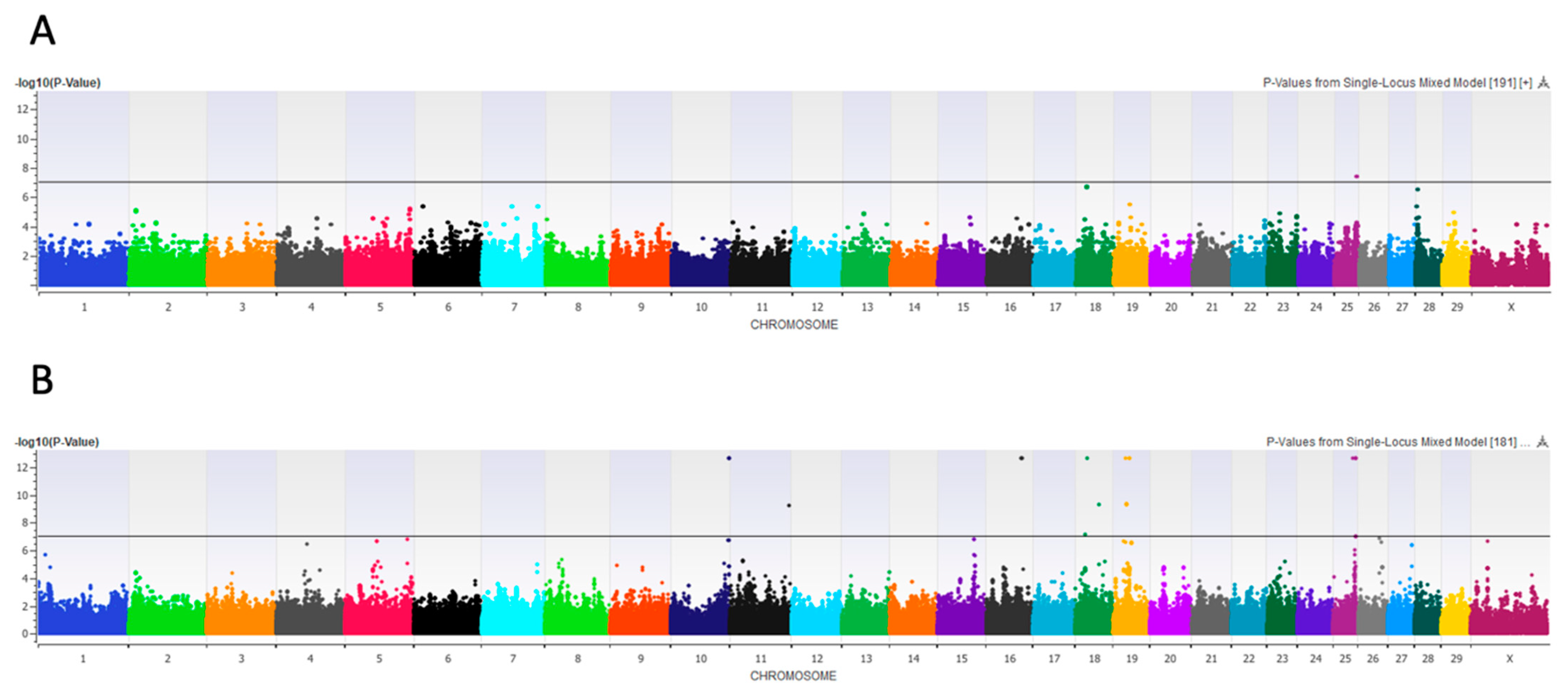
3.2.8. Front Teat Placement
4. Discussion
4.1. Risk PC1
4.2. Fore Udder Attachment
4.3. Rear Teat End Shape
4.4. Rear Teat Width
4.5. Rear Udder Height
4.6. Primiparous Subset
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- IGem, C. Legendairy: Market Analysis. 2016. Available online: http://2016.igem.org/wiki/images/2/21/T--Cornell_NY--MarketAnalysis.pdf (accessed on 1 August 2019).
- Weigel, K.; VanRaden, P.; Norman, H.; Grosu, H. A 100-Year Review: Methods and impact of genetic selection in dairy cattle—From daughter–dam comparisons to deep learning algorithms. J. Dairy Sci. 2017, 100, 10234–10250. [Google Scholar] [CrossRef] [PubMed]
- Ruegg, P.L. A 100-Year Review: Mastitis detection, management, and prevention. J. Dairy Sci. 2017, 100, 10381–10397. [Google Scholar] [CrossRef] [PubMed]
- Miles, A.M.; Huson, H.J. Graduate Student Literature Review: Understanding the genetic mechanisms underlying mastitis. J. Dairy Sci. 2021, 104, 1183–1191. [Google Scholar] [CrossRef] [PubMed]
- O’Bleness, G.; Van Vleck, L.; Henderson, C. Heritabilities of Some Type Appraisal Traits and Their Genetic and Phenotypic Correlations with Production. J. Dairy Sci. 1960, 43, 1490–1498. [Google Scholar] [CrossRef]
- Wiggans, G.; VanRaden, P.; Cooper, T. The genomic evaluation system in the United States: Past, present, future. J. Dairy Sci. 2011, 94, 3202–3211. [Google Scholar] [CrossRef]
- Interbull. Description of National Genetic Evaluation Systems: Conformation 2013. 2013. Available online: https://queries.uscdcb.com/reference/Form_GENO_Conformation_1302.pdf (accessed on 1 August 2019).
- Seykora, A.J.; McDaniel, B.T. Udder and Teat Morphology Related to Mastitis Resistance: A Review. J. Dairy Sci. 1985, 68, 2087–2093. [Google Scholar] [CrossRef]
- Rupp, R.; Boichard, D. Genetic Parameters for Clinical Mastitis, Somatic Cell Score, Production, Udder Type Traits, and Milking Ease in First Lactation Holsteins. J. Dairy Sci. 1999, 82, 2198–2204. [Google Scholar] [CrossRef]
- Gaddis, K.P.; Cole, J.; Clay, J.; Maltecca, C. Genomic selection for producer-recorded health event data in US dairy cattle. J. Dairy Sci. 2014, 97, 3190–3199. [Google Scholar] [CrossRef] [PubMed]
- DeGroot, B.; Keown, J.; Van Vleck, L.; Marotz, E. Genetic Parameters and Responses of Linear Type, Yield Traits, and Somatic Cell Scores to Divergent Selection for Predicted Transmitting Ability for Type in Holsteins. J. Dairy Sci. 2002, 85, 1578–1585. [Google Scholar] [CrossRef]
- Seykora, A.; McDaniel, B. Genetics Statistics and Relationships of Teat and Udder Traits, Somatic Cell Counts, and Milk Production. J. Dairy Sci. 1986, 69, 2395–2407. [Google Scholar] [CrossRef]
- Nash, D.; Rogers, G.; Cooper, J.; Hargrove, G.; Keown, J.; Hansen, L. Heritability of Clinical Mastitis Incidence and Relationships with Sire Transmitting Abilities for Somatic Cell Score, Udder Type Traits, Productive Life, and Protein Yield. J. Dairy Sci. 2000, 83, 2350–2360. [Google Scholar] [CrossRef]
- Pausch, H.; Emmerling, R.; Schwarzenbacher, H.; Fries, R. A multi-trait meta-analysis with imputed sequence variants reveals twelve QTL for mammary gland morphology in Fleckvieh cattle. Genet. Sel. Evol. 2016, 48, 1–9. [Google Scholar] [CrossRef]
- García-Ruiz, A.; Cole, J.B.; VanRaden, P.M.; Wiggans, G.R.; Ruiz-López, F.J.; Van Tassell, C.P. Changes in genetic selection differentials and generation intervals in US Holstein dairy cattle as a result of genomic selection. Proc. Natl. Acad. Sci. USA 2016, 113, E3995–E4004. [Google Scholar] [CrossRef]
- Marete, A.; Lund, M.S.; Boichard, D.; Ramayo-Caldas, Y. A system-based analysis of the genetic determinism of udder conformation and health phenotypes across three French dairy cattle breeds. PLoS ONE 2018, 13, e0199931. [Google Scholar] [CrossRef] [PubMed]
- Tolleson, M.W.; Gill, C.A.; Herring, A.D.; Riggs, P.K.; Sawyer, J.E.; Sanders, J.O.; Riley, D.G. Association of udder traits with single nucleotide polymorphisms in crossbred Bos indicus–Bos taurus cows1,2. J. Anim. Sci. 2017, 95, 2399–2407. [Google Scholar] [CrossRef] [PubMed]
- Miles, A.M.; McArt, J.A.; Yepes, F.A.L.; Stambuk, C.R.; Virkler, P.D.; Huson, H.J. Udder and teat conformational risk factors for elevated somatic cell count and clinical mastitis in New York Holsteins. Prev. Vet. Med. 2019, 163, 7–13. [Google Scholar] [CrossRef] [PubMed]
- Bartlett, M.S. The effect of standardization on a χ2 approximation in factor analysis. Biometrika 1951, 38, 337–344. [Google Scholar] [CrossRef]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.; Daly, M.J.; et al. PLINK: A Tool Set for Whole-Genome Association and Population-Based Linkage Analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef]
- Kang, H.M.; Sul, J.H.; Service, S.K.; Zaitlen, N.A.; Kong, S.-Y.; Freimer, N.B.; Sabatti, C.; Eskin, E. Variance component model to account for sample structure in genome-wide association studies. Nat. Genet. 2010, 42, 348–354. [Google Scholar] [CrossRef]
- Lewis, C.M. Genetic association studies: Design, analysis and interpretation. Brief. Bioinform. 2002, 3, 146–153. [Google Scholar] [CrossRef]
- SNP and Variation Suite. Mixed Model Linear Analysis. 2017. Available online: https://doc.goldenhelix.com/SVS/latest/svsmanual/mixedModelMethods/mlm_analysis.html (accessed on 1 August 2019).
- O’Leary, N.A.; Wright, M.W.; Brister, J.R.; Ciufo, S.; Haddad, D.; McVeigh, R.; Rajput, B.; Robbertse, B.; Smith-White, B.; Ako-Adjei, D.; et al. Reference sequence (RefSeq) database at NCBI: Current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016, 44, D733–D745. [Google Scholar] [CrossRef]
- Hu, Z.-L.; Park, C.A.; Reecy, J.M. Building a livestock genetic and genomic information knowledgebase through integrative developments of Animal QTLdb and CorrDB. Nucleic Acids Res. 2019, 47, D701–D710. [Google Scholar] [CrossRef]
- Bonavita, R.; Walas, D.; Brown, A.K.; Luini, A.; Stephens, D.J.; Colanzi, A. Cep126 is required for pericentriolar satellite localisation to the centrosome and for primary cilium formation. Biol. Cell 2014, 106, 254–267. [Google Scholar] [CrossRef]
- Drake, A.C.; Khoury, M.; Leskov, I.; Iliopoulou, B.P.; Fragoso, M.; Lodish, H.; Chen, J. Human CD34+ CD133+ Hematopoietic Stem Cells Cultured with Growth Factors Including Angptl5 Efficiently Engraft Adult NOD-SCID Il2rγ−/− (NSG) Mice. PLoS ONE 2011, 6, e18382. [Google Scholar] [CrossRef] [PubMed]
- Gebreyesus, G.; Buitenhuis, A.J.; Poulsen, N.A.; Visker, M.H.P.W.; Zhang, Q.; Van Valenberg, H.J.F.; Sun, D.; Bovenhuis, H. Multi-population GWAS and enrichment analyses reveal novel genomic regions and promising candidate genes underlying bovine milk fatty acid composition. BMC Genom. 2019, 20, 1–16. [Google Scholar] [CrossRef]
- Cai, Z.; Guldbrandtsen, B.; Lund, M.S.; Sahana, G. Dissecting closely linked association signals in combination with the mammalian phenotype database can identify candidate genes in dairy cattle. BMC Genet. 2019, 20, 15. [Google Scholar] [CrossRef] [PubMed]
- Kiser, J.N.; Keuter, E.M.; Seabury, C.M.; Neupane, M.; Moraes, J.G.N.; Dalton, J.; Burns, G.W.; Spencer, T.E.; Neibergs, H.L. Validation of 46 loci associated with female fertility traits in cattle. BMC Genom. 2019, 20, 1–13. [Google Scholar] [CrossRef]
- Thrasher, A.J.; Burns, S.O. WASP: A key immunological multitasker. Nat. Rev. Immunol. 2010, 10, 182–192. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Zhang, W.; Lu, Y.; Yan, X.; Yan, X.; Zhu, X.; Liu, W.; Yang, Y.; Zhou, T. NudC regulates actin dynamics and ciliogenesis by stabilizing cofilin 1. Cell Res. 2016, 26, 239–253. [Google Scholar] [CrossRef]
- Lee, S.; Kong, Y.; Weatherbee, S.D. Forward genetics identifies Kdf1/1810019J16Rik as an essential regulator of the proliferation–differentiation decision in epidermal progenitor cells. Dev. Biol. 2013, 383, 201–213. [Google Scholar] [CrossRef]
- Ghahary, A.; Karimi-Busheri, F.; Marcoux, Y.; Li, Y.; Tredget, E.E.; Li, L.; Zheng, J.; Karami, A.; Keller, B.O.; Weinfeld, M.; et al. Keratinocyte-Releasable Stratifin Functions as a Potent Collagenase-Stimulating Factor in Fibroblasts. J. Investig. Dermatol. 2004, 122, 1188–1197. [Google Scholar] [CrossRef]
- Plovsing, R.R.; Berg, R.M.; Munthe-Fog, L.; Konge, L.; Iversen, M.; Møller, K.; Garred, P. Alveolar recruitment of ficolin-3 in response to acute pulmonary inflammation in humans. Immunobiology 2016, 221, 690–697. [Google Scholar] [CrossRef]
- Moller, D.E.; Xia, C.H.; Tang, W.; Zhu, A.X.; Jakubowski, M. Human rsk isoforms: Cloning and characterization of tissue-specific expression. Am. J. Physiol. Physiol. 1994, 266, C351–C359. [Google Scholar] [CrossRef]
- Tian, H.; Miao, J.; Zhang, F.; Xiong, F.; Zhu, F.; Li, J.; Wang, X.; Chen, S.; Chen, J.; Huang, N.; et al. Non-histone nuclear protein HMGN2 differently regulates the urothelium barrier function by altering expression of antimicrobial peptides and tight junction protein genes in UPEC J96-infected bladder epithelial cell monolayer. Acta Biochim. Pol. 2018, 65, 93–100. [Google Scholar] [CrossRef]
- Braga, F.A.V.; Hertoghs, K.M.L.; Kragten, N.A.M.; Doody, G.M.; Barnes, N.A.; Remmerswaal, E.B.M.; Hsiao, C.-C.; Moerland, P.D.; Wouters, D.; Derks, I.A.M.; et al. Blimp-1 homolog Hobit identifies effector-type lymphocytes in humans. Eur. J. Immunol. 2015, 45, 2945–2958. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Zhen, Y.-Z.; Lin, Y.-J.; Liu, J.; Wei, J.; Xu, R.; Hu, G. KNDC1 knockdown protects human umbilical vein endothelial cells from senescence. Mol. Med. Rep. 2014, 10, 82–88. [Google Scholar] [CrossRef]
- Knapp, B.; Wolfrum, U. Adhesion GPCR-Related Protein Networks. Organotypic Models Drug Dev. 2016, 234, 147–178. [Google Scholar] [CrossRef]
- Lamas, B.; Michel, M.-L.; Waldschmitt, N.; Pham, H.-P.; Zacharioudaki, V.; Dupraz, L.; Delacre, M.; Natividad, J.M.; Da Costa, G.; Planchais, J.; et al. Card9 mediates susceptibility to intestinal pathogens through microbiota modulation and control of bacterial virulence. Gut 2017, 67, 1836–1844. [Google Scholar] [CrossRef] [PubMed]
- Siebel, C.; Lendahl, U. Notch Signaling in Development, Tissue Homeostasis, and Disease. Physiol. Rev. 2017, 97, 1235–1294. [Google Scholar] [CrossRef]
- Si, Y.; Wen, H.; Du, S. Genetic Mutations in jamb, jamc, and myomaker Revealed Different Roles on Myoblast Fusion and Muscle Growth. Mar. Biotechnol. 2018, 21, 111–123. [Google Scholar] [CrossRef]
- Bouwman, A.C.; Visker, M.H.; Van Arendonk, J.A.; Bovenhuis, H. Genomic regions associated with bovine milk fatty acids in both summer and winter milk samples. BMC Genet. 2012, 13, 93. [Google Scholar] [CrossRef]
- Zhou, C.; Li, C.; Cai, W.; Liu, S.; Yin, H.; Shi, S.; Zhang, Q.; Zhang, S. Genome-Wide Association Study for Milk Protein Composition Traits in a Chinese Holstein Population Using a Single-Step Approach. Front. Genet. 2019, 10, 72. [Google Scholar] [CrossRef]
- Yan, X.; Habedanck, R.; Nigg, E.A. A Complex of Two Centrosomal Proteins, CAP350 and FOP, Cooperates with EB1 in Microtubule Anchoring. Mol. Biol. Cell 2006, 17, 634–644. [Google Scholar] [CrossRef]
- Yamaguchi, H.; Kurosawa, Y.; Hashimoto, K. Expanded Genomic Organization of Conserved Mammalian MHC Class I-Related Genes, HumanMR1and Its Murine Ortholog. Biochem. Biophys. Res. Commun. 1998, 250, 558–564. [Google Scholar] [CrossRef]
- Nakad, B.; Fares, F.; Azzam, N.; Feiner, B.; Zilberlicht, A.; Abramov, Y. Estrogen receptor and laminin genetic polymorphism among women with pelvic organ prolapse. Taiwan J. Obstet. Gynecol. 2017, 56, 750–754. [Google Scholar] [CrossRef]
- Squire, J.; Zhou, A.; Hassel, B.A.; Nie, H.; Silverman, R.H. Localization of the Interferon-Induced, 2-5A-Dependent RNase Gene (RNS4) to Human Chromosome 1q25. Genomics 1994, 19, 174–175. [Google Scholar] [CrossRef]
- Dempsey, A.; Keating, S.E.; Carty, M.; Bowie, A.G. Poxviral protein E3–altered cytokine production reveals that DExD/H-box helicase 9 controls Toll-like receptor–stimulated immune responses. J. Biol. Chem. 2018, 293, 14989–15001. [Google Scholar] [CrossRef]
- Xu, Q.; Liu, X.; Wang, X.; Hua, Y.; Stoeger, T.; Huang, N.; Chen, J.; Li, J. Growth arrest-specific protein 7 regulates the murine M1 alveolar macrophage polarization. Immunol. Res. 2017, 65, 1065–1073. [Google Scholar] [CrossRef] [PubMed]
- Rolf, M.M.; Taylor, J.F.; Schnabel, R.D.; McKay, S.D.; McClure, M.C.; Northcutt, S.L.; Kerley, M.S.; Weaber, R.L. Genome--wide association analysis for feed efficiency in Angus cattle. Anim. Genet. 2011, 43, 367–374. [Google Scholar] [CrossRef] [PubMed]
- Sahana, G.; Guldbrandtsen, B.; Lund, M. Genome-wide association study for calving traits in Danish and Swedish Holstein cattle. J. Dairy Sci. 2011, 94, 479–486. [Google Scholar] [CrossRef] [PubMed]
- Bennewitz, J.; Reinsch, N.; Guiard, V.; Fritz, S.; Thomsen, H.; Looft, C.; Kühn, C.; Schwerin, M.; Weimann, C.; Erhardt, G.; et al. Multiple Quantitative Trait Loci Mapping with Cofactors and Application of Alternative Variants of the False Discovery Rate in an Enlarged Granddaughter Design. Genetics 2004, 168, 1019–1027. [Google Scholar] [CrossRef]
- Bennewitz, J.; Reinsch, N.; Grohs, C.; Levéziel, H.; Malafosse, A.; Thomsen, H.; Xu, N.; Looft, C.; Kühn, C.; Brockmann, G.A.; et al. Combined analysis of data from two granddaughter designs: A simple strategy for QTL con-firmation and increasing experimental power in dairy cattle. Genet. Sel. Evol. 2003, 35, 319–338. [Google Scholar] [CrossRef]
- Kirkpatrick, B.W.; Byla, B.M.; Gregory, K.E. Mapping quantitative trait loci for bovine ovulation rate. Mamm. Genome 2000, 11, 136–139. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Leach, R.J.; Craigmile, S.C.; Knott, S.A.; Williams, J.L.; Glass, E.J. Quantitative trait loci for variation in immune response to a Foot-and-Mouth Disease virus peptide. BMC Genet. 2010, 11, 107–114. [Google Scholar] [CrossRef] [PubMed]
- Leal, M.F.; Ribeiro, H.F.; Rey, J.A.; Pinto, G.R.; Smith, M.C.; Moreira-Nunes, C.A.; Assumpção, P.P.; Lamarão, L.M.; Calcagno, D.Q.; Montenegro, R.C.; et al. YWHAE silencing induces cell proliferation, invasion and migration through the up-regulation of CDC25B and MYC in gastric cancer cells: New insights about YWHAE role in the tumor development and metastasis process. Oncotarget 2016, 7, 85393–85410. [Google Scholar] [CrossRef] [PubMed]
- Burrows, K.; Antignano, F.; Bramhall, M.; Chenery, A.; Scheer, S.; Korinek, V.; Underhill, T.M.; Zaph, C. The transcriptional repressor HIC1 regulates intestinal immune homeostasis. Mucosal Immunol. 2017, 10, 1518–1528. [Google Scholar] [CrossRef]
- Yamagata, M.; Sanes, J.R. Dscam and Sidekick proteins direct lamina-specific synaptic connections in vertebrate retina. Nat. Cell Biol. 2008, 451, 465–469. [Google Scholar] [CrossRef]
- Li, B.; Fang, L.; Null, D.; Hutchison, J.; Connor, E.; VanRaden, P.; VandeHaar, M.; Tempelman, R.; Weigel, K.; Cole, J. High-density genome-wide association study for residual feed intake in Holstein dairy cattle. J. Dairy Sci. 2019, 102, 11067–11080. [Google Scholar] [CrossRef]
- Kühnemuth, B.; Mühlberg, L.; Schipper, M.; Griesmann, H.; Neesse, A.; Milosevic, N.; Wissniowski, T.; Buchholz, M.; Gress, T.M.; Michl, P. CUX1 modulates polarization of tumor-associated macrophages by antagonizing NF-κB signaling. Oncogene 2013, 34, 177–187. [Google Scholar] [CrossRef]
- Xu, H.; He, J.; Xu, S.; Xie, S.; Ma, L.; Zhang, Y.; Zhou, H.; Qu, L. A group of tissue--specific microRNAs contribute to the silencing of CUX1 in different cell lineages during development. J. Cell. Biochem. 2018, 119, 6238–6248. [Google Scholar] [CrossRef]
- Oltz, E.M.; Yancopoulos, G.D.; Morrow, M.A.; Rolink, A.; Lee, G.; Wong, F.; Kaplan, K.; Gillis, S.; Melchers, F.; Alt, F.W. A novel regulatory myosin light chain gene distinguishes pre-B cell subsets and is IL-7 inducible. EMBO J. 1992, 11, 2759–2767. [Google Scholar] [CrossRef]
- Chen, Y.; Zhao, J.; Li, D.; Hao, J.; He, P.; Wang, H.; Zhang, M. TRIM56 Suppresses Multiple Myeloma Progression by Activating TLR3/TRIF Signaling. Yonsei Med. J. 2018, 59, 43–50. [Google Scholar] [CrossRef]
- Patel, N.; Khan, A.O.; Mansour, A.; Mohamed, J.Y.; Al-Assiri, A.; Haddad, R.; Jia, X.; Xiong, Y.; Mégarbané, A.; Traboulsi, E.I.; et al. Mutations in ASPH Cause Facial Dysmorphism, Lens Dislocation, Anterior-Segment Abnormalities, and Spontaneous Filtering Blebs, or Traboulsi Syndrome. Am. J. Hum. Genet. 2014, 94, 755–759. [Google Scholar] [CrossRef]
- Fox, M.D.; Carson, V.J.; Feng, H.-Z.; Lawlor, M.W.; Gray, J.T.; Brigatti, K.W.; Jin, J.-P.; Strauss, K.A. TNNT1 nemaline myopathy: Natural history and therapeutic frontier. Hum. Mol. Genet. 2018, 27, 3272–3282. [Google Scholar] [CrossRef]
- Michenet, A.; Barbat, M.; Saintilan, R.; Venot, E.; Phocas, F. Detection of quantitative trait loci for maternal traits using high-density genotypes of Blonde d’Aquitaine beef cattle. BMC Genet. 2016, 17, 1–13. [Google Scholar] [CrossRef]
- Jones, J.D.G.; Vance, R.E.; Dangl, J.L. Intracellular innate immune surveillance devices in plants and animals. Science 2016, 354, aaf6395. [Google Scholar] [CrossRef]
- Schäfgen, J.; Cremer, K.; Becker, J.; Wieland, T.; Zink, A.M.; Kim, S.; Windheuser, I.C.; Kreiß, M.; Aretz, S.; Strom, T.M.; et al. De novo nonsense and frameshift variants of TCF20 in individuals with intellectual disability and postnatal overgrowth. Eur. J. Hum. Genet. 2016, 24, 1739–1745. [Google Scholar] [CrossRef] [PubMed]
- Asa, S.L.; Ramyar, L.; Murphy, P.R.; Li, A.W.; Ezzat, S. The Endogenous Fibroblast Growth Factor-2 Antisense Gene Product Regulates Pituitary Cell Growth and Hormone Production. Mol. Endocrinol. 2001, 15, 589–599. [Google Scholar] [CrossRef] [PubMed]
- Koledova, Z.; Zhang, X.; Streuli, C.; Clarke, R.B.; Klein, O.D.; Werb, Z.; Lu, P. SPRY1 regulates mammary epithelial morphogenesis by modulating EGFR-dependent stromal paracrine signaling and ECM remodeling. Proc. Natl. Acad. Sci. USA 2016, 113, E5731–E5740. [Google Scholar] [CrossRef] [PubMed]
- Ashwell, M.; Heyen, D.; Weller, J.; Ron, M.; Sonstegard, T.; Van Tassell, C.; Lewin, H. Detection of Quantitative Trait Loci Influencing Conformation Traits and Calving Ease in Holstein-Friesian Cattle. J. Dairy Sci. 2005, 88, 4111–4119. [Google Scholar] [CrossRef]
- Plante, Y.; Gibson, J.; Nadesalingam, J.; Mehrabani-Yeganeh, H.; Lefebvre, S.; Vandervoort, G.; Jansen, G. Detection of Quantitative Trait Loci Affecting Milk Production Traits on 10 Chromosomes in Holstein Cattle. J. Dairy Sci. 2001, 84, 1516–1524. [Google Scholar] [CrossRef]

| Trait | N | Model Type | Inheritance | Pseudo-Lambda 1 | Pseudo-Heritability | FDR 2 | Bonferroni 2 |
|---|---|---|---|---|---|---|---|
| Front teat length | 458 | Linear | Recessive | 1.00 | 0.33 | 5 | 2 |
| Front teat width | 458 | Case–control 3 | Recessive | 1.01 | 0.06 | 4 | 4 |
| Fore udder attachment | 288 | Case–control 4 | Additive | 1.00 | 0.45 | 4 | 3 |
| Risk PC 5 1 | 458 | Linear | Dominant | 1.01 | 0.07 | 2 | 1 |
| Rear teat length | 458 | Case–control 6 | Additive | 1.00 | 0.42 | 1 | 1 |
| Rear teat end shape | 227 | Case–control 7 | Recessive | 1.02 | 0.50 | 1 | 1 |
| Rear teat width | 458 | Case–control 3 | Recessive | 1.02 | 0.03 | 3 | 1 |
| Rear teat width | 458 | Linear | Recessive | 0.98 | 0.18 | 50 | 23 |
| Udder depth | 265 | Case–control 8 | Dominant | 0.99 | 0.99 | 1 | 1 |
| Udder height | 458 | Case–control 9 | Recessive | 1.00 | 0.02 | 129 | 32 |
| Udder width | 458 | Case–control 10 | Dominant | 1.00 | 0.30 | 1 | 1 |
| Front teat placement 11 | 144 | Linear | Recessive | 1.02 | 0.38 | 10 | 7 |
| Udder depth 11 | 144 | Linear | Recessive | 1.05 | 0.62 | 10 | 0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Miles, A.M.; Posbergh, C.J.; Huson, H.J. Direct Phenotyping and Principal Component Analysis of Type Traits Implicate Novel QTL in Bovine Mastitis through Genome-Wide Association. Animals 2021, 11, 1147. https://doi.org/10.3390/ani11041147
Miles AM, Posbergh CJ, Huson HJ. Direct Phenotyping and Principal Component Analysis of Type Traits Implicate Novel QTL in Bovine Mastitis through Genome-Wide Association. Animals. 2021; 11(4):1147. https://doi.org/10.3390/ani11041147
Chicago/Turabian StyleMiles, Asha M., Christian J. Posbergh, and Heather J. Huson. 2021. "Direct Phenotyping and Principal Component Analysis of Type Traits Implicate Novel QTL in Bovine Mastitis through Genome-Wide Association" Animals 11, no. 4: 1147. https://doi.org/10.3390/ani11041147
APA StyleMiles, A. M., Posbergh, C. J., & Huson, H. J. (2021). Direct Phenotyping and Principal Component Analysis of Type Traits Implicate Novel QTL in Bovine Mastitis through Genome-Wide Association. Animals, 11(4), 1147. https://doi.org/10.3390/ani11041147







