Molecular Detection and Phylogeny of Anaplasma phagocytophilum and Related Variants in Small Ruminants from Turkey
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
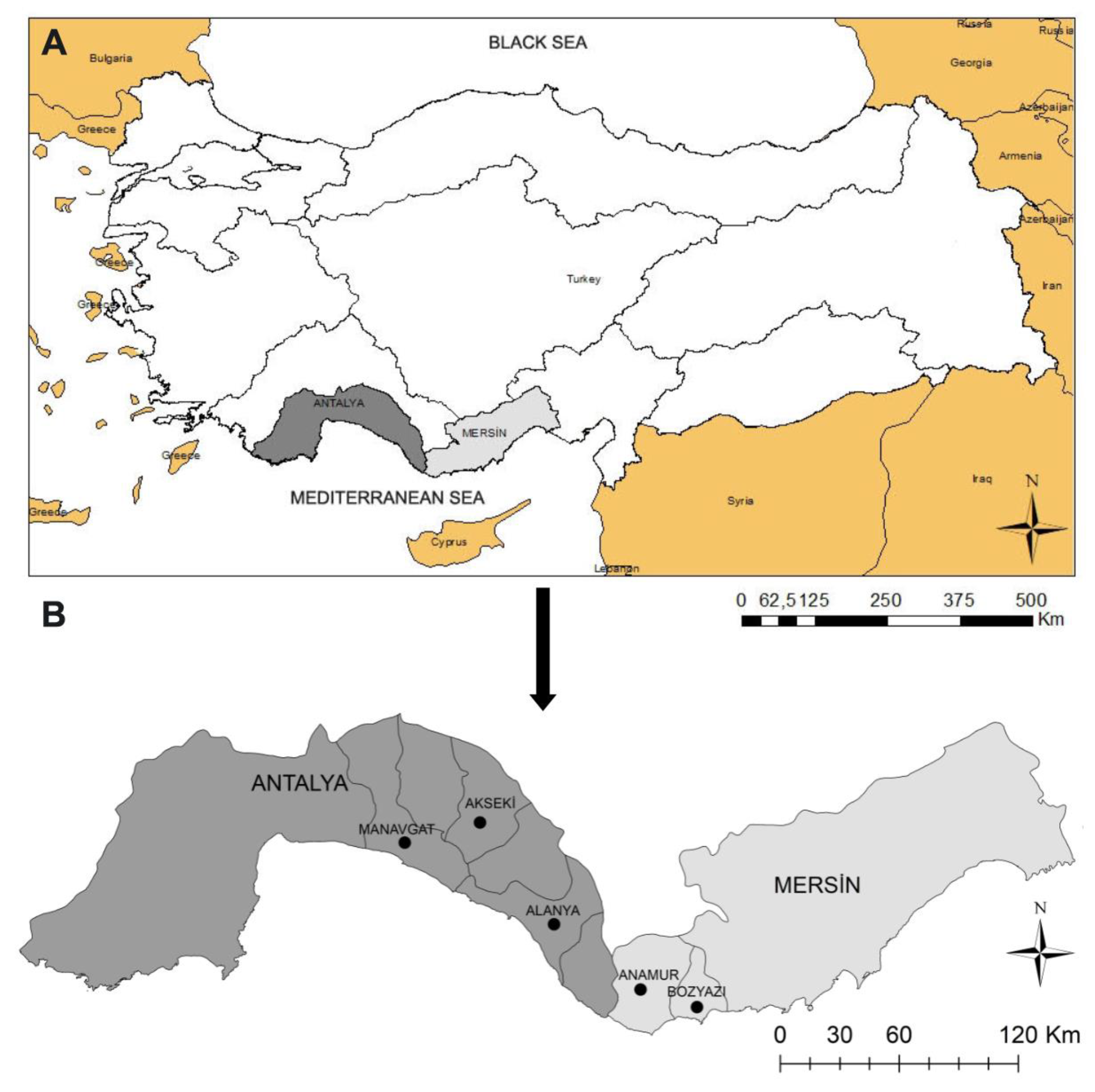
2.1. Study Region and Sample Collection
2.2. DNA Extraction and Amplification of 16S rRNA Gene
2.3. Restriction Fragment Length Polymorphism (RFLP)
2.4. GroEL PCR
2.5. Sequencing and Phylogenetic Analyses
2.6. Statistical Analysis
3. Results
3.1. Tick Infestation
3.2. Prevalence and Distribution of Anaplasma spp.
3.3. Molecular and Phylogenetic Analyses
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Woldehiwet, Z. The natural history of Anaplasma phagocytophilum. Vet. Parasitol. 2010, 167, 108–112. [Google Scholar] [CrossRef]
- Stuen, S.; Granquist, E.G.; Silaghi, C. Anaplasma phagocytophilum-a widespread multi-host pathogen with highly adaptive strategies. Front. Cell. Infect. Microbiol. 2013, 3, 31. [Google Scholar] [CrossRef]
- Kocan, K.M.; de la Fuente, J.; Cabezas-Cruz, A. The genus Anaplasma: New challenges after reclassification. Rev. Sci. Technol. 2015, 34, 577–586. [Google Scholar] [CrossRef]
- Langenwalder, D.B.; Schmidt, S.; Gilli, U.; Pantchev, N.; Ganter, M.; Silaghi, C.; Aardema, M.L.; von Loewenich, F.D. Genetic characterization of Anaplasma phagocytophilum strains from goats (Capra aegagrus hircus) and water buffalo (Bubalus bubalis) by 16S rRNA gene, ankA gene and multilocus sequence typing. Ticks Tick Borne Dis. 2019, 10, 101267. [Google Scholar] [CrossRef] [PubMed]
- Aktas, M.; Vatansever, Z.; Altay, K.; Aydin, M.F.; Dumanli, N. Molecular evidence for Anaplasma phagocytophilum in Ixodes ricinus from Turkey. Trans. R. Soc. Trop. Med. Hyg. 2010, 104, 10–15. [Google Scholar] [CrossRef] [PubMed]
- Aktas, M.; Ozubek, S. Bovine anaplasmosis in Turkey: First laboratory confirmed clinical cases caused by Anaplasma phagocytophilum. Vet. Microbiol. 2015, 178, 246–251. [Google Scholar] [CrossRef]
- Ben Said, M.; Belkahia, H.; Alberti, A.; Zobba, R.; Bousrih, M.; Yahiaoui, M.; Daaloul-Jedidi, M.; Mamlouk, A.; Gharbi, M.; Messadi, L. Molecular survey of Anaplasma species in small ruminants reveals the presence of novel strains closely related to A. phagocytophilum in Tunisia. Vector Borne Zoonotic Dis. 2015, 15, 580–590. [Google Scholar] [CrossRef]
- Ben Said, M.; Belkahia, H.; El Mabrouk, N.; Saidani, M.; Ben Hassen, M.; Alberti, A.; Zobba, R.; Bouattour, S.; Bouattour, A.; Messadi, L. Molecular typing and diagnosis of Anaplasma spp. closely related to Anaplasma phagocytophilum in ruminants from Tunisia. Ticks Tick Borne Dis. 2017, 8, 412–422. [Google Scholar] [CrossRef] [PubMed]
- Ben Said, M.; Belkahia, H.; Messadi, L. Anaplasma spp. in North Africa: A review on molecular epidemiology, associated risk factors and genetic characteristics. Ticks Tick Borne Dis. 2018, 9, 543–555. [Google Scholar] [CrossRef] [PubMed]
- Kawahara, M.; Rikihisa, Y.; Lin, Q.; Isogai, E.; Tahara, K.; Itagaki, A.; Hiramitsu, Y.; Tajima, T. Novel genetic variants of Anaplasma phagocytophilum, Anaplasma bovis, Anaplasma centrale, and a novel Ehrlichia sp. in wild deer and ticks on two major islands in Japan. Appl. Environ. Microbiol. 2006, 72, 1102–1109. [Google Scholar] [CrossRef]
- Jilintai, S.N.; Hayakawa, D.; Suzuki, M.; Hata, H.; Kondo, S.; Matsumoto, K.; Yokoyam, N.; Inokuma, H. Molecular survey for Anaplasma bovis and Anaplasma phagocytophilum infection in cattle in pastureland where sika deer appear in Hokkaido, Japan. Jpn. J. Infect. Dis. 2009, 62, 73–75. [Google Scholar]
- Ohashi, N.; Inayoshi, M.; Kitamura, K.; Kawamori, F.; Kawaguchi, D.; Nishimura, Y.; Naitou, H.; Hiroi, M.; Masuzawa, T. Anaplasma phagocytophilum-infected ticks, Japan. Emerg. Infect. Dis. 2005, 11, 1780–1783. [Google Scholar] [CrossRef] [PubMed]
- Yoshimoto, K.; Matsuyama, Y.; Matsuda, H.; Sakamoto, L.; Matsumoto, K.; Yokoyama, N.; Inokuma, H. Detection of Anaplasma bovis and Anaplasma phagocytophilum DNA from Haemaphysalis megaspinosa in Hokkaido, Japan. Vet. Parasitol. 2010, 168, 170–172. [Google Scholar] [CrossRef] [PubMed]
- Kang, Y.J.; Diao, X.N.; Zhao, G.Y.; Chen, M.H.; Xiong, Y.; Shi, M.; Fu, W.M.; Guo, Y.J.; Pan, B.; Chen, X.P.; et al. Extensive diversity of Rickettsiales bacteria in two species of ticks from China and the evolution of the Rickettsiales. BMC Evol. Biol. 2014, 14, 167. [Google Scholar] [CrossRef]
- Yang, J.; Li, Y.; Liu, Z.; Liu, J.; Niu, Q.; Ren, Q.; Chen, Z.; Guan, G.; Luo, J.; Yin, H. Molecular detection and characterization of Anaplasma spp. in sheep and cattle from Xinjiang, northwest China. Parasit. Vectors 2015, 19, 108. [Google Scholar] [CrossRef] [PubMed]
- Seo, M.G.; Ouh, I.-O.; Kwon, O.-D.; Kwak, D. Molecular detection of Anaplasma phagocytophilum-like Anaplasma spp. and pathogenic A. Phagocytophilum in cattle from South Korea. Mol. Phyl. Evol. 2018, 126, 23–30. [Google Scholar] [CrossRef]
- Zobba, R.; Ben Said, M.; Belkahia, H.; Pittaua, M.; Cacciotto, C.; Parpagliaa, M.L.P.; Messadic, L.; Alberto, A. Molecular epidemiology of Anaplasma spp. related to A. phagocytophilum in Mediterranean small ruminants. Acta Tropica. 2020, 202, 105286. [Google Scholar] [CrossRef]
- Gokce, H.I.; Genc, O.; Akca, A.; Vatansever, Z.; Unver, A.; Erdogan, H.M. Molecular and serological evidence of Anaplasma phagocytophilum infection of farm animals in the Black Sea Region of Turkey. Acta Vet. Hung. 2008, 56, 281–292. [Google Scholar] [CrossRef] [PubMed]
- Aktas, M.; Altay, K.; Ozubek, S.; Dumanli, N. A survey of ixodid ticks feeding on cattle and prevalence of tick-borne pathogens in the BlackSea Region of Turkey. Vet. Parasitol. 2012, 187, 567–571. [Google Scholar] [CrossRef]
- Aktas, M. A survey of ixodid tick species and molecular identification of tick-borne pathogens. Vet. Parasitol. 2014, 200, 276–283. [Google Scholar] [CrossRef]
- Altay, K.; Dumanli, N.; Aktas, M.; Ozubek, S. Survey of Anaplasma infections in small ruminants from East part of Turkey. Kafkas Univ. Vet. Fak. Derg. 2014, 20, 1–4. [Google Scholar] [CrossRef]
- Estrada-Peña, A.; Bouattour, A.; Camicas, J.L.; Walker, A.R. Ticks of Domestic Animals in the Mediterranean Region. A Guide of Identification of Species; University of Zaragoza Press: Zaragoza, Spain, 2004. [Google Scholar]
- Alberti, A.; Zobba, R.; Chessa, B.; Addis, M.F.; Sparagano, O.; Pinna Parpaglia, M.L.; Cubeddu, T.; Pintori, G.; Pittau, M. Equine and canine Anaplasma phagocytophilum strains isolated on the island of Sardinia (Italy) are phylogenetically related to pathogenic strains from the United States. Appl. Environ. Microbiol. 2005, 71, 6418–6422. [Google Scholar] [CrossRef]
- Ybañez, A.P.; Matsumoto, K.; Kishimoto, T.; Inokuma, H. Molecular analyses of a potentially novel Anaplasma species closely related to Anaplasma phagocytophilum detected in sika deer (Cervus nippon yesoensis) in Japan. Vet. Microbiol. 2012, 157, 232–236. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: A multiple sequence alignment method with reduced time and space complexity. BMC Bioinform. 2004, 5, 113. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Posada, D. jModelTest: Phylogenetic model averaging. Mol. Biol. Evol. 2008, 25, 1253–1256. [Google Scholar] [CrossRef] [PubMed]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef]
- Belkahia, H.; Ben Said, M.; Ghribi, R.; Selmi, R.; Ben Asker, A.; Yahiaoui, M.; Bousrih, M.; Daaloul-Jedidi, M.; Messadi, L. Molecular detection, genotyping and phylogeny of Anaplasma spp. in Rhipicephalus ticks from Tunisia. Acta Trop. 2019, 191, 38–49. [Google Scholar] [CrossRef]
- Benedicto, B.; Ceylan, O.; Moumouni, P.F.A.; Lee, S.H.; Tumwebaze, M.A.; Li, X.; Galoni, E.M.; Li, Y.; Ji, S.; Ringo, A.; et al. Molecular detection and assessment of risk factors for tick-borne diseases in Sheep and Goats from Turkey. Acta Parasit. 2020, 65, 723–732. [Google Scholar] [CrossRef] [PubMed]
- Movilla, R.; García, C.; Siebert, S.; Roura, X. Countrywide serological evaluation of canine prevalence for Anaplasma spp., Borrelia burgdorferi (sensu lato), Dirofilaria immitis and Ehrlichia canis in Mexico. Parasit. Vectors 2016, 9, 421. [Google Scholar] [CrossRef]
- Dreher, U.M.; de la Fuente, J.; Hofmann-Lehmann, R.; Meli, M.L.; Pusterla, N.; Kocan, K.M.; Woldehiwet, Z.; Braun, U.; Regula, G.; Staerk, K.D.; et al. Serologic cross-reactivity between Anaplasma marginale and Anaplasma phagocytophilum. Clin. Diagn. Lab. Immunol. 2005, 12, 1177–1183. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Yang, J.; Liu, Z.; Niu, Q.; Liu, J.; Xie, J.; Chen, Q.; Chen, Z.; Guan, G.; Liu, G.; Luo, J.; et al. Evaluation of different nested PCRs for detection of Anaplasma phagocytophilum in ruminants and ticks. BMC Vet. Res. 2016, 12, 35. [Google Scholar] [CrossRef] [PubMed]
- Aydin, L.; Bakırcı, S. Geographical distribution of ticks in Turkey. Parasitol. Res. 2007, 101, 163–166. [Google Scholar] [CrossRef] [PubMed]
- Sarih, M.; M’ghirbi, Y.; Bouattour, A.; Gern, L.; Baranton, G.; Postic, D. Detection and identification of Ehrlichia spp. in ticks collected in Tunisia andMorocco. J. Clin. Microbiol. 2005, 43, 1127–1132. [Google Scholar] [CrossRef] [PubMed]

| Target Gene | Specificity | Primer Name | Oligonucleotide Dequence (5′-3′) | Annealing | Amplicon Size (bp) | Reference |
|---|---|---|---|---|---|---|
|
-
- - 16S rRNA | All Anaplasma/Ehrlichia - - A. phagocytophilum and related variants | EC9 EC12a - SSAP2f SSAP2r | TACCTTGTTACGACTT TGATCCTGGCTCAGAACGAACG - GCTGAATGTGGGGATAATTTAT ATGGCTGCTTCCTTTCGGTTA | 54 - 53 | 1462 - 641–642 | [10] |
|
-
groEL | - A. phagocytophilum | EphplgroEL(569)F EphgroEL(1142)R - EphplgroEL(569)F EphgroEL(1142)R | ATGGTATGCAGTTTGATCGC TTGAGTACAGCAACACCACCGGAA - ATGGTATGCAGTTTGATCGC TTGAGTACAGCAACACCACCGGAA | 54 - - 54 | 624 - - 573 | [23] |
| groEL | A. phagocytophilum-like 1 | EEGro1F AnaGroe712R AnaGroe240F | GAGTTCGACGGTAAGAAGTTCA ATTAGY *AAGCCTTATGGGTC CCGCGATCAAACTGCATACC | 52 - 57 | 670 - 432 | [25] |
| Host | District/Province | 16S rRNA PCR+/No. of Samples | 16S rRNA PCR + RFLP | groEL+/16S+ | groEL PCR | |||
|---|---|---|---|---|---|---|---|---|
| - | - | - | AP | AP-like 1 | AP-like 2 | AP | AP-like 1 | |
| Goat | Akseki/Antalya | 13/56 (23.2%) | 2 | 11 | 0 | 12/13 | 2 | 10 |
| - | Manavgat/Antalya | 26/111 (23.4%) | 3 | 23 | 0 | 23/26 | 3 | 20 |
| - | Alanya/Antalya | 24/55 (43.6%) | 0 | 24 | 0 | 24/24 | 24 | |
| - | Anamur/Mersin | 15/44 (34.1%) | 0 | 15 | 0 | 12/15 | 12 | |
| - | Bozyazı/Mersin | 5/30 (16.7%) | 0 | 5 | 0 | 5/5 | 5 | |
| Goat Total | - | 83/296 (28%) | 5 | 78 | 0 | 76/83 (91.5%) | 5 | 71 |
| - | - | - | - | - | - | - | - | - |
| Sheep | Akseki/Antalya | 5/9 (55.6%) | 1 | 4 | 0 | 3/5 | 1 | 2 |
| - | Manavgat/Antalya | 18/103 (17.5%) | 0 | 18 | 0 | 17/18 | - | 17 |
| - | Alanya/Antalya | 9/9 (100%) | 0 | 9 | 0 | 9/9 | - | 9 |
| - | Anamur/Mersin | 6/16 (37.5%) | 0 | 6 | 0 | 5/6 | - | 5 |
| Sheep Total | - | 38/137 (27.7%) | 1 | 37 | 0 | 34/38 (89.4%) | 1 | 33 |
| Grand Total | - | 121/433 (27.9%) | 6 (1.4%) | 115 (26.5%) | 0 | 110/115 (95.6%) | 6 | 104 |
| Species | Presence of Ticks on the Animals | |||
|---|---|---|---|---|
| Goats n (%) | Sheep n (%) | No n (%) | Yes n (%) | |
| Number | 296 | 137 | 243 | 190 |
| Positive | 83 (28) | 38 (27.7) | 51 (20.9) | 70 (36.8) |
| Negative | 213 (72) | 99 (72.3) | 192 (79.1) | 120 (63.2) |
| p-Value | 0.9603 | 0.0003 | ||
| Host | Genetic Variant a | Country | GenBank | Nucleotide Positions b | Reference | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| - | - | - | - | 823 | 830 | 1011 | 1109 | 1111 | 1113 | 1120 | 1137 | 1148 | 1237 | 1239 | 1240 | 1260 | 1291 | - |
| Human | Webster | USA | NR_044762 | T | T | A | G | T | A | C | A | T | T | T | C | G | C | Unpublished |
| Horse | Camawi | USA | AF172167 | * | * | * | * | * | * | * | * | * | * | * | * | * | * | Unpublished |
| Dog | Dog2 | USA | CP006618 | * | * | * | * | * | * | * | * | * | * | * | * | * | * | Unpublished |
| Deer | Clone 1 | Japan | JN055357 | C | * | * | * | A | - | * | G | C | * | * | * | * | * | [24] |
| Goat | Aplike1GGo1 | Tunisia | KM285226 | C | * | * | * | A | - | * | G | C | * | * | * | * | * | [7] |
| Goat | Aplike1GGo2 | Tunisia | KM285227 | C | * | * | * | A | T | * | G | * | * | * | * | * | * | [7] |
| Sheep | Aplike1GOv1 | Tunisia | KM285230 | C | * | * | * | A | - | * | G | C | * | * | * | * | * | [7] |
| Cattle and Goat | Aplike1BvCp1 | Tunisia | KX702974 | C | * | * | * | A | - | * | G | C | * | * | * | * | * | [8] |
| Sheep | Aplike1Ov1 | Tunisia | KX702978 | C | * | * | * | A | T | * | G | * | * | * | * | * | * | [8] |
| Sheep and Goat | Aplike2OvCp1 | Tunisia | KX702980 | C | A | G | * | A | T | T | G | C | C | C | T | A | T | [8] |
| Cattle | Aplike1Bv | Turkey | MT338494 | C | * | * | * | A | T | * | G | * | * | * | * | * | * | Unpublished |
| Sheep and Goat | Aplike1OvineCaprine | Turkey | MT881655 | C | * | * | * | A | T | * | G | * | * | * | * | * | * | Present study |
| Sheep | Aphaakseki11 | Turkey | MT881656 | * | * | * | * | * | * | * | * | * | * | * | T | * | * | Present study |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aktaş, M.; Özübek, S.; Uluçeşme, M.C. Molecular Detection and Phylogeny of Anaplasma phagocytophilum and Related Variants in Small Ruminants from Turkey. Animals 2021, 11, 814. https://doi.org/10.3390/ani11030814
Aktaş M, Özübek S, Uluçeşme MC. Molecular Detection and Phylogeny of Anaplasma phagocytophilum and Related Variants in Small Ruminants from Turkey. Animals. 2021; 11(3):814. https://doi.org/10.3390/ani11030814
Chicago/Turabian StyleAktaş, Münir, Sezayi Özübek, and Mehmet Can Uluçeşme. 2021. "Molecular Detection and Phylogeny of Anaplasma phagocytophilum and Related Variants in Small Ruminants from Turkey" Animals 11, no. 3: 814. https://doi.org/10.3390/ani11030814
APA StyleAktaş, M., Özübek, S., & Uluçeşme, M. C. (2021). Molecular Detection and Phylogeny of Anaplasma phagocytophilum and Related Variants in Small Ruminants from Turkey. Animals, 11(3), 814. https://doi.org/10.3390/ani11030814







