CircAgtpbp1 Acts as a Molecular Sponge of miR-543-5p to Regulate the Secretion of GH in Rat Pituitary Cells
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Animals and Cell Culture
2.3. Short Interfering RNA (siRNA) Synthesis and Plasmid Construction
2.4. Cell Transfection
2.5. RNA Isolation, RNase R Treatment and RT–qPCR
2.6. Identification of Target Genes by Target Prediction
2.7. RIP Assay
2.8. FISH
2.9. GH Detection
2.10. Western Blot Analysis
3. Results
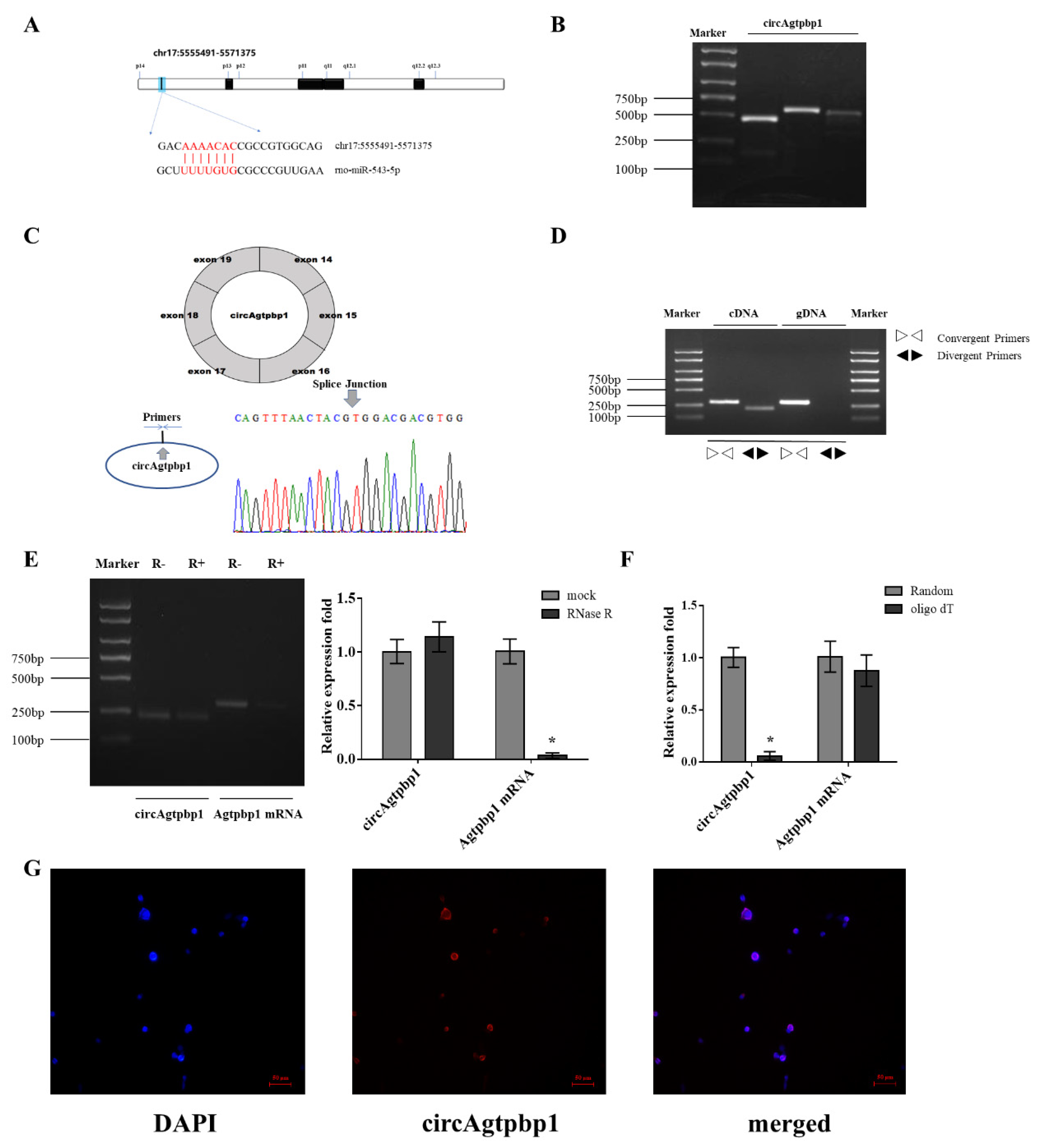
3.1. Identification and Validation of the circRNA
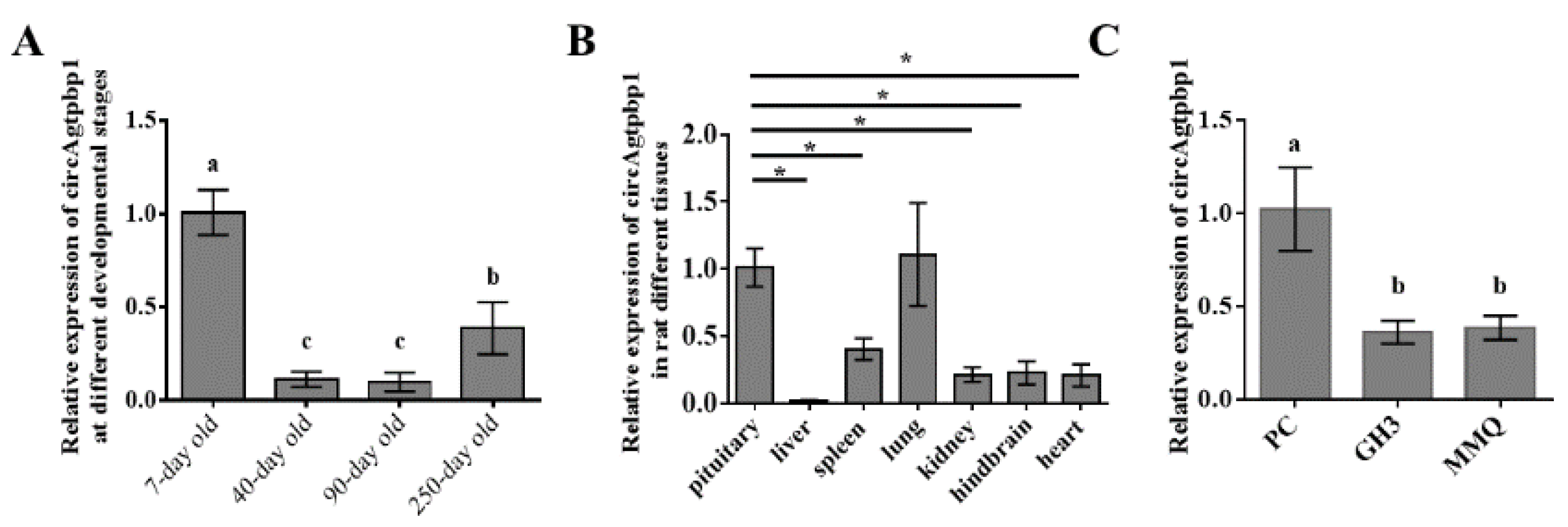
3.2. CircAgtpbp1 Expression Pattern in the Rat Anterior Pituitary
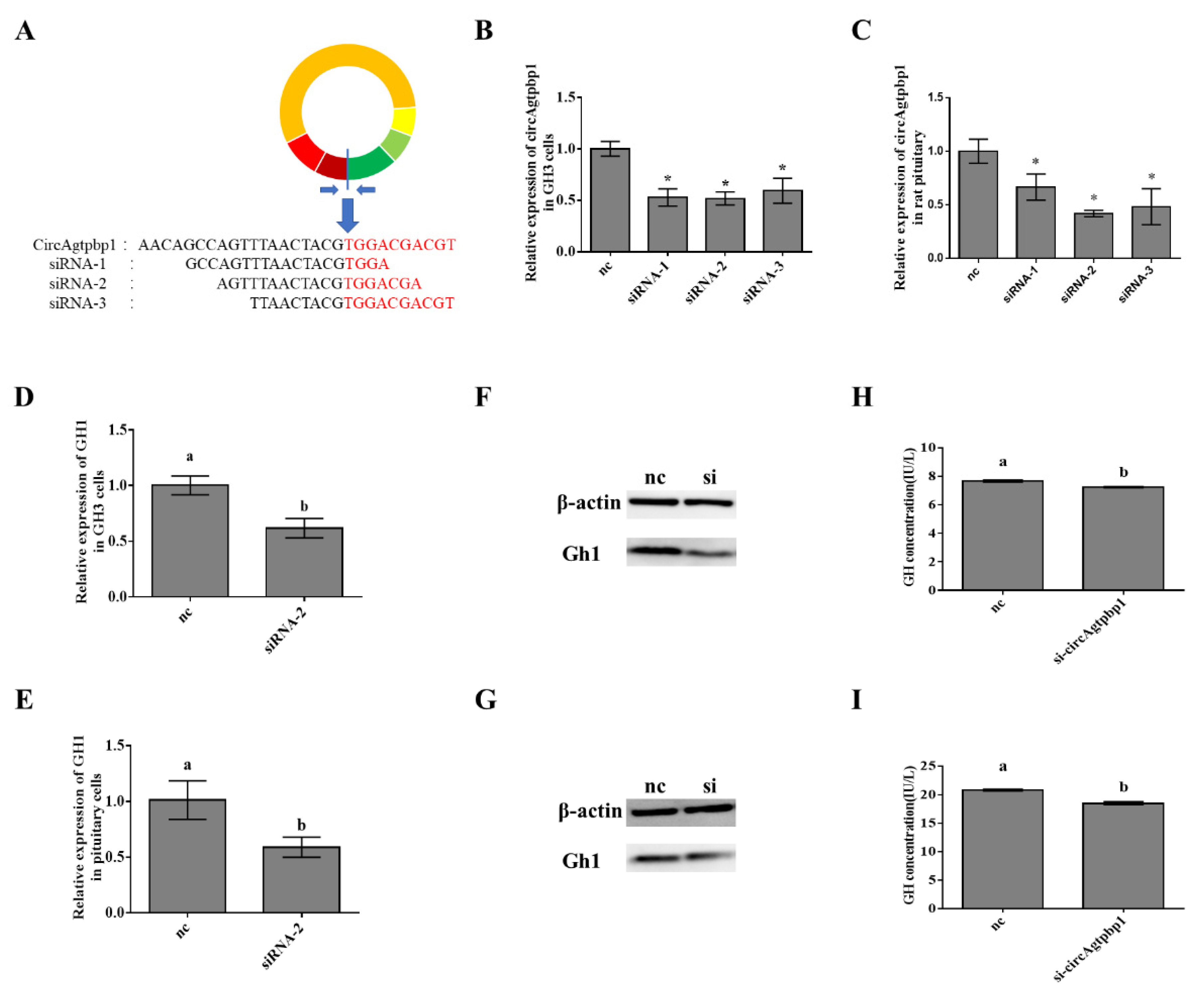
3.3. Effects of CircAgtpbp1 Knockdown on Gh1 Transcription
3.4. Effects of CircAgtpbp1 Overexpression on Gh1 Transcription
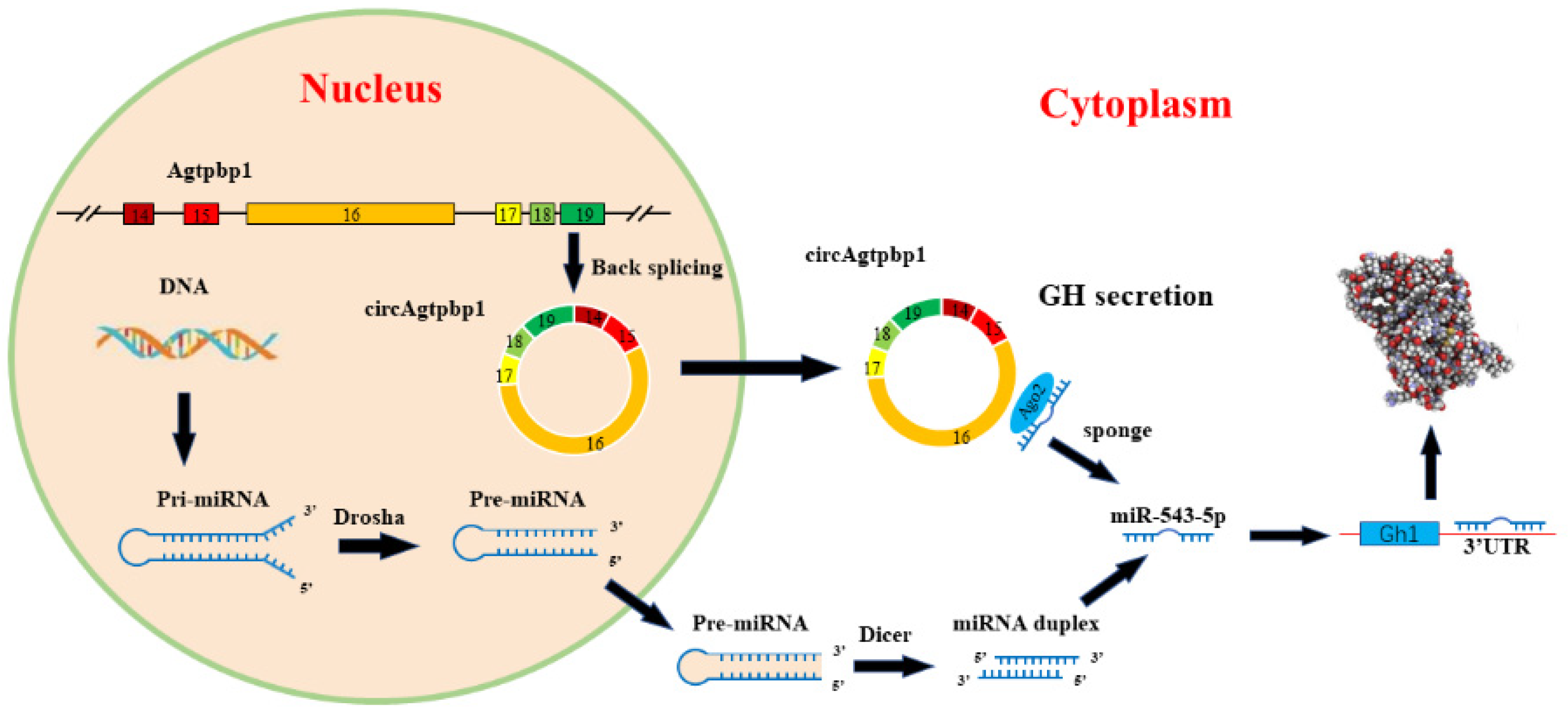
3.5. CircAgtpbp1 Sponges miR-543-5p in Primary Rat Pituitary Cells
3.6. CircAgtpbp1 Regulates Gh1 Expression and GH Secretion by Targeting miR-543-5p
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Edwards, W.; Raetzman, L.T. Complex integration of intrinsic and peripheral signaling is required for pituitary gland development. Biol. Reprod. 2018, 99, 504–513. [Google Scholar] [CrossRef] [PubMed]
- Brändli-Baiocco, A.; Balme, E.; Bruder, M.; Chandra, S.; Hellmann, J.; Hoenerhoff, M.J.; Kambara, T.; Landes, C.; Lenz, B.; Mense, M.; et al. Nonproliferative and Proliferative Lesions of the Rat and Mouse Endocrine System. J. Toxicol. Pathol. 2018, 31, 1S–95S. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Gleiberman, A.S.; Rosenfeld, M.G. Molecular Physiology of Pituitary Development: Signaling and Transcriptional Networks. Physiol. Rev. 2007, 87, 933–963. [Google Scholar] [CrossRef]
- Xie, Y.; Dorsky, R.I. Development of the hypothalamus: Conservation, modification and innovation. Development 2017, 144, 1588–1599. [Google Scholar] [CrossRef]
- Zhang, H.; Qi, Q.; Chen, T.; Luo, J.; Xi, Q.; Jiang, Q.; Sun, J.; Zhang, Y. Age-Related Changes in MicroRNA in the Rat Pituitary and Potential Role in GH Regulation. Int. J. Mol. Sci. 2018, 19, 2058. [Google Scholar] [CrossRef] [PubMed]
- Pierce, A.L.; Breves, J.P.; Moriyama, S.; Hirano, T.; Grau, E.G. Differential regulation of Igf1 and Igf2 mRNA levels in tilapia hepatocytes: Effects of insulin and cortisol on GH sensitivity. J. Endocrinol. 2011, 211, 201–210. [Google Scholar] [CrossRef] [PubMed]
- Caruso, M.A.; Sheridan, M.A. New insights into the signaling system and function of insulin in fish. Gen. Comp. Endocrinol. 2011, 173, 227–247. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.L.; Feng, M.; Tan, X.; Yan, G.Y.; Sun, C. The role of SOCS2 in recombinant human growth hormone (rhGH) regulating lipid metabolism in high-fat-diet-induced obesity mice. Mol. Biol. Rep. 2013, 40, 2319–2326. [Google Scholar] [CrossRef] [PubMed]
- Bergan, H.E.; Kittilson, J.D.; Sheridan, M.A. PKC and ERK mediate GH-stimulated lipolysis. J. Mol. Endocrinol. 2013, 51, 213–224. [Google Scholar] [CrossRef][Green Version]
- Brooks, A.J.; Waters, M.J. The growth hormone receptor: Mechanism of activation and clinical implications. Nat. Rev. Endocrinol. 2010, 6, 515–525. [Google Scholar] [CrossRef] [PubMed]
- Kato, Y.; Murakami, Y.; Sohmiya, M.; Nishiki, M. Regulation of Human Growth Hormone Secretion and Its Disorders. Intern. Med. 2002, 41, 7–13. [Google Scholar] [CrossRef] [PubMed]
- Pollak, M. The insulin and insulin-like growth factor receptor family in neoplasia: An update. Nat. Rev. Cancer 2012, 12, 159–169. [Google Scholar] [CrossRef]
- Tatar, M.; Bartke, A.; Antebi, A. The Endocrine Regulation of Aging by Insulin-like Signals. Science 2003, 299, 1346–1351. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.; Li, C.; Li, C.; Zhang, X. Growth Hormone Receptor Mutations Related to Individual Dwarfism. Int. J. Mol. Sci. 2018, 19, 1433. [Google Scholar] [CrossRef] [PubMed]
- Alzhanov, D.; Mukherjee, A.; Rotwein, P. Identifying growth hormone-regulated enhancers in the Igf1 locus. Physiol. Genom. 2015, 47, 559–568. [Google Scholar] [CrossRef]
- Goldenberg, N.; Barkan, A. Factors Regulating Growth Hormone Secretion in Humans. Endocrinol. Metab. Clin. N. Am. 2007, 36, 37–55. [Google Scholar] [CrossRef] [PubMed]
- Ohlsson, C.; Bengtsson, B.-A.; Isaksson, O.G.P.; Andreassen, T.T.; Slootweg, M.C. Growth Hormone and Bone. Endocr. Rev. 1998, 19, 55–79. [Google Scholar] [CrossRef][Green Version]
- Müller, E.E.; Locatelli, V.; Cocchi, D. Neuroendocrine Control of Growth Hormone Secretion. Physiol. Rev. 1999, 79, 511–607. [Google Scholar] [CrossRef]
- Khatib, N.; Gaidhane, S.; Gaidhane, A.M.; Khatib, M.; Simkhada, P.; Gode, D.; Zahiruddin, Q.S. Ghrelin: Ghrelin as a Regulatory Peptide in Growth Hormone Secretion. J. Clin. Diagn. Res. 2014, 8, MC13–MC17. [Google Scholar] [CrossRef]
- Wu, J.; Jiang, Z.; Chen, C.; Hu, Q.; Fu, Z.; Chen, J.; Wang, Z.; Wang, Q.; Li, A.; Marks, J.R.; et al. CircIRAK3 sponges miR-3607 to facilitate breast cancer metastasis. Cancer Lett. 2018, 430, 179–192. [Google Scholar] [CrossRef]
- Liu, J.; Song, S.; Lin, S.; Zhang, M.; Du, Y.; Zhang, D.; Xu, W.; Wang, H. Circ-SERPINE2 promotes the development of gastric carcinoma by sponging miR-375 and modulating YWHAZ. Cell Prolif. 2019, 52, e12648. [Google Scholar] [CrossRef]
- Cai, X.; Zhao, Z.; Dong, J.; Lv, Q.; Yun, B.; Liu, J.; Shen, Y.; Kang, J.; Li, J. Circular RNA circBACH2 plays a role in papillary thyroid carcinoma by sponging miR-139-5p and regulating LMO4 expression. Cell Death Dis. 2019, 10, 184. [Google Scholar] [CrossRef]
- Chen, L.-Y.; Zhi, Z.; Wang, L.; Zhao, Y.Y.; Deng, M.; Liu, Y.H.; Qin, Y.; Tian, M.M.; Liu, Y.; Shen, T.; et al. NSD2 circular RNA promotes metastasis of colorectal cancer by targeting miR-199b-5p-mediated DDR1 and JAG1 signalling. J. Pathol. 2019, 248, 103–115. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; He, P.; Pan, H.; Long, J.; Wang, J.; Li, Z.; Liu, H.; Jiang, W.; Zheng, Z. Circular RNA circ-4099 is induced by TNF-α and regulates ECM synthesis by blocking miR-616-5p inhibition of Sox9 in intervertebral disc degeneration. Exp. Mol. Med. 2018, 50, 27. [Google Scholar] [CrossRef]
- Xie, F.; Li, Y.; Wang, M.; Huang, C.; Tao, D.; Zheng, F.; Zhang, H.; Zeng, F.; Xiao, X.; Jiang, G. Circular RNA BCRC-3 suppresses bladder cancer proliferation through miR-182-5p/p27 axis. Mol. Cancer 2018, 17, 144. [Google Scholar] [CrossRef]
- Venø, M.T.; Hansen, T.B.; Venø, S.T.; Clausen, B.H.; Grebing, M.; Finsen, B.; Holm, I.E.; Kjems, J. Spatio-temporal regulation of circular RNA expression during porcine embryonic brain development. Genome Biol. 2015, 16, 245. [Google Scholar] [CrossRef] [PubMed]
- Salzman, J.; Chen, R.E.; Olsen, M.N.; Wang, P.L.; Brown, P.O. Cell-Type Specific Features of Circular RNA Expression. PLoS Genet. 2013, 9, e1003777. [Google Scholar] [CrossRef]
- Yang, C.; Yuan, W.; Yang, X.; Li, P.; Wang, J.; Han, J.; Tao, J.; Li, P.; Yang, H.; Lv, Q.; et al. Circular RNA circ-ITCH inhibits bladder cancer progression by sponging miR-17/miR-224 and regulating p21, PTEN expression. Mol. Cancer 2018, 17, 19. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Tang, D.; Wang, W.; Yang, Y.; Wu, X.; Wang, L.; Wang, D. circLMTK2 acts as a sponge of miR-150-5p and promotes proliferation and metastasis in gastric cancer. Mol. Cancer 2019, 18, 162. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Yang, Z.; Liu, C.; Wang, L.; Yang, J.; Chen, L.; Li, W. Circ_0078767 suppresses non-small-cell lung cancer by protecting RASSF1A expression via sponging miR-330-3p. Cell Prolif. 2019, 52, e12548. [Google Scholar] [CrossRef]
- Zhou, L.; Li, Y.; Jiang, W.; Zhang, H.; Wen, Z.; Su, Y.; Wu, F.; Zhi, Z.; Shen, Q.; Li, H.; et al. Down-regulation of circ-PRKCI inhibits cell migration and proliferation in Hirschsprung disease by suppressing the expression of miR-1324 target PLCB1. Cell Cycle 2018, 17, 1092–1101. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Li, C.; Liu, Z.; Ni, W.; Yao, R.; Xu, Y.; Quan, R.; Zhang, M.; Li, H.; Liu, L.; et al. Circular RNA circ-FoxO3 Inhibits Myoblast Cells Differentiation. Cells 2019, 8, 616. [Google Scholar] [CrossRef]
- Chen, K.; Hou, J.; An, X.; Song, Y.; Zhang, X.; Liu, Y.; Zhang, G.; Wen, K.; Ma, H.; Li, G.; et al. Chi-miR-3031 regulates beta-casein via the PI3K/AKT-mTOR signaling pathway in goat mammary epithelial cells (GMECs). BMC Vet. Res. 2018, 14, 369. [Google Scholar] [CrossRef]
- Han, D.-X.; Sun, X.-L.; Zhang, J.-B.; Wang, C.-J.; Yu, Z.-W.; Zheng, Y.; Huang, Y.-J.; Wang, W.-H.; Jiang, H.; Gao, Y.; et al. Differentially expressed lncRNA-m433s1 regulates FSH secretion by functioning as a miRNA sponge in male rat anterior pituitary cells. Biol. Reprod. 2019, 101, 416–425. [Google Scholar] [CrossRef]
- Yu, Z.-W.; Gao, W.; Feng, X.-Y.; Zhang, J.-Y.; Guo, H.-X.; Wang, C.-J.; Chen, J.; Hu, J.-P.; Ren, W.-Z.; Yuan, B. Roles of differential expression of miR-543-5p in GH regulation in rat anterior pituitary cells and GH3 cells. PLoS ONE 2019, 14, e0222340. [Google Scholar] [CrossRef] [PubMed]
- Rehmsmeier, M.; Steffen, P.; Höchsmann, M.; Giegerich, R. Fast and effective prediction of microRNA/target duplexes. RNA 2004, 10, 1507–1517. [Google Scholar] [CrossRef] [PubMed]
- Miranda, K.C.; Huynh, T.; Tay, Y.; Ang, Y.-S.; Tam, W.-L.; Thomson, A.M.; Lim, B.; Rigoutsos, I. A Pattern-Based Method for the Identification of MicroRNA Binding Sites and Their Corresponding Heteroduplexes. Cell 2006, 126, 1203–1217. [Google Scholar] [CrossRef] [PubMed]
- Han, D.-X.; Wang, C.-J.; Sun, X.-L.; Liu, J.-B.; Jiang, H.; Gao, Y.; Chen, C.-Z.; Yuan, B.; Zhang, J.-B. Identification of circular RNAs in the immature and mature rat anterior pituitary. J. Endocrinol. 2019, 240, 393–402. [Google Scholar] [CrossRef] [PubMed]
- Qu, S.; Yang, X.; Li, X.; Wang, J.; Gao, Y.; Shang, R.; Sun, W.; Dou, K.; Li, H. Circular RNA: A new star of noncoding RNAs. Cancer Lett. 2015, 365, 141–148. [Google Scholar] [CrossRef] [PubMed]
- Salzman, J. Circular RNA Expression: Its Potential Regulation and Function. Trends Genet. 2016, 32, 309–316. [Google Scholar] [CrossRef]
- Li, Z.; Huang, C.; Bao, C.; Chen, L.; Lin, M.; Wang, X.; Zhong, G.; Yu, B.; Hu, W.; Dai, L.; et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat. Struct. Mol. Biol. 2015, 22, 256–264. [Google Scholar] [CrossRef]
- Memczak, S.; Jens, M.; Loewer, A.; Ziebold, U.; Landthaler, M.; Kocks, C.; Le Noble, F.; Rajewsky, N.; Elefsinioti, A.; Torti, F.; et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013, 495, 333–338. [Google Scholar] [CrossRef]
- Hansen, T.B.; Jensen, T.I.; Clausen, B.H.; Bramsen, J.B.; Finsen, B.; Damgaard, C.K.; Kjems, J. Natural RNA circles function as efficient microRNA sponges. Nature 2013, 495, 384–388. [Google Scholar] [CrossRef]
- Guo, J.U.; Agarwal, V.; Guo, H.; Bartel, D.P. Expanded identification and characterization of mammalian circular RNAs. Genome Biol. 2014, 15, 409. [Google Scholar] [CrossRef]
- Szabo, L.A.; Morey, R.; Palpant, N.J.; Wang, P.L.; Afari, N.; Jiang, C.; Parast, M.M.; Murry, C.E.; Laurent, L.C.; Salzman, J. Statistically based splicing detection reveals neural enrichment and tissue-specific induction of circular RNA during human fetal development. Genome Biol. 2015, 16, 126. [Google Scholar] [CrossRef] [PubMed]
- Qi, Q.-E.; Xi, Q.-Y.; Ye, R.-S.; Chen, T.; Cheng, X.; Li, C.-Y.; Zhu, X.-T.; Shu, G.; Wang, L.-N.; Jiang, Q.-Y.; et al. Alteration of the miRNA expression profile in male porcine anterior pituitary cells in response to GHRH and CST and analysis of the potential roles for miRNAs in regulating GH. Growth Horm. IGF Res. 2015, 25, 66–74. [Google Scholar] [CrossRef] [PubMed]
- Beltrutti, D.; Debernardi, F.; Pelosi, G. Analgesic effects of natural growth hormone release inhibitors and their synthetic analogs. Intraspinal administration. Minerva Med. 1997, 88, 39–47. [Google Scholar]
- Müller, E.E.; Cella, S.G.; De Gennaro Colonna, V.; Parenti, M.; Cocchi, D.; Locatelli, V. Aspects of the neuroendocrine control of growth hormone secretion in ageing mammals. J. Reprod. Fertil. Suppl. 1993, 46, 99–114. [Google Scholar] [PubMed]
- Crew, M.D.; Spindler, S.R.; Walford, R.L.; Koizumi, A. Age-Related Decrease of Growth Hormone and Prolactin Gene Expression in the Mouse Pituitary. Endocrinology 1987, 121, 1251–1255. [Google Scholar] [CrossRef] [PubMed]
- Sonntag, W.E.; Steger, R.W.; Forman, L.J.; Meites, J. Decreased Pulsatile Release of Growth Hormone in Old Male Rats*. Endocrinol. 1980, 107, 1875–1879. [Google Scholar] [CrossRef] [PubMed]
- Berryman, D.E.; Christiansen, J.S.; Johannsson, G.; Thorner, M.O.; Kopchick, J.J. Role of the GH/IGF-1 axis in lifespan and healthspan: Lessons from animal models. Growth Horm. IGF Res. 2008, 18, 455–471. [Google Scholar] [CrossRef]
- Besson, A.; Salemi, S.; Gallati, S.; Jenal, A.; Horn, R.; Mullis, P.S.; Mullis, P.E. Reduced Longevity in Untreated Patients with Isolated Growth Hormone Deficiency. J. Clin. Endocrinol. Metab. 2003, 88, 3664–3667. [Google Scholar] [CrossRef]
- Bartke, A. Can Growth Hormone (GH) Accelerate Aging? Evidence from GH-Transgenic Mice. Neuroendocrinology 2003, 78, 210–216. [Google Scholar] [CrossRef]
- Orme, S.M.; McNally, R.J.Q.; Cartwright, R.A.; Belchetz, P.E. Mortality and Cancer Incidence in Acromegaly: A Retrospective Cohort Study. J. Clin. Endocrinol. Metab. 1998, 83, 2730–2734. [Google Scholar] [CrossRef] [PubMed]
- Sheppard, M.C. GH and mortality in acromegaly. J. Endocrinol. Invest. 2005, 28, 75–77. [Google Scholar] [PubMed]
- Lou, A.; Jin, T.; Zhang, R.; Ji, J.; Xiang, S.; Cui, C.; Yu, L.; Guan, L. The effect of miR-93 on GH secretion in pituitary cells of Yanbian yellow cattle. Anim. Biotechnol. 2019, 10, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Rui, Q.H.; Ma, J.B.; Liao, Y.F.; Dai, J.H.; Cai, Z.Y. Effect of lncRNA HULC knockdown on rat secreting pituitary adenoma GH3 cells. Braz. J. Med Biol. Res. 2019, 52, e7728. [Google Scholar] [CrossRef]
- Zheng, Q.; Bao, C.; Guo, W.; Li, S.; Chen, J.; Chen, B.; Luo, Y.; Lyu, D.; Li, Y.; Shi, G.; et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat. Commun. 2016, 7, 11215. [Google Scholar] [CrossRef]
- Ebbesen, K.K.; Hansen, T.B.; Kjems, J. Insights into circular RNA biology. RNA Biol. 2017, 14, 1035–1045. [Google Scholar] [CrossRef]
- Ashwal-Fluss, R.; Meyer, M.; Pamudurti, N.R.; Ivanov, A.; Bartok, O.; Hanan, M.; Evantal, N.; Memczak, S.; Rajewsky, N.; Kadener, S. circRNA Biogenesis Competes with Pre-mRNA Splicing. Mol. Cell 2014, 56, 55–66. [Google Scholar] [CrossRef]
- Schneider, T.; Hung, L.-H.; Schreiner, S.; Starke, S.; Eckhof, H.; Rossbach, O.; Reich, S.; Medenbach, J.; Bindereif, A. CircRNA-protein complexes: IMP3 protein component defines subfamily of circRNPs. Sci. Rep. 2016, 6, 31313. [Google Scholar] [CrossRef]
- Liang, M.; Huang, G.; Liu, Z.; Wang, Q.; Yu, Z.; Liu, Z.; Lin, H.; Li, M.; Zhou, X.; Zheng, Y. Elevated levels of hsa_circ_006100 in gastric cancer promote cell growth and metastasis via miR-195/GPRC5A signalling. Cell Prolif. 2019, 52, e12661. [Google Scholar] [CrossRef] [PubMed]
- Lu, Q.; Liu, T.; Feng, H.; Yang, R.; Zhao, X.; Chen, W.; Jiang, B.; Qin, H.; Guo, X.; Liu, M.; et al. Circular RNA circSLC8A1 acts as a sponge of miR-130b/miR-494 in suppressing bladder cancer progression via regulating PTEN. Mol. Cancer 2019, 18, 111. [Google Scholar] [CrossRef]
- Li, S.; Gu, H.; Huang, Y.; Peng, Q.; Zhou, R.; Yi, P.; Chen, R.; Huang, Z.; Hu, X.; Huang, Y.; et al. Circular RNA 101368/miR-200a axis modulates the migration of hepatocellular carcinoma through HMGB1/RAGE signaling. Cell Cycle 2018, 17, 2349–2359. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Nan, A.; Zhang, N.; Jia, Y.; Li, X.; Ling, Y.; Dai, J.; Zhang, S.; Yang, Q.; Yi, Y.; et al. Circular RNA 100146 functions as an oncogene through direct binding to miR-361-3p and miR-615-5p in non-small cell lung cancer. Mol. Cancer 2019, 18, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Ma, H.-B.; Yao, Y.-N.; Yu, J.-J.; Chen, X.-X.; Li, H.-F. Extensive profiling of circular RNAs and the potential regulatory role of circRNA-000284 in cell proliferation and invasion of cervical cancer via sponging miR-506. Am. J. Transl. Res. 2018, 10, 592–604. [Google Scholar]
- Bi, W.; Huang, J.; Nie, C.; Liu, B.; He, G.; Han, J.; Pang, R.; Ding, Z.; Xu, J.; Zhang, J. CircRNA circRNA_102171 promotes papillary thyroid cancer progression through modulating CTNNBIP1-dependent activation of β-catenin pathway. J. Exp. Clin. Cancer Res. 2018, 37, 275. [Google Scholar] [CrossRef]
- He, J.H.; Han, Z.P.; Zhou, J.B.; Chen, W.M.; Lv, Y.B.; He, M.L.; Li, Y.G. MiR-145 affected the circular RNA expression in prostate cancer LNCaP cells. J. Cell. Biochem. 2018, 119, 9168–9177. [Google Scholar] [CrossRef]
- Chen, X.; Su, X.; Zhu, C.; Zhou, J. Knockdown of hsa_circ_0023028 inhibits cell proliferation, migration, and invasion in laryngeal cancer by sponging miR-194-5p. Biosci. Rep. 2019, 39. [Google Scholar] [CrossRef] [PubMed]
- Peng, S.; Song, C.; Chen, H.; Li, H.; Cao, X.; Ma, Y.; Wang, X.; Huang, Y.; Lan, X.; Lei, C.; et al. Circular RNA SNX29 Sponges miR-744 to Regulate Proliferation and Differentiation of Myoblasts by Activating the Wnt5a/Ca2+ Signaling Pathway. Mol. Ther. Nucleic Acids 2019, 16, 481–493. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Wei, X.; Lin, F.; Bai, Y.; Chen, H.; Yang, J.; Dong, D.; Hao, D.; Huang, Y.; Lan, X.; et al. circFGFR4 Promotes Differentiation of Myoblasts via Binding miR-107 to Relieve Its Inhibition of Wnt3a. Mol. Ther. Nucleic Acids 2018, 11, 272–283. [Google Scholar] [CrossRef] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, Z.; Ren, W.; Wang, T.; Zhang, W.; Wang, C.; Wang, H.; Gao, F.; Yuan, B. CircAgtpbp1 Acts as a Molecular Sponge of miR-543-5p to Regulate the Secretion of GH in Rat Pituitary Cells. Animals 2021, 11, 558. https://doi.org/10.3390/ani11020558
Yu Z, Ren W, Wang T, Zhang W, Wang C, Wang H, Gao F, Yuan B. CircAgtpbp1 Acts as a Molecular Sponge of miR-543-5p to Regulate the Secretion of GH in Rat Pituitary Cells. Animals. 2021; 11(2):558. https://doi.org/10.3390/ani11020558
Chicago/Turabian StyleYu, ZeWen, WenZhi Ren, Tian Wang, WeiDi Zhang, ChangJiang Wang, HaoQi Wang, Fei Gao, and Bao Yuan. 2021. "CircAgtpbp1 Acts as a Molecular Sponge of miR-543-5p to Regulate the Secretion of GH in Rat Pituitary Cells" Animals 11, no. 2: 558. https://doi.org/10.3390/ani11020558
APA StyleYu, Z., Ren, W., Wang, T., Zhang, W., Wang, C., Wang, H., Gao, F., & Yuan, B. (2021). CircAgtpbp1 Acts as a Molecular Sponge of miR-543-5p to Regulate the Secretion of GH in Rat Pituitary Cells. Animals, 11(2), 558. https://doi.org/10.3390/ani11020558




