Application of Next-Generation Sequencing for the Determination of the Bacterial Community in the Gut Contents of Brackish Copepod Species (Acartia hudsonica, Sinocalanus tenellus, and Pseudodiaptomus inopinus)
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
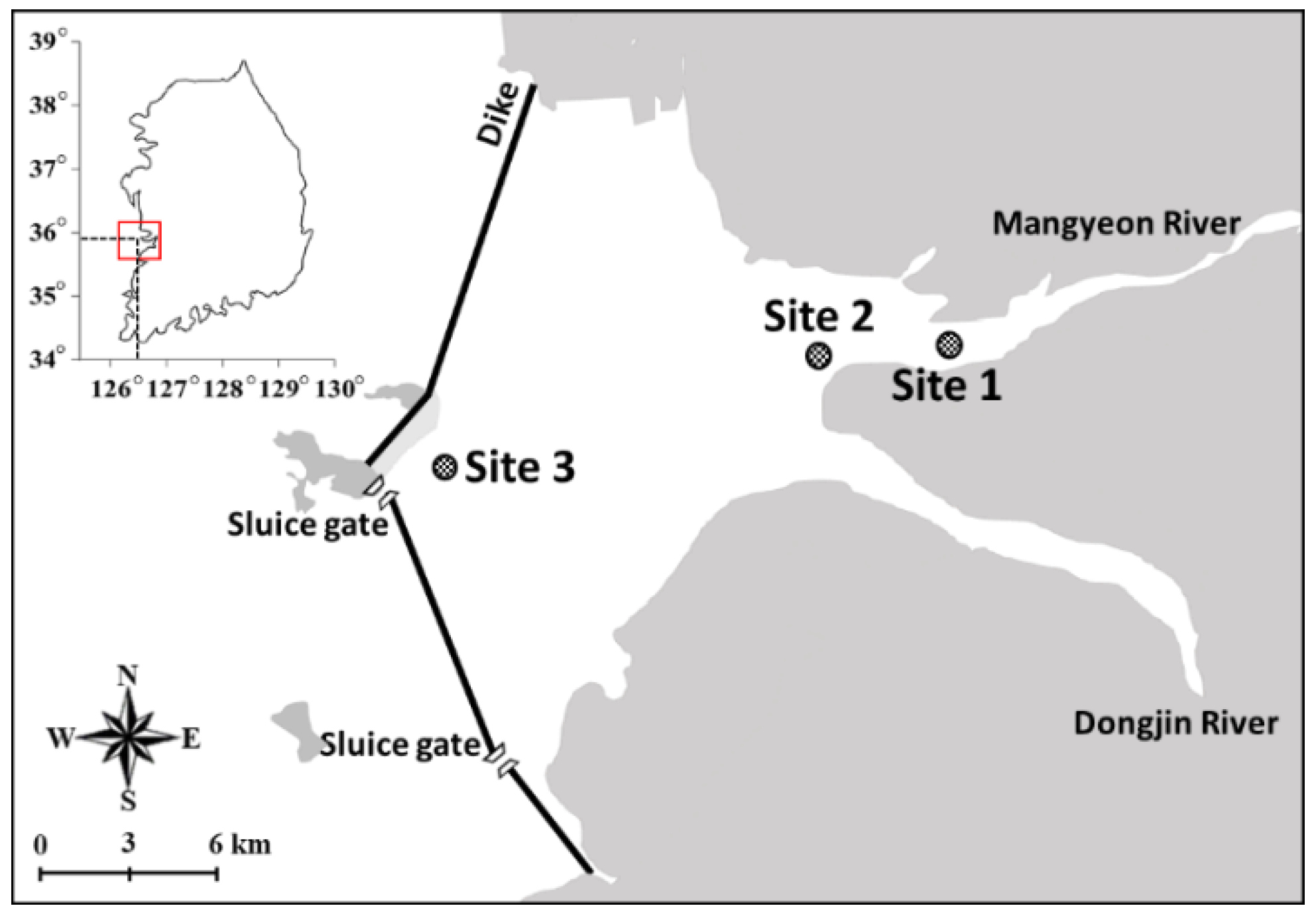
2.1. Sampling Site and Period
2.2. Environmental Characteristics: Water Quality and Plankton Compositions
2.3. Sample Collection and Treatment for DNA Analysis
2.4. DNA Extraction and 16S rRNA Amplicon Library Generation
2.5. Data Analyses
3. Results and Discussion
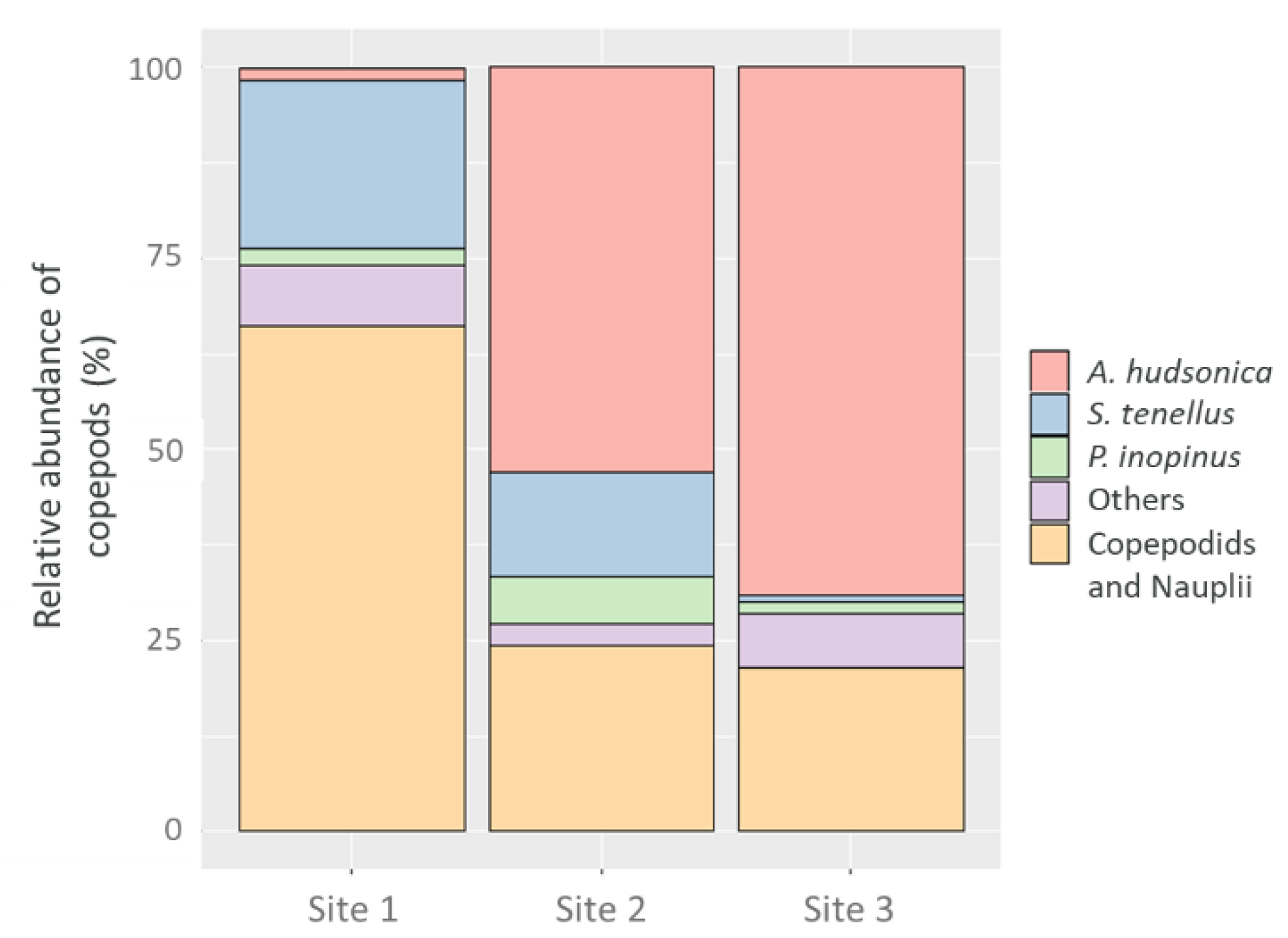
3.1. Environmental Conditions and the Copepod Community Composition by Microscopic Examination
3.2. Comparison of Copepod Gut Bacterial Communities Based on the NGS Analysis
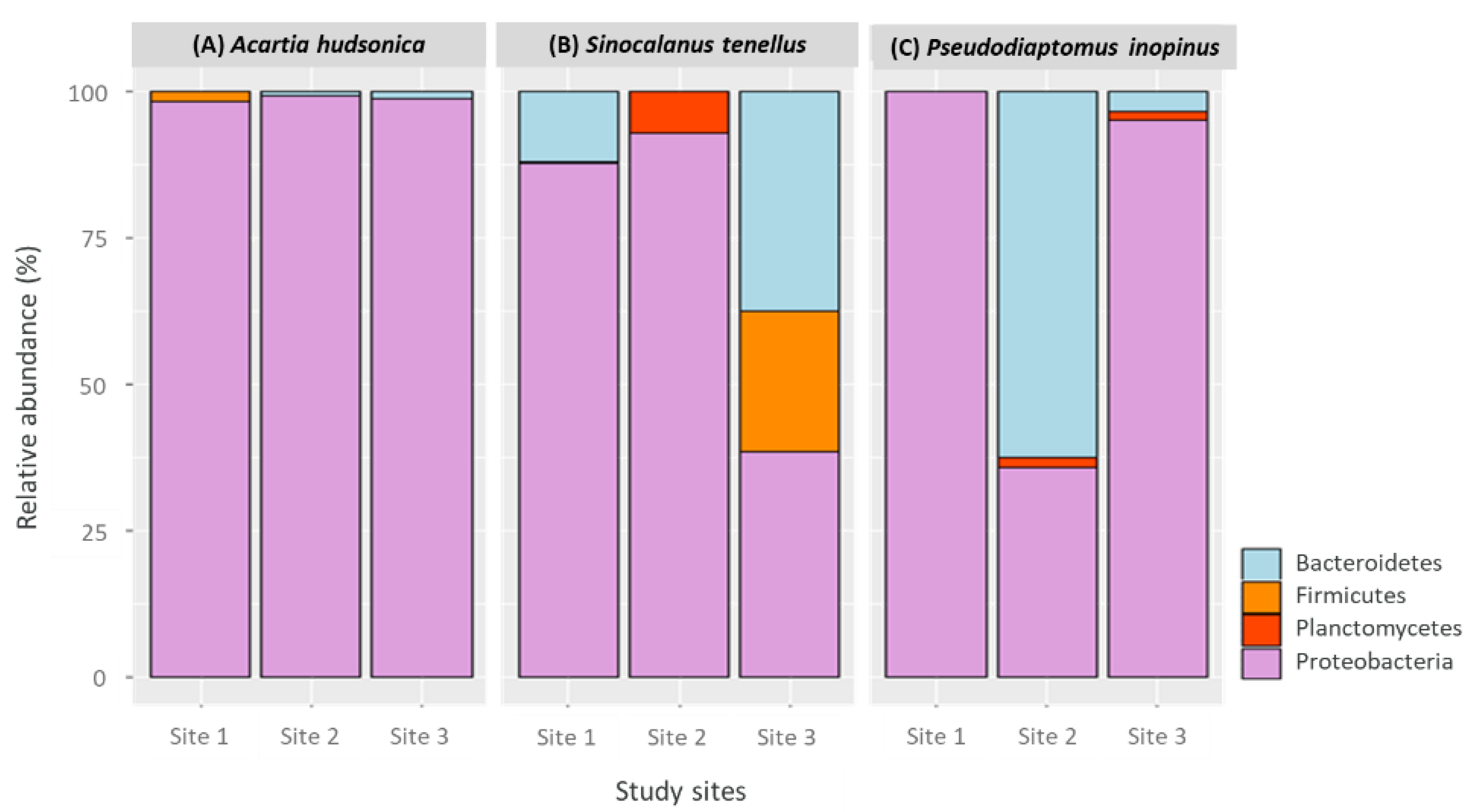
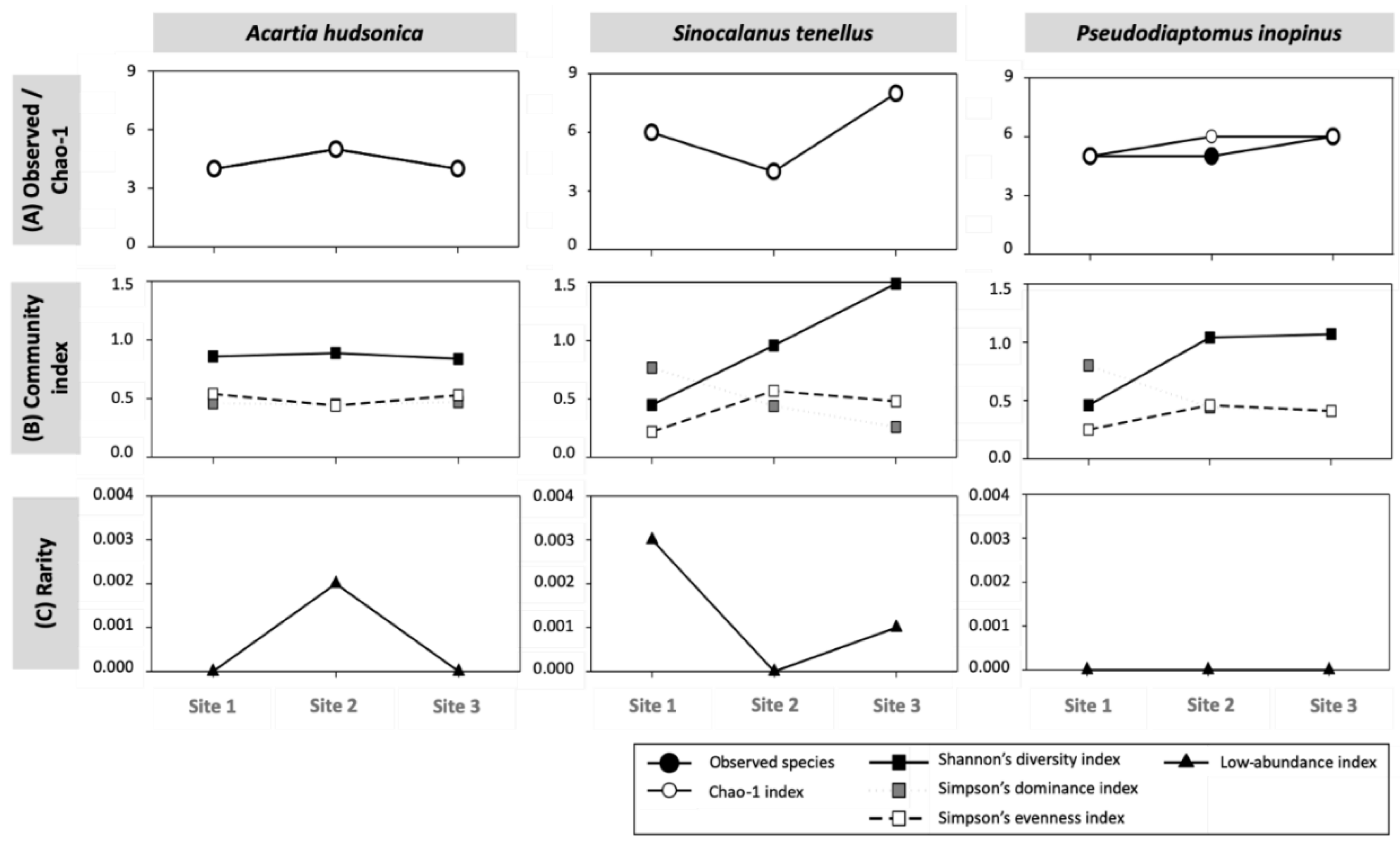
3.2.1. Community Composition and Diversity Indices
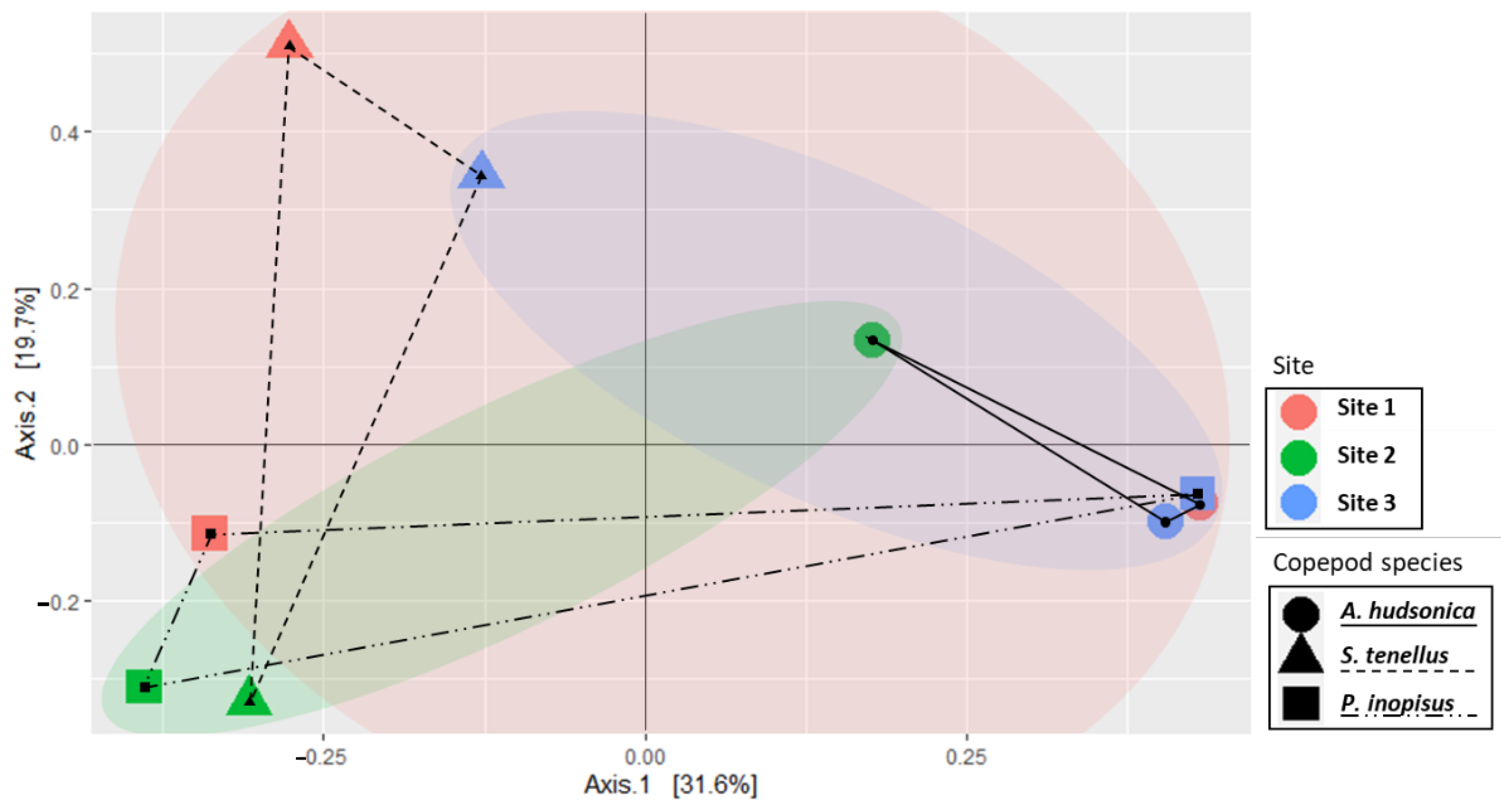
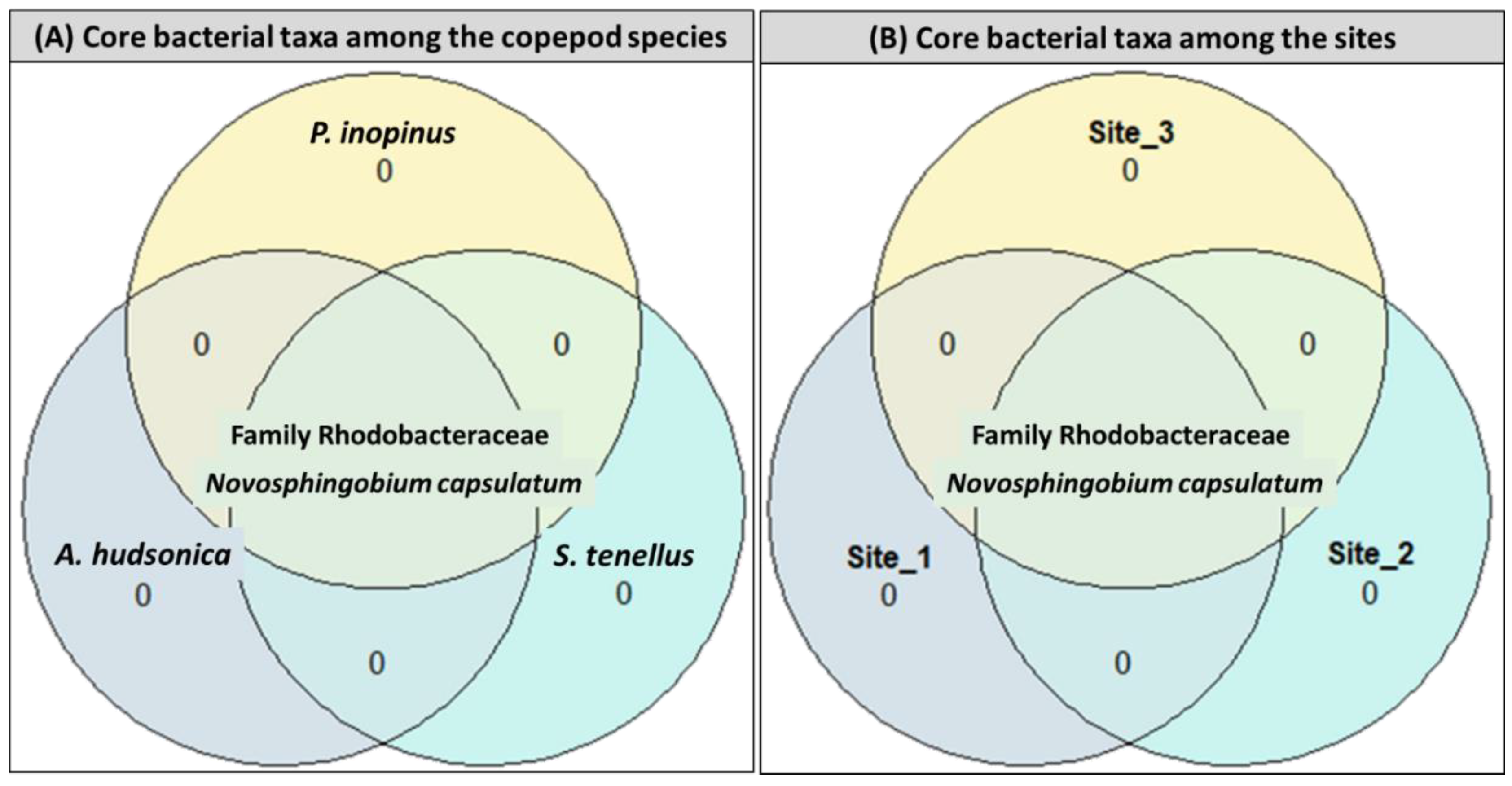
3.2.2. Community Similarity and Core Bacterial Taxa
3.2.3. Core Bacterial Species in Copepod Gut and Free-Living Bacterial Communities
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rakhesh, M.; Raman, A.V.; Ganesh, T.; Chandramohan, P.; Dehairs, F. Small copepods structuring mesozooplankton community dynamics in a tropical estuary-coastal system. Estuar. Coast. Shelf Sci. 2013, 126, 7–22. [Google Scholar] [CrossRef]
- Thor, P.; Dam, H.G.; Rogers, D.R. Fate of organic carbon released from decomposing copepod fecal pellets in relation to bacterial production and ectoenzymatic activity. Aquat. Microb. Ecol. 2003, 33, 279–288. [Google Scholar] [CrossRef]
- Møller, E.F.; Thor, P.; Nielsen, T.G. Production of DOC by Calanus finmarchicus, C. glacialis and C. hyperboreus through sloppy feeding and leakage from fecal pellets. Mar. Ecol. Prog. Ser. 2003, 262, 185–191. [Google Scholar] [CrossRef]
- Chevalier, C.; Stojanović, O.; Colin, D.J.; Suarez-Zamorano, N.; Tarallo, V.; Veyrat-Durebex, C.; Rigo, D.; Fabbiano, S.; Stevanović, A.; Hagemann, S. Gut microbiota orchestrates energy homeostasis during cold. Cell 2015, 163, 1360–1374. [Google Scholar] [CrossRef] [PubMed]
- Macke, E.; Callens, M.; De Meester, L.; Decaestecker, E. Host-genotype dependent gut microbiota drives zooplankton tolerance to toxic cyanobacteria. Nat. Commun. 2017, 8, 1608. [Google Scholar] [CrossRef]
- Tang, K.W.; Turk, V.; Grossart, H.-P. Linkage between crustacean zooplankton and aquatic bacteria. Aquat. Microb. Ecol. 2010, 61, 261–277. [Google Scholar] [CrossRef]
- Glud, R.N.; Grossart, H.P.; Larsen, M.; Tang, K.W.; Arendt, K.E.; Rysgaard, S. Copepod carcasses as microbial hot spots for pelagic denitrification. Limnol. Oceanogr. 2015, 60, 2026–2036. [Google Scholar] [CrossRef]
- Stief, P.; Lundgaard, A.B.; Morales-Ramirez, A.; Thamdrup, B.; Glud, R.N. Fixed-nitrogen loss associated with sinking zooplankton carcasses in a coastal oxygen minimum zone (Golfo Dulce, Costa Rica). Front. Mar. Sci. 2017, 4, 152. [Google Scholar] [CrossRef]
- Nuester, J.; Shema, S.; Vermont, A.; Fields, D.M.; Twining, B.S. The regeneration of highly bioavailable iron by meso- and microzooplankton. Limnol. Oceanogr. 2014, 59, 1399–1409. [Google Scholar] [CrossRef]
- Schmidt, K.; Schlosser, C.; Atkinson, A.; Fielding, S.; Venables, H.J.; Waluda, C.M.; Achterberg, E.P. Zooplankton gut passage mobilizes lithogenic iron for ocean productivity. Curr. Biol. 2016, 26, 2667–2673. [Google Scholar] [CrossRef]
- Li, Y.; Xu, Z.; Liu, H. Nutrient-imbalanced Conditions Shift the Interplay Between Zooplankton and Gut Microbiota. BMC Bioinform. 2021, 22, 1–18. [Google Scholar]
- Mioduchowska, M.; Zając, K.; Bartoszek, K.; Madanecki, P.; Kur, J.; Zając, T. 16S rRNA-based metagenomic analysis of the gut microbial community associated with the DUI species Unio crassus (Bivalvia: Unionidae). J. Zoolog. Syst. Evol. Res. 2020, 1–9. [Google Scholar] [CrossRef]
- Su, S.; Munganga, B.P.; Du, F.; Yu, J.; Li, J.; Yu, F.; Wang, M.; He, X.; Li, X.; Bouzoualegh, R.; et al. Relationship between the fatty acid profiles and gut bacterial communities of the Chinese Mitten Crab (Eriocheir sinensis) from ecologically different habitats. Front. Microbiol. 2020, 11. [Google Scholar] [CrossRef]
- Jing, T.Z.; Qi, F.H.; Wang, Z.Y. Most dominant roles of insect gut bacteria: Digestion, detoxification, or essential nutrient provision? Microbiome 2020, 8, 1–20. [Google Scholar] [CrossRef]
- Muratore, M.; Sun, Y.; Prather, C. Environmental nutrients alter bacterial and fungai gut microbiomes in the common medow katydid, Orchelimum vulgare. Front. Microbiol. 2020, 11, 2644. [Google Scholar] [CrossRef]
- Datta, M.S.; Almada, A.A.; Baumgartner, M.F.; Mincer, T.J.; Tarrant, A.M.; Polz, M.F. Inter-individual variability in copepod microbiomes reveals bacterial networks linked to host physiology. ISME J. 2018, 12, 2103–2113. [Google Scholar] [CrossRef]
- Grossart, H.-P.; Dziallas, C.; Leunert, F.; Tang, K.W. Bacteria dispersal by hitchhiking on zooplankton. Proc. Natl. Acad. Sci. USA 2010, 107, 11959–11964. [Google Scholar] [CrossRef]
- Shoemaker, K.M.; Duhamel, S.; Moisander, P.H. Copepods promote bacterial community changes in surrounding seawater through farming and nutrient enrichment. Environ. Microbiol. 2019, 21, 3737–3750. [Google Scholar] [CrossRef]
- Ho, T.W.; Hwang, J.S.; Cheung, M.K.; Kwan, H.S.; Wong, C.K. DNA-based study of the diet of the marine calanoid copepod Calanus sinicus. J. Exp. Mar. Biol. Ecol. 2017, 494, 1–9. [Google Scholar] [CrossRef]
- Hirai, J.; Hamamoto, Y.; Honda, D.; Hidaka, K. Possible aplanochytrid (Labyrinthulea) prey detected using 18S metagenetic diet analysis in the key copepod species Calanus sinicus in the coastal waters of the subtropical western North Pacific. Plankton Benthos Res. 2018, 13, 75–82. [Google Scholar] [CrossRef]
- Yeh, H.D.; Questel, J.M.; Maas, K.R.; Bucklin, A. Metabarcoding analysis of regional variation in gut contents of the copepod Calanus finmarchicus in the North Atlantic Ocean. Deep. Res. Part II Top. Stud. Oceanogr. 2020, 51, 104738. [Google Scholar] [CrossRef]
- Oh, H.J.; Krogh, P.H.; Jeong, H.G.; Joo, G.J.; Kwak, I.S.; Hwang, S.J.; Gim, J.S.; Chang, K.H.; Jo, H. Pretreatment method for DNA barcoding to analyze gut contents of rotifers. Appl. Sci. 2020, 10, 1064. [Google Scholar] [CrossRef]
- Majumder, S.; Dhua, R.P.; Kar, S.; Mishra, T.; Mahapatra, S.S.; Shit, S.; Patra, A. Zooplankton diversity influenced by hydro biological parameters in some ponds of south eastern part of Bankura town of WB, India. Int. J. Adv. Res. 2015, 3, 354–368. [Google Scholar]
- Oda, Y.; Nakano, S.; Suh, J.M.; Oh, H.J.; Jin, M.Y.; Kim, Y.J.; Chang, K.H. Spatiotemporal variability in a copepod community associated with fluctuations in salinity and trophic state in an artificial brackish reservoir at Saemangeum, Korea. PLoS ONE 2018, 13, e0209403. [Google Scholar] [CrossRef]
- Lie, H.J.; Cho, C.H.; Lee, S.; Kim, E.S.; Koo, B.J.; Noh, J.H. Changes in marine environment by a large coastal development of the Saemangeum reclamation project in Korea. Ocean. Polar Res. 2008, 30, 475–484. [Google Scholar] [CrossRef]
- Ryu, J.; Khim, J.S.; Choi, J.W.; Shin, H.C.; An, S.; Park, J. Environmentally associated spatial changes of a macrozoobenthic community in the Saemangeum tidal flat, Korea. J. Sea Res. 2011, 65, 390–400. [Google Scholar] [CrossRef]
- Lee, H.J.; Lee, S.H. Geological consequences of the Saemangeum Dyke, mid–west coast of korea: A review. Ocean. Sci. J. 2012, 47, 395–410. [Google Scholar] [CrossRef]
- Ministry of Environment. Water Pollution Process Standard; Korean literature; Ministry of Environment: Sejong, Korea, 2018. [Google Scholar]
- Ministry of Oceans and Fisheries. Marine Environmental Process Standards; Korean literature; Ministry of Oceans and Fisheries: Sejong, Korea, 2018. [Google Scholar]
- Hendey, N.I. The permanganate method for cleaning using diatoms. Nova Hedwig. Beih. 1974, 64, 305–323. [Google Scholar]
- Bennke, C.M.; Pollehne, F.; Müller, A.; Hansen, R.; Kreikemeyer, B.; Labrenz, M. The distribution of phytoplankton in the Baltic Sea assessed by a prokaryotic 16S rRNA gene primer system. J. Plankton Res. 2018, 40, 244–254. [Google Scholar] [CrossRef]
- Edgar, R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010, 26, 2460–2461. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335. [Google Scholar] [CrossRef] [PubMed]
- Lahti, L.; Shetty, S. Tools for Microbiome Analysis in R. Version 1.5.28. 2017. Available online: http://microbiome.github.com/microbiome (accessed on 10 November 2020).
- Lance, J. Respiration and osmotic behaviour of the copepod Acartia tonsa in diluted sea water. Comp. Biochem. Physiol. 1965, 14, 155–165. [Google Scholar] [CrossRef]
- Tester, P.A.; Turner, J.T. Why is Acartia tonsa restricted to estuarine habitats. In Proceedings of the Fourth International Conference Copepoda, Karuizawa, Japan, 16-20 September 1990; pp. 603–611. [Google Scholar]
- Cervetto, G.; Gaudy, R.; Pagano, M. Influence of salinity on the distribution of Acartia tonsa (Copepoda, Calanoida). J. Exp. Mar. Biol. Ecol. 1999, 239, 33–45. [Google Scholar] [CrossRef]
- Lu, K.; Lu, Y.; Lin, X.; Zheng, Z.; Yao, G. Effects of the experimental factors on filtering and grazing rates of Sinocalanus tenellus. Mar. Sci. Haiyang Kexue 2001, 25, 44–47. [Google Scholar]
- Cordell, J.R.; Rassmussen, M.; Bollens, S.M. Biology of the invasive copepod Pseudodiaptomus inopinus in a northeast Pacific estuary. Mar. Ecol. Prog. Ser. 2007, 333, 213–227. [Google Scholar] [CrossRef]
- Grossart, H.P.; Dziallas, C.; Tang, K.W. Bacterial diversity associated with freshwater zooplankton. Environ. Microbiol. Rep. 2009, 1, 50–55. [Google Scholar] [CrossRef]
- Bickel, S.L.; Tang, K.W.; Grossart, H.P. Structure and function of zooplankton-associated bacterial communities in a temperate estuary change more with time than with zooplankton species. Aquat. Microb. Ecol. 2014, 72, 1–15. [Google Scholar] [CrossRef]
- Tang, K.; Dziallas, C.; Hutalle-Schmelzer, K.; Grossart, H.P. Effects of food on bacterial community composition associated with the copepod Acartia tonsa Dana. Biol. Lett. 2009, 5, 54. [Google Scholar] [CrossRef]
- Bickel, S.L.; Tang, K.W. Carbon substrate usage by zooplankton-associated bacteria, phytoplankton-associated bacteria, and free-living bacteria under aerobic and anaerobic conditions. Mar. Biol. 2014, 161, 2233–2242. [Google Scholar] [CrossRef]
- Bradford-Grieve, J.M.; Boxshall, G.A.; Ahyong, S.T.; Ohtsuka, S. Cladistic analysis of the calanoid copepoda. Invertebr. Syst. 2010, 24, 291–321. [Google Scholar] [CrossRef][Green Version]
- Rollwagen Bollens, G.C.; Penry, D.L. Feeding dynamics of Acartia spp. copepods in a large, temperate estuary (San Francisco Bay, CA). Mar. Ecol. Prog. Ser. 2003, 257, 139–158. [Google Scholar] [CrossRef]
- Hada, A.; Uye, S. Cannibalistic feeding behavior of the brackish-water copepod Sinocalanus tenellus. J. Plankton Res. 1991, 13, 155–166. [Google Scholar] [CrossRef]
- Chen, M.; Kim, D.; Liu, H.; Kang, C.K. Variability in copepod trophic levels and feeding selectivity based on stable isotope analysis in Gwangyang Bay of the southern coast of the Korean Peninsula. Biogeosciences 2018, 15, 2055–2073. [Google Scholar] [CrossRef]
- De Corte, D.; Lekunberri, I.; Sintes, E.; Garcia, J.A.L.; Gonzales, S.; Herndl, G.J. Linkage between copepods and bacteria in the North Atlantic Ocean. Aquat. Microb. Ecol. 2014, 72, 215–225. [Google Scholar] [CrossRef]
- Haggerty, J.M.; Dinsdale, E.A. Distinct biogeographical patterns of marine bacterial taxonomy and functional genes. Glob. Ecol. Biogeogr. 2017, 26, 177–190. [Google Scholar] [CrossRef]
- Pohlner, M.; Dlugosch, L.; Wemheuer, B.; Mills, H.; Engelen, B.; Reese, B.K. The majority of active Rhodobacteraceae in marine sediments belong to uncultured genera: A molecular approach to link their distribution to environmental conditions. Front. Microbiol. 2019, 10, 659. [Google Scholar] [CrossRef]
- Gan, H.M.; Hudson, A.O.; Rahman, A.Y.A.; Chan, K.G.; Savka, M.A. Comparative genomic analysis of six bacteria belonging to the genus Novosphingobium: Insights into marine adaptation, cell-cell signaling and bioremediation. BMC Genom. 2013, 14, 431. [Google Scholar] [CrossRef]
| Factors | Site 1 | Site 2 | Site 3 | ||
|---|---|---|---|---|---|
| Water quality | Water depths | (m) | 2 | 4.5 | 22.5 |
| Temperature | °C | 14.6 | 13.7 | 13.0 | |
| Salinity | (‰) | 4.1 | 5.1 | 12.9 | |
| pH | 8.4 | 8.5 | 8.4 | ||
| Dissolved Oxygen | (mg/L) | 10.7 | 11.1 | 9.2 | |
| Chemical Oxygen Demand | (mg/L) | 5.2 | 8.4 | 6.8 | |
| Electrical Conductivity | (S/m) | 5898 | 6795 | 15,665 | |
| Chlorophyll-a | (μg/L) | 5.4 | 7.0 | 4.9 | |
| Total Nitrogen | (mg/L) | 3.391 | 3.023 | 1.240 | |
| Total Phosphorus | (mg/L) | 0.084 | 0.039 | 0.045 | |
| Phytoplankton composition | Chlorophyta | (%) | 38.75 | 24.82 | 7.32 |
| Bacillariophyta | (%) | 61.25 | 75.08 | 92.68 | |
| Sample | Dominant and Subdominant Bacterial Species | ||||
|---|---|---|---|---|---|
| Phylum | Class | Family/Genus/Species | OTUs (%) | ||
| (A) A. hudsonica | Site 1 | Proteobacteria | Alphaproteobacteria | Novosphingobium capsulatum | 752 (48.9) |
| Rhodobacteraceae | 725 (47.1) | ||||
| Site 2 | Proteobacteria | Alphaproteobacteria | N. capsulatum | 6813 (51.1) | |
| Rhodobacteraceae | 5799 (43.5) | ||||
| Site 3 | Proteobacteria | Alphaproteobacteria | Rhodobacteraceae | 671 (54.1) | |
| N. capsulatum | 524 (42.2) | ||||
| (B) S. tenellus | Site 1 | Proteobacteria | Gammaproteobacteria | Aeromonas hydrophila | 13,024 (86.7) |
| Bacteroidetes | Bacteroidia | Muribaculum sp. | 1366 (9.1) | ||
| Site 2 | Proteobacteria | Alphaproteobacteria | Rhodobacteraceae | 54 (54.5) | |
| N. capsulatum | 36 (36.4) | ||||
| Site 3 | Bacteroidetes Firmicutes | Cytophagia Bacilli | Sporocytophaga sp. | 1546 (37.3) | |
| Bacillus velezensis | 1005 (24.2) | ||||
| (C) P. inopinus | Site 1 | Bacteroidetes | Flavobacteriia | Polaribacter sp. | 41 (61.2) |
| Proteobacteria | Alphaproteobacteria | Rhodobacteraceae | 12 (17.9) | ||
| N. capsulatum | |||||
| Site 2 | Proteobacteria | Alphaproteobacteria | Brevundimonas denitrificans | 373 (89.2) | |
| Rhodobacteraceae | 25 (6.0) | ||||
| Site 3 | Proteobacteria | Alphaproteobacteria | Rhodobacteraceae | 1131 (48.6) | |
| N. capsulatum | 958 (41.2) | ||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chae, Y.-J.; Oh, H.-J.; Chang, K.-H.; Kwak, I.-S.; Jo, H. Application of Next-Generation Sequencing for the Determination of the Bacterial Community in the Gut Contents of Brackish Copepod Species (Acartia hudsonica, Sinocalanus tenellus, and Pseudodiaptomus inopinus). Animals 2021, 11, 542. https://doi.org/10.3390/ani11020542
Chae Y-J, Oh H-J, Chang K-H, Kwak I-S, Jo H. Application of Next-Generation Sequencing for the Determination of the Bacterial Community in the Gut Contents of Brackish Copepod Species (Acartia hudsonica, Sinocalanus tenellus, and Pseudodiaptomus inopinus). Animals. 2021; 11(2):542. https://doi.org/10.3390/ani11020542
Chicago/Turabian StyleChae, Yeon-Ji, Hye-Ji Oh, Kwang-Hyeon Chang, Ihn-Sil Kwak, and Hyunbin Jo. 2021. "Application of Next-Generation Sequencing for the Determination of the Bacterial Community in the Gut Contents of Brackish Copepod Species (Acartia hudsonica, Sinocalanus tenellus, and Pseudodiaptomus inopinus)" Animals 11, no. 2: 542. https://doi.org/10.3390/ani11020542
APA StyleChae, Y.-J., Oh, H.-J., Chang, K.-H., Kwak, I.-S., & Jo, H. (2021). Application of Next-Generation Sequencing for the Determination of the Bacterial Community in the Gut Contents of Brackish Copepod Species (Acartia hudsonica, Sinocalanus tenellus, and Pseudodiaptomus inopinus). Animals, 11(2), 542. https://doi.org/10.3390/ani11020542







