Highly Fermentable Fiber Alters Fecal Microbiota and Mitigates Swine Dysentery Induced by Brachyspira hyodysenteriae
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals, Housing, and Experimental Design
2.2. Histopathology
2.3. Fecal DNA Extraction and 16S Library Preparation
2.4. Bioinformatics
2.5. Statistical Analysis
3. Results
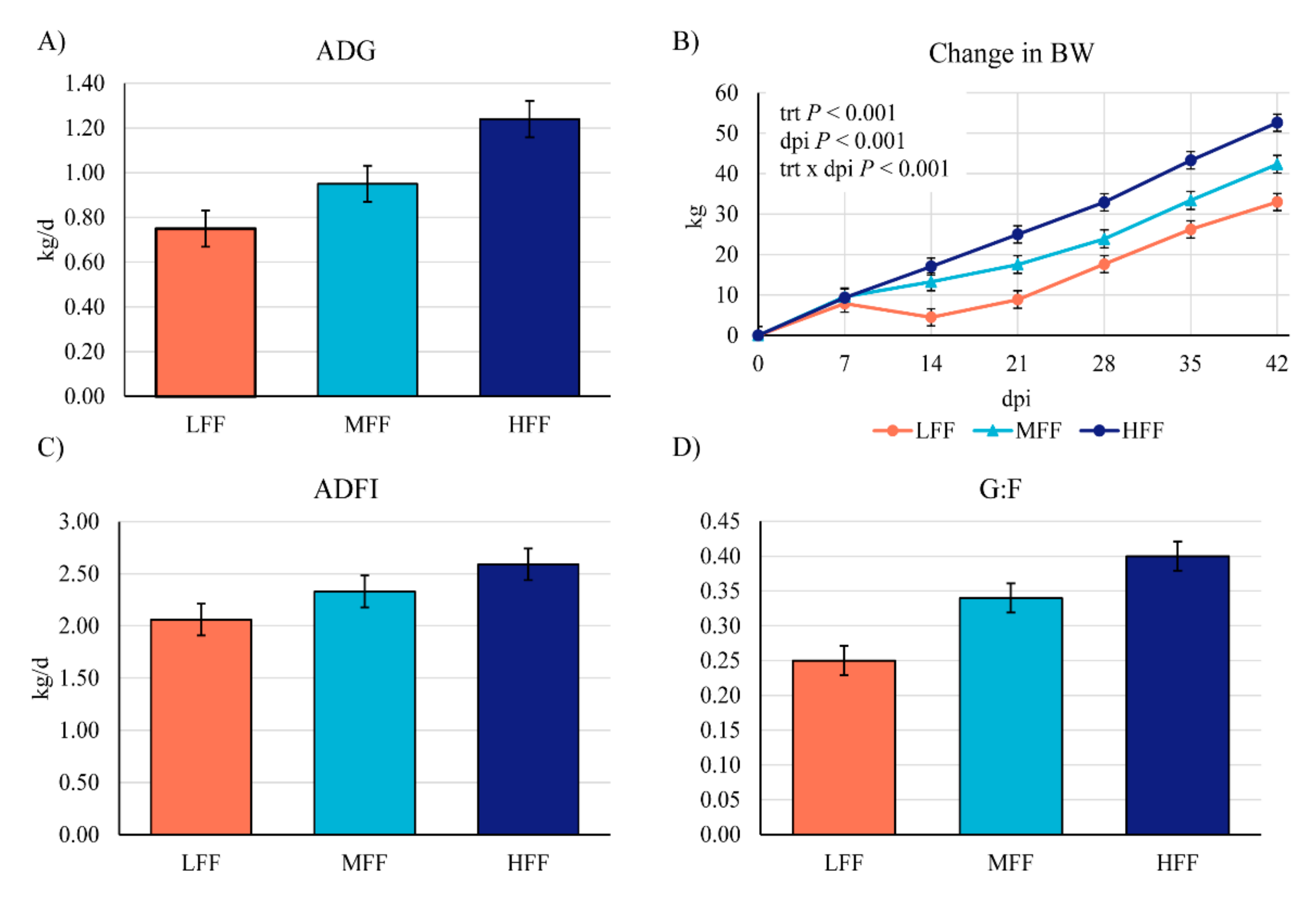
3.1. Clinical Observations, Diagnostics, and Growth Performance
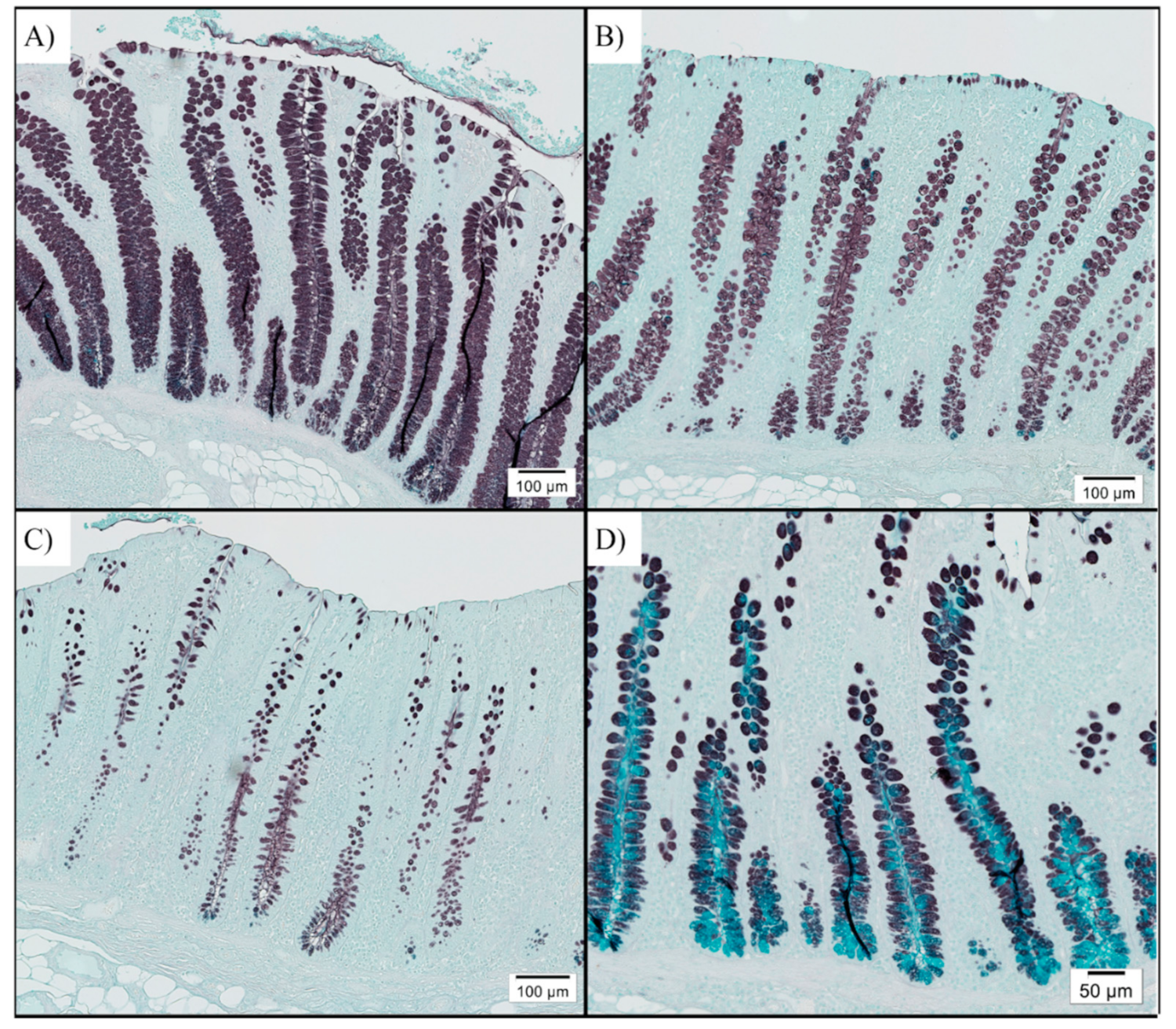
3.2. Digesta pH and Colon Goblet Cell Counts
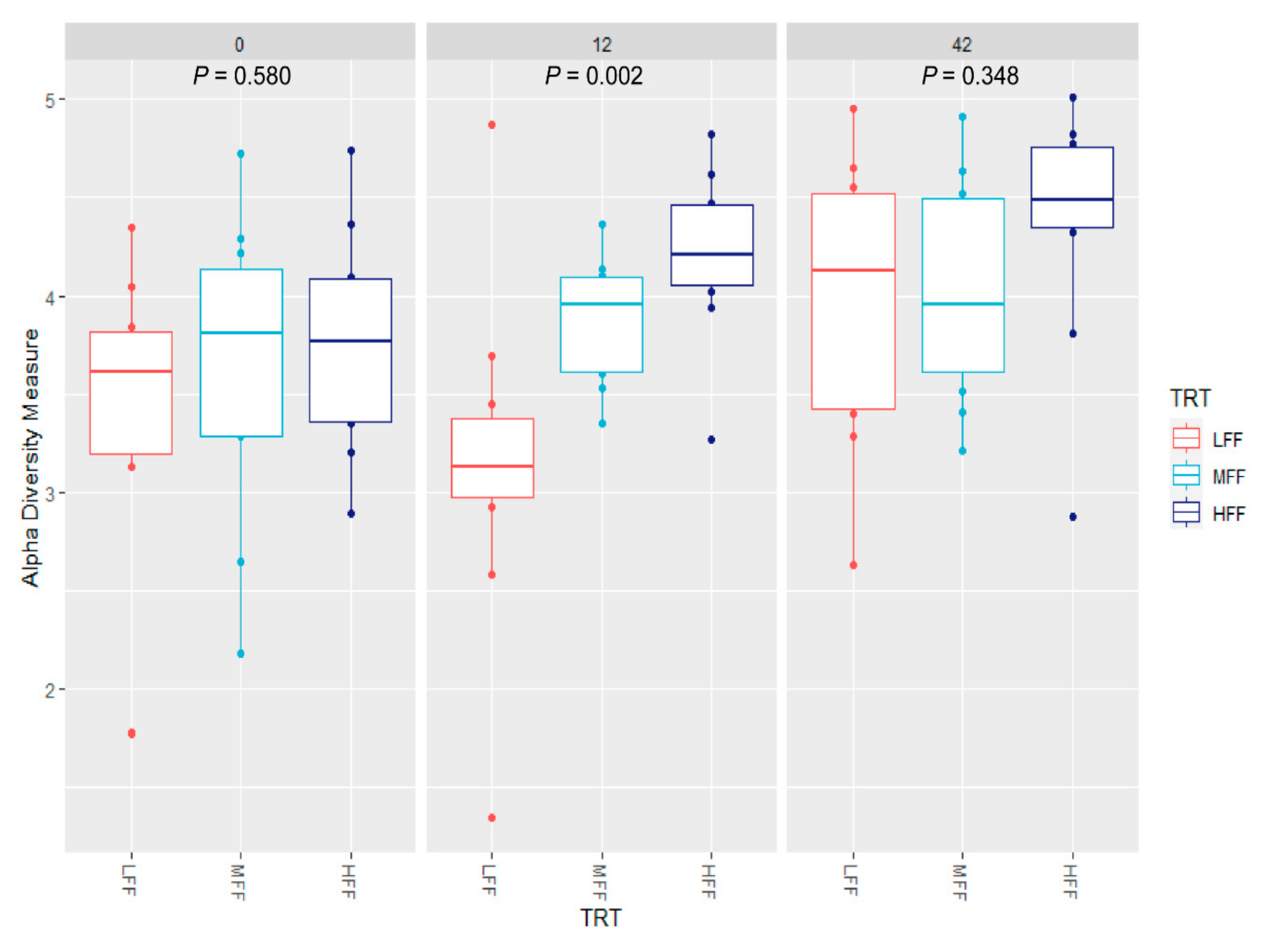
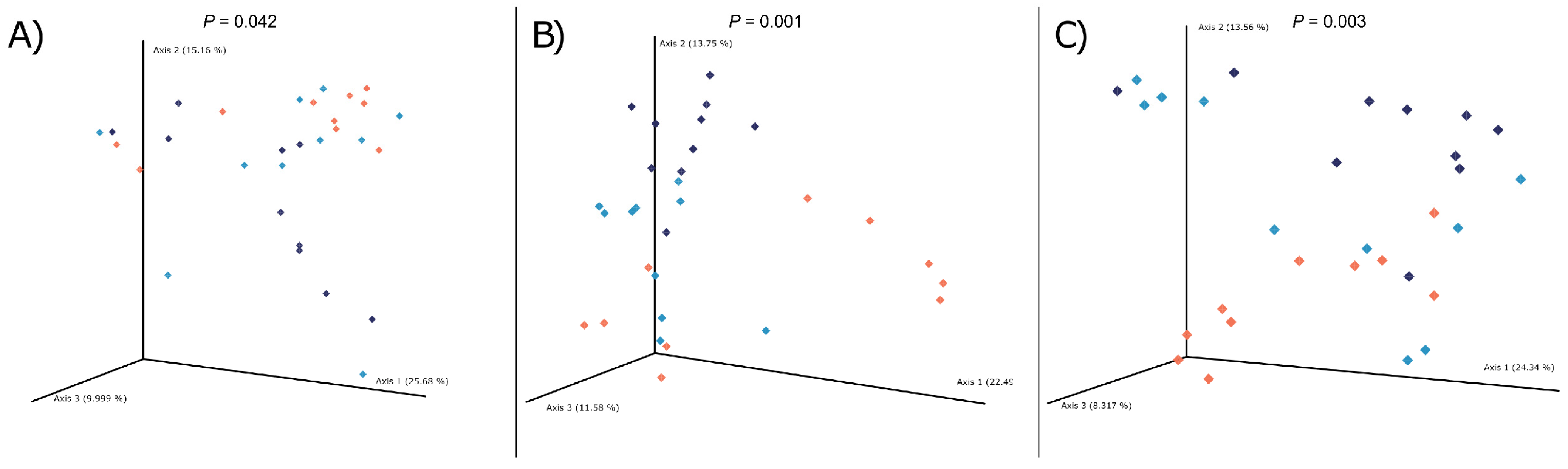
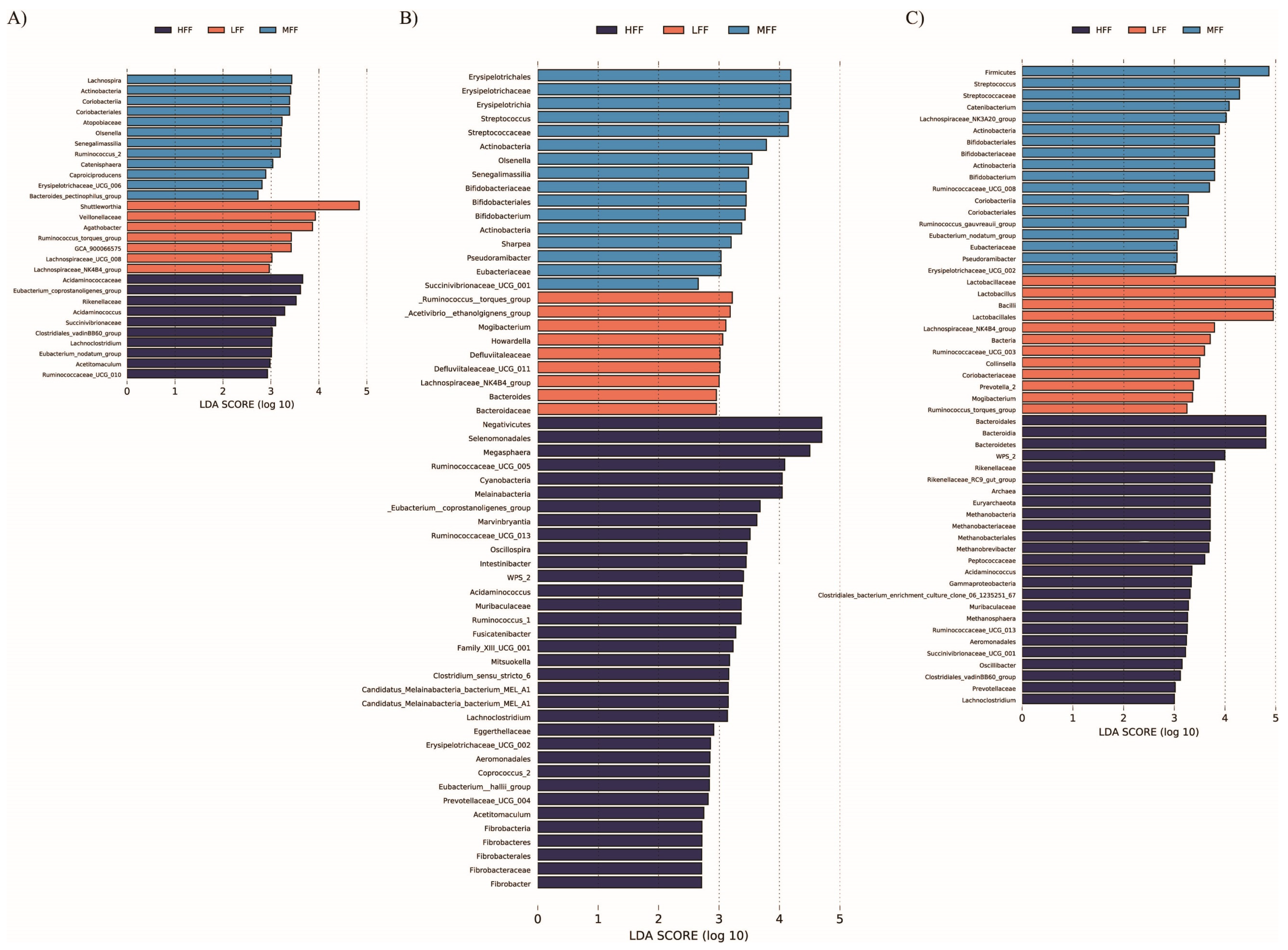
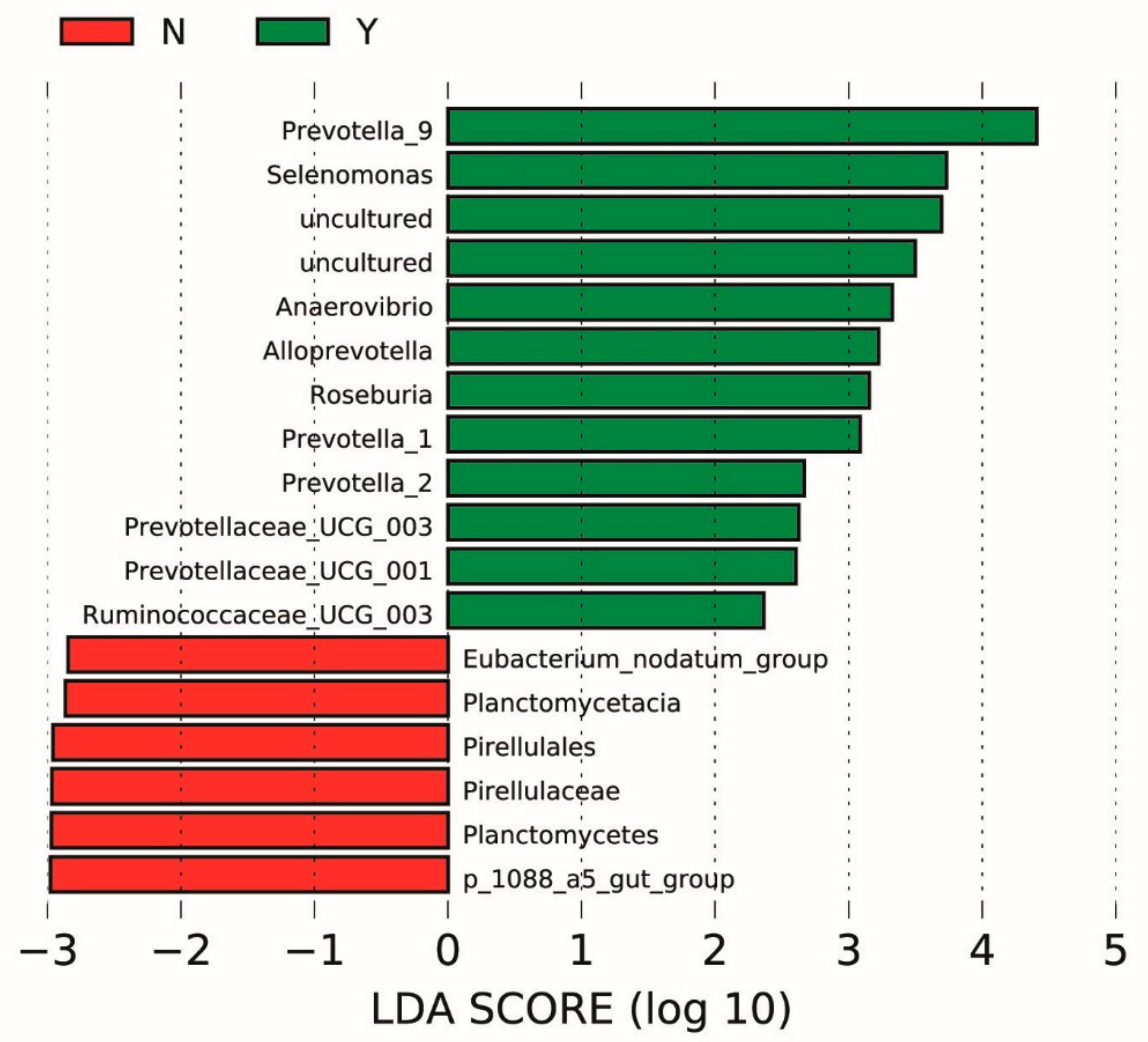
3.3. Microbial Communities
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Burrough, E.R. Swine Dysentery. Vet. Pathol. 2017, 54, 22–31. [Google Scholar] [CrossRef] [PubMed]
- Pluske, J.R.; Durmic, Z.; Pethick, D.W.; Mullan, B.P.; Hampson, D.J. Confirmation of the role of rapidly fermentable carbohydrates in the expression of swine dysentery in pigs after experimental infection. J. Nutr. 1998, 128, 1737–1744. [Google Scholar] [CrossRef] [PubMed]
- Hansen, C.F.; Phillips, N.D.; La, T.; Hernandez, A.; Mansfield, J.; Kim, J.C.; Mullan, B.P.; Hampson, D.J.; Pluske, J.R. Diets containing inulin but not lupins help to prevent swine dysentery in experimentally challenged pigs. J. Anim. Sci. 2010, 88, 3327–3336. [Google Scholar] [CrossRef] [PubMed]
- Wilberts, B.L.; Arruda, P.H.; Kinyon, J.M.; Frana, T.S.; Wang, C.; Magstadt, D.R.; Madson, D.M.; Patience, J.F.; Burrough, E.R. Investigation of the impact of increased dietary insoluble fiber through the feeding of distillers dried grains with solubles (DDGS) on the incidence and severity of Brachyspira-associated colitis in pigs. PLoS ONE 2014, 9, e114741. [Google Scholar] [CrossRef] [PubMed]
- Siba, P.M.; Pethick, D.W.; Hampson, D.J. Pigs experimentally infected with Serpulina hyodysenteriae can be protected from developing swine dysentery by feeding them a highly digestible diet. Epidemiol. Infect. 1996, 116, 207–216. [Google Scholar] [CrossRef] [PubMed]
- Hansen, C.F.; Hernandez, A.; Mansfield, J.; Hidalgo, A.; La, T.; Phillips, N.D.; Hampson, D.J.; Pluske, J.R. A high dietary concentration of inulin is necessary to reduce the incidence of swine dysentery in pigs experimentally challenged with Brachyspira hyodysenteriae. Br. J. Nutr. 2011, 106, 1506–1513. [Google Scholar] [CrossRef] [PubMed]
- Montagne, L.; Piel, C.; Lalles, J.P. Effect of diet on mucin kinetics and composition: Nutrition and health implications. Nutr. Rev. 2004, 62, 105–114. [Google Scholar] [CrossRef] [PubMed]
- Montagne, L.; Pluske, J.R.; Hampson, D.J. A review of interactions between dietary fibre and the intestinal mucosa, and their consequences on digestive health in young non-ruminant animals. Anim. Feed Sci. Technol. 2003, 108, 95–117. [Google Scholar] [CrossRef]
- Quintana-Hayashi, M.P.; Venkatakrishnan, V.; Haesebrouck, F.; Linden, S. Role of Sialic Acid in Brachyspira hyodysenteriae Adhesion to Pig Colonic Mucins. Infect. Immun. 2019, 87. [Google Scholar] [CrossRef]
- AACC. The Definition of Dietary Fiber; American Association of Cereal Chemists: Eagan, MN, USA, 2001; pp. 112–126. [Google Scholar]
- Cummings, J.H.; Stephen, A.M. Carbohydrate terminology and classification. Eur. J. Clin. Nutr. 2007, 61 (Suppl. 1), S5–S18. [Google Scholar] [CrossRef]
- Mølbak, L.; Thomsen, L.; Jensen, T.K.; Bach Knudsen, K.; Boye, M. Increased amount of Bifidobacterium thermacidophilum and Megasphaera elsdenii in the colonic microbiota of pigs fed a swine dysentery preventive diet containing chicory roots and sweet lupine. J. Appl. Microbiol. 2007, 103, 1853–1867. [Google Scholar] [CrossRef]
- Trachsel, J.; Briggs, C.; Gabler, N.K.; Allen, H.K.; Loving, C.L. Dietary Resistant Potato Starch Alters Intestinal Microbial Communities and Their Metabolites, and Markers of Immune Regulation and Barrier Function in Swine. Front. Immunol. 2019, 10, 1381. [Google Scholar] [CrossRef] [PubMed]
- Burrough, E.R.; Arruda, B.L.; Patience, J.F.; Plummer, P.J. Alterations in the Colonic Microbiota of Pigs Associated with Feeding Distillers Dried Grains with Solubles. PLoS ONE 2015, 10, e0141337. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Burrough, E.R.; Arruda, B.L.; Plummer, P.J. Comparison of the Luminal and Mucosa-Associated Microbiota in the Colon of Pigs with and without Swine Dysentery. Front. Vet. Sci. 2017, 4, 139. [Google Scholar] [CrossRef]
- Helm, E.T.; Lin, S.J.; Gabler, N.K.; Burrough, E.R. Brachyspira hyodysenteriae Infection Reduces Digestive Function but Not Intestinal Integrity in Growing Pigs While Disease Onset Can Be Mitigated by Reducing Insoluble Fiber. Front. Vet. Sci. 2020, 7, 587926. [Google Scholar] [CrossRef]
- National Research Council. Nutrient Requirements of Swine: Eleventh Revised Edition; The National Academies Press: Washington, DC, USA, 2012. [Google Scholar]
- Horwitz, W.; AOAC. Official Methods of Analysis, 18th ed.; Association of Official Analytical Chemists: Washington, DC, USA, 2005; Volume 222. [Google Scholar]
- Curry, S.M.; Schwartz, K.J.; Yoon, K.J.; Gabler, N.K.; Burrough, E.R. Effects of porcine epidemic diarrhea virus infection on nursery pig intestinal function and barrier integrity. Vet. Microbiol. 2017, 211, 58–66. [Google Scholar] [CrossRef]
- Kozich, J.J.; Westcott, S.L.; Baxter, N.T.; Highlander, S.K.; Schloss, P.D. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl. Environ. Microbiol. 2013, 79, 5112–5120. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef] [PubMed]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef] [PubMed]
- McMurdie, P.J.; Holmes, S. Waste Not, Want Not: Why Rarefying Microbiome Data Is Inadmissible. PLoS Comput. Biol. 2014, 10, e1003531. [Google Scholar] [CrossRef] [PubMed]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Pluske, J.R.; Siba, P.M.; Pethick, D.W.; Durmic, Z.; Mullan, B.P.; Hampson, D.J. The incidence of swine dysentery in pigs can be reduced by feeding diets that limit the amount of fermentable substrate entering the large intestine. J. Nutr. 1996, 126, 2920–2933. [Google Scholar] [CrossRef] [PubMed]
- Schweer, W.P.; Burrough, E.R.; Patience, J.F.; Kerr, B.J.; Gabler, N.K. Impact of Brachyspira hyodysenteriae on intestinal amino acid digestibility and endogenous amino acid losses in pigs. J. Anim. Sci. 2019, 97, 257–268. [Google Scholar] [CrossRef] [PubMed]
- Kirkwood, R.N.; Huang, S.X.; McFall, M.; Aherne, F.X. Dietary factors do not influence the clinical expression of swine dysentery. J. Swine Health Prod. 2000, 8, 73–76. [Google Scholar]
- Zohoun, E.V.; Ndindeng, S.A.; Soumanou, M.M.; Tang, E.N.; Bigoga, J.; Manful, J.; Sanyang, S.; Akissoe, N.H.; Futakuchi, K. Appropriate parboiling steaming time at atmospheric pressure and variety to produce rice with weak digestive properties. Food Sci. Nutr. 2018, 6, 757–764. [Google Scholar] [CrossRef]
- Serena, A.; Knudsen, K.E.B. Chemical and physicochemical characterisation of co-products from the vegetable food and agro industries. Anim. Feed Sci. Technol. 2007, 139, 109–124. [Google Scholar] [CrossRef]
- Nugent, A.P. Health properties of resistant starch. Nutr. Bull. 2005, 30, 27–54. [Google Scholar] [CrossRef]
- Satchithanandam, S.; Vargofcak-Apker, M.; Calvert, R.J.; Leeds, A.R.; Cassidy, M.M. Alteration of gastrointestinal mucin by fiber feeding in rats. J. Nutr. 1990, 120, 1179–1184. [Google Scholar] [CrossRef]
- Rieger, J.; Drewes, B.; Hunigen, H.; Plendl, J. Mucosubstances in the porcine gastrointestinal tract: Fixation, staining and quantification. Eur. J. Histochem. 2019, 63. [Google Scholar] [CrossRef]
- Wilberts, B.L. Investigation of Swine Dysentery Associated with “Brachyspira hampsonii” Strain EB107 and Comparison of Diagnostic Methods; Iowa State University: Ames, IA, USA, 2014. [Google Scholar]
- Kleessen, B.; Hartmann, L.; Blaut, M. Fructans in the diet cause alterations of intestinal mucosal architecture, released mucins and mucosa-associated bifidobacteria in gnotobiotic rats. Br. J. Nutr. 2003, 89, 597–606. [Google Scholar] [CrossRef]
- Harris, D.L.; Alexander, T.J.; Whipp, S.C.; Robinson, I.M.; Glock, R.D.; Matthews, P.J. Swine dysentery: Studies of gnotobiotic pigs inoculated with Treponema hyodysenteriae, Bacteroides vulgatus, and Fusobacterium necrophorum. J. Am. Vet. Med. Assoc. 1978, 172, 468–471. [Google Scholar]
- Whipp, S.; Robinson, I.; Harris, D.; Glock, R.; Matthews, P.; Alexander, T. Pathogenic synergism between Treponema hyodysenteriae and other selected anaerobes in gnotobiotic pigs. Infect. Immun. 1979, 26, 1042–1047. [Google Scholar] [CrossRef]
- Costa, M.O.; Fernando, C.; Nosach, R.; Harding, J.C.S.; Hill, J.E. Infection of porcine colon explants with “Brachyspira hampsonii” leads to increased epithelial necrosis and catarrhal exudate. Pathog. Dis. 2017, 75. [Google Scholar] [CrossRef] [PubMed]
- Thorell, K.; Inganäs, L.; Backhans, A.; Agréus, L.; Öst, Å.; Walker, M.M.; Talley, N.J.; Kjellström, L.; Andreasson, A.; Engstrand, L. Isolates from Colonic Spirochetosis in Humans Show High Genomic Divergence and Potential Pathogenic Features but Are Not Detected Using Standard Primers for the Human Microbiota. J. Bacteriol. 2019, 201. [Google Scholar] [CrossRef]
- Cole, J.R.; Wang, Q.; Fish, J.A.; Chai, B.; McGarrell, D.M.; Sun, Y.; Brown, C.T.; Porras-Alfaro, A.; Kuske, C.R.; Tiedje, J.M. Ribosomal Database Project: Data and tools for high throughput rRNA analysis. Nucleic Acids Res. 2014, 42, D633–D642. [Google Scholar] [CrossRef]
- Hong, S.; Bunge, J.; Leslin, C.; Jeon, S.; Epstein, S.S. Polymerase chain reaction primers miss half of rRNA microbial diversity. ISME J. 2009, 3, 1365–1373. [Google Scholar] [CrossRef] [PubMed]
- Tremblay, J.; Singh, K.; Fern, A.; Kirton, E.; He, S.; Woyke, T.; Lee, J.; Chen, F.; Dangl, J.; Tringe, S. Primer and platform effects on 16S rRNA tag sequencing. Front. Microbiol. 2015, 6. [Google Scholar] [CrossRef] [PubMed]
- Balvočiūtė, M.; Huson, D.H. SILVA, RDP, Greengenes, NCBI and OTT—How do these taxonomies compare? BMC Genom. 2017, 18, 1–8. [Google Scholar] [CrossRef]
- Li, Q.Y.; Peng, X.Y.; Burrough, E.R.; Sahin, O.; Gould, S.A.; Gabler, N.K.; Loving, C.L.; Dorman, K.S.; Patience, J.F. Dietary Soluble and Insoluble Fiber with or without Enzymes Altered the Intestinal Microbiota in Weaned Pigs Challenged with Enterotoxigenic E. coli F18. Front. Microbiol. 2020, 11, 1110. [Google Scholar] [CrossRef]
- Riviere, A.; Selak, M.; Lantin, D.; Leroy, F.; De Vuyst, L. Bifidobacteria and Butyrate-Producing Colon Bacteria: Importance and Strategies for Their Stimulation in the Human Gut. Front. Microbiol. 2016, 7, 979. [Google Scholar] [CrossRef]
- Lee, K.C.; Kil, D.Y.; Sul, W.J. Cecal microbiome divergence of broiler chickens by sex and body weight. J. Microbiol. 2017, 55, 939–945. [Google Scholar] [CrossRef]
- O’Hara, E.; Kelly, A.; McCabe, M.S.; Kenny, D.A.; Guan, L.; Waters, S.M. Effect of a butyrate-fortified milk replacer on gastrointestinal microbiota and products of fermentation in artificially reared dairy calves at weaning. Sci. Rep. 2018, 8, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Ye, H.; Liu, J.; Mao, S. High-grain diets altered rumen fermentation and epithelial bacterial community and resulted in rumen epithelial injuries of goats. Appl. Microbiol. Biotechnol. 2017, 101, 6981–6992. [Google Scholar] [CrossRef]
- Downes, J.; Munson, M.A.; Radford, D.R.; Spratt, D.A.; Wade, W.G. Shuttleworthia satelles gen. nov., sp. nov., isolated from the human oral cavity. Int. J. Syst. Evol. Microbiol. 2002, 52, 1469–1475. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, S.; Wu, C.; Chen, X.; Duan, Z.; Xu, Q.; Jiang, W.; Xu, L.; Wang, T.; Su, L.; et al. Oral Microbiome Alterations Associated with Early Childhood Caries Highlight the Importance of Carbohydrate Metabolic Activities. mSystems 2019, 4. [Google Scholar] [CrossRef] [PubMed]
- Nakazawa, F.; Sato, M.; Poco, S.E.; Hashimura, T.; Ikeda, T.; Kalfas, S.; Sundqvist, G.; Hoshino, E. Description of Mogibacterium pumilum gen. nov., sp. nov. and Mogibacterium vescum gen. nov., sp. nov., and reclassification of Eubacterium timidum (Holdeman et al. 1980) as Mogibacterium timidum gen. nov., comb. nov. Int. J. Syst. Evol. Microbiol. 2000, 50 Pt 2, 679–688. [Google Scholar] [CrossRef]
- Cook, A.R.; Riley, P.W.; Murdoch, H.; Evans, P.N.; McDonald, I.R. Howardella ureilytica gen. nov., sp. nov., a Gram-positive, coccoid-shaped bacterium from a sheep rumen. Int. J. Syst. Evol. Microbiol. 2007, 57, 2940–2945. [Google Scholar] [CrossRef]
- Jabari, L.; Gannoun, H.; Cayol, J.-L.; Hamdi, M.; Fauque, G.; Ollivier, B.; Fardeau, M.-L. Characterization of Defluviitalea saccharophila gen. nov., sp. nov., a thermophilic bacterium isolated from an upflow anaerobic filter treating abattoir wastewaters, and proposal of Defluviitaleaceae fam. nov. Int. J. Syst. Evol. Microbiol. 2012, 62, 550–555. [Google Scholar] [CrossRef] [PubMed]
- Larsen, J.M. The immune response to Prevotella bacteria in chronic inflammatory disease. Immunology 2017, 151, 363–374. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Chen, D.; Tian, G.; Zheng, P.; Mao, X.; Yu, J.; He, J.; Huang, Z.; Luo, Y.; Luo, J.; et al. Soluble Fiber and Insoluble Fiber Regulate Colonic Microbiota and Barrier Function in a Piglet Model. BioMed Res. Int. 2019, 2019, 7809171. [Google Scholar] [CrossRef] [PubMed]
- Costabile, A.; Kolida, S.; Klinder, A.; Gietl, E.; Bauerlein, M.; Frohberg, C.; Landschutze, V.; Gibson, G.R. A double-blind, placebo-controlled, cross-over study to establish the bifidogenic effect of a very-long-chain inulin extracted from globe artichoke (Cynara scolymus) in healthy human subjects. Br. J. Nutr. 2010, 104, 1007–1017. [Google Scholar] [CrossRef] [PubMed]
- Precup, G.; Vodnar, D.C. Gut Prevotella as a possible biomarker of diet and its eubiotic versus dysbiotic roles: A comprehensive literature review. Br. J. Nutr. 2019, 122, 131–140. [Google Scholar] [CrossRef] [PubMed]
- Roberton, A.M.; McKenzie, C.G.; Sharfe, N.; Stubbs, L.B. A glycosulphatase that removes sulphate from mucus glycoprotein. Biochem. J. 1993, 293 Pt 3, 683–689. [Google Scholar] [CrossRef]
- Lennon, G.; Balfe, A.; Earley, H.; Devane, L.A.; Lavelle, A.; Winter, D.C.; Coffey, J.C.; O’Connell, P.R. Influences of the colonic microbiome on the mucous gel layer in ulcerative colitis. Gut Microbes 2014, 5, 277–476. [Google Scholar] [CrossRef]
| Ingredient, % | LFF | MFF | HFF |
|---|---|---|---|
| Corn | 57.68 | 55.20 | 51.00 |
| Soybean meal | 18.40 | 20.60 | 23.70 |
| Soybean oil | 1.00 | 1.27 | 2.37 |
| Salt | 0.35 | 0.35 | 0.35 |
| Monocalcium phosphate, 21% | 0.63 | 0.78 | 0.94 |
| Limestone | 1.24 | 1.03 | 0.85 |
| L-lysine HCl | 0.36 | 0.34 | 0.29 |
| L-threonine | 0.04 | 0.07 | 0.09 |
| DL-methionine | - | 0.06 | 0.11 |
| Vitamin premix 1 | 0.15 | 0.15 | 0.15 |
| Trace mineral premix 2 | 0.15 | 0.15 | 0.15 |
| Corn DDGS 3 | 20.00 | 10.00 | - |
| Resistant potato starch | - | 5.00 | 10.00 |
| Sugar beet pulp | - | 5.00 | 10.00 |
| Calculated composition | |||
| Metabolizable energy, kcal/kg | 3341 | 3300 | 3301 |
| Crude protein, % | 19.36 | 17.97 | 16.82 |
| SID lysine 4, % | 0.98 | 0.98 | 0.98 |
| SID methionine + cysteine, % | 0.55 | 0.55 | 0.56 |
| SID threonine, % | 0.60 | 0.60 | 0.58 |
| SID tryptophan, % | 0.16 | 0.16 | 0.16 |
| Analyzed composition | |||
| DM, % | 88.5 | 88.6 | 88.8 |
| Crude protein, % | 17.19 | 16.72 | 14.72 |
| Gross energy, kcal/kg | 3957 | 3884 | 3875 |
| Acid detergent fiber, % | 4.47 | 4.31 | 4.24 |
| Neutral detergent fiber, % | 11.93 | 9.52 | 8.30 |
| Insoluble dietary fiber, % | 13.56 | 10.99 | 8.94 |
| Total dietary fiber, % | 13.95 | 11.33 | 9.20 |
| Resistant starch | 3.45 | 6.83 | 10.02 |
| Treatment 1 | p-Value | |||
|---|---|---|---|---|
| LFF | MFF | HFF | ||
| Number of pigs with clinical disease 2 | 11/13 | 6/13 | 2/13 | 0.002 |
| Median days to clinical disease 2 | 9 | 20 | - | <0.001 |
| Median days with clinical disease 2 | 6 | 2 | 0 | 0.001 |
| Number of pigs positive 3 | 12/13 | 12/13 | 11/13 | 0.588 |
| Item | Treatment 1 | SEM | p-Value | ||
|---|---|---|---|---|---|
| LFF | MFF | HFF | |||
| Pre-Challenge (dpi −14 to 0) | |||||
| ADG, kg/d | 0.79 | 0.84 | 0.89 | 0.039 | 0.197 |
| ADFI, kg/d | 1.83 | 1.84 | 1.74 | 0.068 | 0.506 |
| G:F | 0.43 b | 0.46 b | 0.50 a | 0.012 | 0.001 |
| dpi 0 to 7 | |||||
| ADG, kg/d | 1.12 b | 1.37 a | 1.33 a | 0.056 | 0.007 |
| ADFI, kg/d | 2.33 | 2.43 | 2.24 | 0.075 | 0.221 |
| G:F | 0.50 b | 0.57 a | 0.59 a | 0.018 | 0.002 |
| dpi 8 to 14 | |||||
| ADG, kg/d | −0.48 b | 0.38 a | 1.10 a | 0.231 | <0.001 |
| ADFI, kg/d | 1.00 b | 2.06 a | 2.30 a | 0.212 | <0.001 |
| G:F | −1.97 b | 0.02 a | 0.44 a | 0.422 | 0.001 |
| dpi 15 to 21 | |||||
| ADG, kg/d | 0.63 | 0.61 | 1.14 | 0.249 | 0.232 |
| ADFI, kg/d | 1.39 b | 1.96 ab | 2.49 a | 0.261 | 0.014 |
| G:F | 0.32 | 0.33 | 0.40 | 0.149 | 0.703 |
| dpi 22 to 28 | |||||
| ADG, kg/d | 1.25 | 1.04 | 1.13 | 0.158 | 0.625 |
| ADFI, kg/d | 2.41 | 2.24 | 2.79 | 0.250 | 0.277 |
| G:F | 0.53 | 0.43 | 0.40 | 0.062 | 0.233 |
| dpi 29 to 35 | |||||
| ADG, kg/d | 1.34 | 1.36 | 1.48 | 0.106 | 0.562 |
| ADFI, kg/d | 2.97 | 3.00 | 3.12 | 0.167 | 0.766 |
| G:F | 0.42 | 0.48 | 0.49 | 0.053 | 0.558 |
| dpi 36 to 42 | |||||
| ADG, kg/d | 0.97 b | 1.41 a | 1.33 a | 0.129 | 0.033 |
| ADFI, kg/d | 2.88 b | 3.57 a | 3.48 a | 0.172 | 0.009 |
| G:F | 0.33 | 0.40 | 0.38 | 0.043 | 0.458 |
| dpi 0 BW, kg | 40.0 | 40.6 | 41.9 | 1.410 | 0.623 |
| dpi 42 BW, kg | 73.0 b | 83.3 b | 94.6 a | 3.150 | <0.001 |
| Item | Treatment 1 | SEM | p-Value | ||
|---|---|---|---|---|---|
| LFF | MFF | HFF | |||
| Cecum pH | 5.92 a | 5.71 b | 5.68 b | 0.058 | 0.013 |
| Colon pH | 6.71 | 6.59 | 6.56 | 0.056 | 0.139 |
| Item | Treatment 1 | SEM | p-Value | ||
|---|---|---|---|---|---|
| LFF | MFF | HFF | |||
| Sulfomucin-positive goblet cells/10,000 μm2 | 12.5 a | 10.0 a,b | 9.0 b | 0.979 | 0.034 |
| Sialomucin-positive goblet cells/10,000 μm2 | 0.04 | 0.05 | 0.05 | 0.066 | 0.990 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Helm, E.T.; Gabler, N.K.; Burrough, E.R. Highly Fermentable Fiber Alters Fecal Microbiota and Mitigates Swine Dysentery Induced by Brachyspira hyodysenteriae. Animals 2021, 11, 396. https://doi.org/10.3390/ani11020396
Helm ET, Gabler NK, Burrough ER. Highly Fermentable Fiber Alters Fecal Microbiota and Mitigates Swine Dysentery Induced by Brachyspira hyodysenteriae. Animals. 2021; 11(2):396. https://doi.org/10.3390/ani11020396
Chicago/Turabian StyleHelm, Emma T., Nicholas K. Gabler, and Eric. R. Burrough. 2021. "Highly Fermentable Fiber Alters Fecal Microbiota and Mitigates Swine Dysentery Induced by Brachyspira hyodysenteriae" Animals 11, no. 2: 396. https://doi.org/10.3390/ani11020396
APA StyleHelm, E. T., Gabler, N. K., & Burrough, E. R. (2021). Highly Fermentable Fiber Alters Fecal Microbiota and Mitigates Swine Dysentery Induced by Brachyspira hyodysenteriae. Animals, 11(2), 396. https://doi.org/10.3390/ani11020396






