Comparable Number of Genes Having Experienced Positive Selection among Great Ape Species
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Identification of Orthologs in Six Primates
2.2. Building the Gene Family
2.3. Positive Selection of Genes
2.4. Population Analysis of PSGs
2.5. Expression Analysis of PSGs
2.6. Association Analysis of PSGs in Great Apes
3. Results
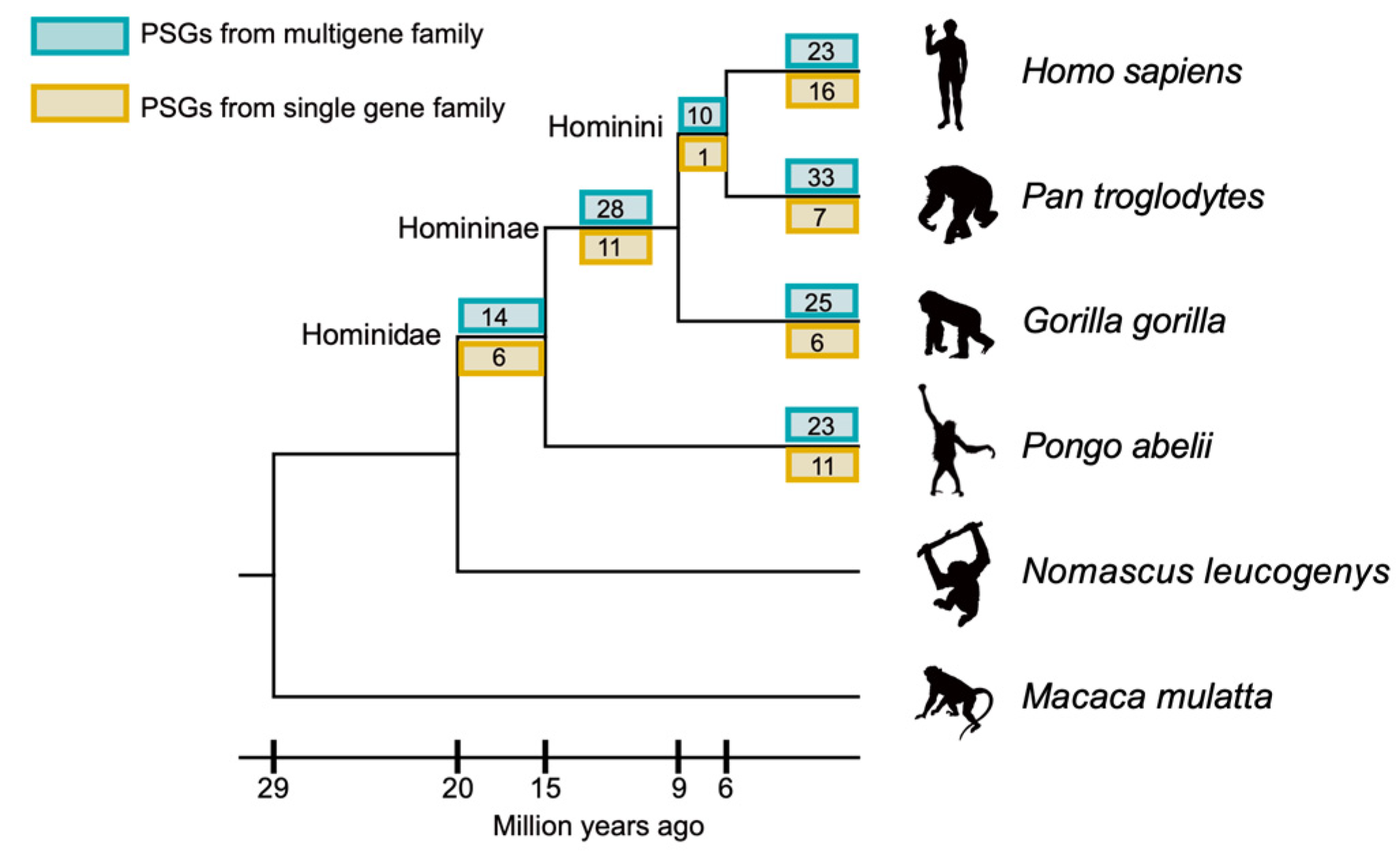
3.1. Comparable Number of Genes Had Experienced Positive Selection among Great Ape Species
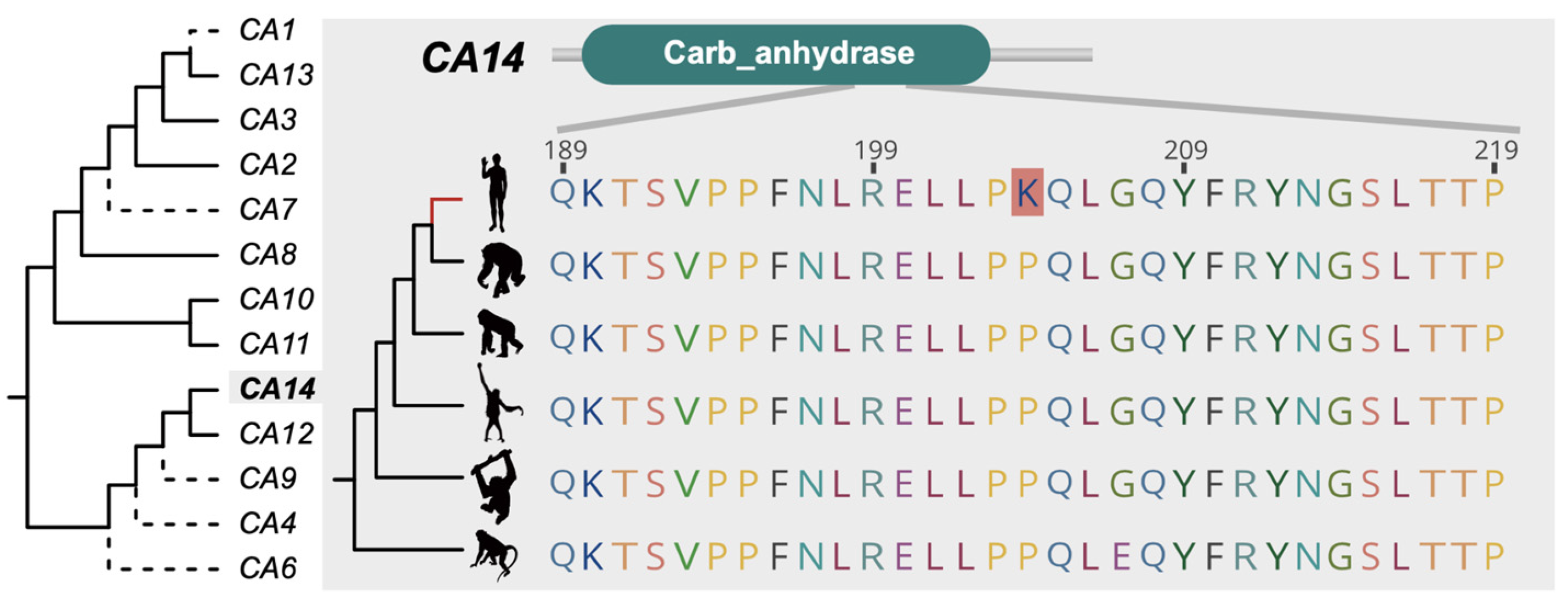
3.2. The PSGs Contributed to Functional Evolution of Great Ape
3.3. The Expression Pattern of Positively Selected Genes
3.4. PSG and Disease Evolution
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Darwin, C. The Descent of Man, and Selection in Relation to Sex; Princeton University Press: Princeton, NJ, USA, 2008. [Google Scholar]
- Sabeti, P.C.; Varilly, P.; Fry, B.; Lohmueller, J.; Hostetter, E.; Cotsapas, C.; Xie, X.; Byrne, E.H.; McCarroll, S.A.; Gaudet, R.; et al. Genome-Wide Detection and Characterization of Positive Selection in Human Populations. Nature 2007, 449, 913–918. [Google Scholar] [CrossRef]
- Vallender, E.J.; Lahn, B.T. Positive Selection on the Human Genome. Hum. Mol. Genet. 2004, 13, R245–R254. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leigh, S.R.; Shea, B.T. Ontogeny and the Evolution of Adult Body Size Dimorphism in Apes. Am. J. Primatol. 1995, 36, 37–60. [Google Scholar] [CrossRef]
- Smith, R.J.; Jungers, W.L. Body Mass in Comparative Primatology. J. Hum. Evol. 1997, 32, 523–559. [Google Scholar] [CrossRef] [PubMed]
- Barton, R.A.; Venditti, C. Rapid Evolution of the Cerebellum in Humans and Other Great Apes. Curr. Biol. 2014, 24, 2440–2444. [Google Scholar] [CrossRef] [Green Version]
- Wong, K.M.; Suchard, M.A.; Huelsenbeck, J.P. Alignment Uncertainty and Genomic Analysis. Science 2008, 319, 473–476. [Google Scholar] [CrossRef] [Green Version]
- Fletcher, W.; Yang, Z. The Effect of Insertions, Deletions, and Alignment Errors on the Branch-Site Test of Positive Selection. Mol. Biol. Evol. 2010, 27, 2257–2267. [Google Scholar] [CrossRef] [Green Version]
- Privman, E.; Penn, O.; Pupko, T. Improving the Performance of Positive Selection Inference by Filtering Unreliable Alignment Regions. Mol. Biol. Evol. 2012, 29, 1–5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moretti, S.; Laurenczy, B.; Gharib, W.H.; Castella, B.; Kuzniar, A.; Schabauer, H.; Studer, R.A.; Valle, M.; Salamin, N.; Stockinger, H.; et al. Selectome Update: Quality Control and Computational Improvements to a Database of Positive Selection. Nucleic Acids Res. 2014, 42, D917–D921. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Nielsen, R.; Yang, Z. Evaluation of an Improved Branch-Site Likelihood Method for Detecting Positive Selection at the Molecular Level. Mol. Biol. Evol. 2005, 22, 2472–2479. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Z. PAML 4: Phylogenetic Analysis by Maximum Likelihood. Mol. Biol. Evol. 2007, 24, 1586–1591. [Google Scholar] [CrossRef] [Green Version]
- Bakewell, M.A.; Shi, P.; Zhang, J. More Genes Underwent Positive Selection in Chimpanzee Evolution than in Human Evolution. Proc. Natl. Acad. Sci. USA 2007, 104, 7489–7494. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van der Lee, R.; Wiel, L.; van Dam, T.J.P.; Huynen, M.A. Genome-Scale Detection of Positive Selection in Nine Primates Predicts Human-Virus Evolutionary Conflicts. Nucleic Acids Res. 2017, 45, 10634–10648. [Google Scholar] [CrossRef] [Green Version]
- Yang, Z. Molecular Evolution: A Statistical Approach; Oxford University Press: Oxford, UK, 2014. [Google Scholar]
- Kronenberg, Z.N.; Fiddes, I.T.; Gordon, D.; Murali, S.; Cantsilieris, S.; Meyerson, O.S.; Underwood, J.G.; Nelson, B.J.; Chaisson, M.J.P.; Dougherty, M.L.; et al. High-Resolution Comparative Analysis of Great Ape Genomes. Science 2018, 360, eaar6343. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Emms, D.M.; Kelly, S. OrthoFinder: Phylogenetic Orthology Inference for Comparative Genomics. Genome Biol. 2019, 20, 238. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, H.; Coghlan, A.; Ruan, J.; Coin, L.J.; Hériché, J.-K.; Osmotherly, L.; Li, R.; Liu, T.; Zhang, Z.; Bolund, L.; et al. TreeFam: A Curated Database of Phylogenetic Trees of Animal Gene Families. Nucleic Acids Res. 2006, 34, D572–D580. [Google Scholar] [CrossRef]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A New Generation of Protein Database Search Programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [Green Version]
- Sela, I.; Ashkenazy, H.; Katoh, K.; Pupko, T. GUIDANCE2: Accurate Detection of Unreliable Alignment Regions Accounting for the Uncertainty of Multiple Parameters. Nucleic Acids Res. 2015, 43, W7–W14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wertheim, J.O.; Murrell, B.; Smith, M.D.; Kosakovsky Pond, S.L.; Scheffler, K. RELAX: Detecting Relaxed Selection in a Phylogenetic Framework. Mol. Biol. Evol. 2015, 32, 820–832. [Google Scholar] [CrossRef] [Green Version]
- Prado-Martinez, J.; Sudmant, P.H.; Kidd, J.M.; Li, H.; Kelley, J.L.; Lorente-Galdos, B.; Veeramah, K.R.; Woerner, A.E.; O’Connor, T.D.; Santpere, G.; et al. Great Ape Genetic Diversity and Population History. Nature 2013, 499, 471–475. [Google Scholar] [CrossRef] [Green Version]
- Harris, R.S. Improved Pairwise Alignment of Genomic DNA. Ph.D. Thesis, Pennsylvania State University, State College, PA, USA, 2007. [Google Scholar]
- Kent, W.J.; Baertsch, R.; Hinrichs, A.; Miller, W.; Haussler, D. Evolution’s Cauldron: Duplication, Deletion, and Rearrangement in the Mouse and Human Genomes. Proc. Natl. Acad. Sci. USA 2003, 100, 11484–11489. [Google Scholar] [CrossRef] [Green Version]
- Yanai, I.; Benjamin, H.; Shmoish, M.; Chalifa-Caspi, V.; Shklar, M.; Ophir, R.; Bar-Even, A.; Horn-Saban, S.; Safran, M.; Domany, E.; et al. Genome-Wide Midrange Transcription Profiles Reveal Expression Level Relationships in Human Tissue Specification. Bioinformatics 2005, 21, 650–659. [Google Scholar] [CrossRef] [Green Version]
- Amberger, J.S.; Bocchini, C.A.; Schiettecatte, F.; Scott, A.F.; Hamosh, A. OMIM.org: Online Mendelian Inheritance in Man (OMIM®), an Online Catalog of Human Genes and Genetic Disorders. Nucleic Acids Res. 2015, 43, D789–D798. [Google Scholar] [CrossRef] [Green Version]
- Howe, K.L.; Achuthan, P.; Allen, J.; Allen, J.; Alvarez-Jarreta, J.; Amode, M.R.; Armean, I.M.; Azov, A.G.; Bennett, R.; Bhai, J.; et al. Ensembl 2021. Nucleic Acids Res. 2021, 49, D884–D891. [Google Scholar] [CrossRef] [PubMed]
- Long, M.; VanKuren, N.W.; Chen, S.; Vibranovski, M.D. New Gene Evolution: Little Did We Know. Annu. Rev. Genet. 2013, 47, 307–333. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, J. Evolution by Gene Duplication: An Update. Trends Ecol. Evol. 2003, 18, 292–298. [Google Scholar] [CrossRef] [Green Version]
- Pegueroles, C.; Laurie, S.; Albà, M.M. Accelerated Evolution after Gene Duplication: A Time-Dependent Process Affecting Just One Copy. Mol. Biol. Evol. 2013, 30, 1830–1842. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Y.; Zhou, B.; Pache, L.; Chang, M.; Khodabakhshi, A.H.; Tanaseichuk, O.; Benner, C.; Chanda, S.K. Metascape Provides a Biologist-Oriented Resource for the Analysis of Systems-Level Datasets. Nat. Commun. 2019, 10, 1523. [Google Scholar] [CrossRef]
- Jeong, J.; Eide, D.J. The SLC39 Family of Zinc Transporters. Mol. Asp. Med. 2013, 34, 612–619. [Google Scholar] [CrossRef] [Green Version]
- Dufner-Beattie, J.; Weaver, B.P.; Geiser, J.; Bilgen, M.; Larson, M.; Xu, W.; Andrews, G.K. The Mouse Acrodermatitis Enteropathica Gene Slc39a4 (Zip4) Is Essential for Early Development and Heterozygosity Causes Hypersensitivity to Zinc Deficiency. Hum. Mol. Genet. 2007, 16, 1391–1399. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qian, J.; Xu, K.; Yoo, J.; Chen, T.T.; Andrews, G.; Noebels, J.L. Knockout of Zn Transporters Zip-1 and Zip-3 Attenuates Seizure-Induced CA1 Neurodegeneration. J. Neurosci. 2011, 31, 97–104. [Google Scholar] [CrossRef] [Green Version]
- Takeda, A.; Tamano, H. Insight into Zinc Signaling from Dietary Zinc Deficiency. Brain Res. Rev. 2009, 62, 33–44. [Google Scholar] [CrossRef]
- Takeda, A.; Tamano, H. Proposed Glucocorticoid-Mediated Zinc Signaling in the Hippocampus. Metallomics 2012, 4, 614. [Google Scholar] [CrossRef] [PubMed]
- Cabrera, A.; Neculai, D.; Kain, K.C. CD36 and Malaria: Friends or Foes? A Decade of Data Provides Some Answers. Trends Parasitol. 2014, 30, 436–444. [Google Scholar] [CrossRef] [PubMed]
- Aitman, T.J.; Cooper, L.D.; Norsworthy, P.J.; Wahid, F.N.; Gray, J.K.; Curtis, B.R.; McKeigue, P.M.; Kwiatkowski, D.; Greenwood, B.M.; Snow, R.W.; et al. Malaria Susceptibility and CD36 Mutation. Nature 2000, 405, 1015–1016. [Google Scholar] [CrossRef] [Green Version]
- Omi, K.; Ohashi, J.; Patarapotikul, J.; Hananantachai, H.; Naka, I.; Looareesuwan, S.; Tokunaga, K. CD36 Polymorphism Is Associated with Protection from Cerebral Malaria. Am. J. Hum. Genet. 2003, 72, 364–374. [Google Scholar] [CrossRef] [Green Version]
- Tsang, C.K.; Liu, Y.; Thomas, J.; Zhang, Y.; Zheng, X.F.S. Superoxide Dismutase 1 Acts as a Nuclear Transcription Factor to Regulate Oxidative Stress Resistance. Nat. Commun. 2014, 5, 3446. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Watanabe, K.; Shibuya, S.; Koyama, H.; Ozawa, Y.; Toda, T.; Yokote, K.; Shimizu, T. Sod1 Loss Induces Intrinsic Superoxide Accumulation Leading to p53-Mediated Growth Arrest and Apoptosis. Int. J. Mol. Sci. 2013, 14, 10998–11010. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Chen, Y.; Zhao, J.; Shi, J.; Wang, M.; Qiu, S.; Hu, Y.; Xu, Y.; Cui, Y.; Liu, C.; et al. The Specific Inhibition of SOD1 Selectively Promotes Apoptosis of Cancer Cells via Regulation of the ROS Signaling Network. Oxidative Med. Cell. Longev. 2019, 2019, 9706792. [Google Scholar] [CrossRef]
- Muñoz-Espín, D.; Serrano, M. Cellular Senescence: From Physiology to Pathology. Nat. Rev. Mol. Cell Biol. 2014, 15, 482–496. [Google Scholar] [CrossRef]
- Parkes, T.L.; Elia, A.J.; Dickinson, D.; Hilliker, A.J.; Phillips, J.P.; Boulianne, G.L. Extension of Drosophila Lifespan by Overexpression of Human SOD1 in Motorneurons. Nat. Genet. 1998, 19, 171–174. [Google Scholar] [CrossRef]
- Tacutu, R.; Thornton, D.; Johnson, E.; Budovsky, A.; Barardo, D.; Craig, T.; Diana, E.; Lehmann, G.; Toren, D.; Wang, J.; et al. Human Ageing Genomic Resources: New and Updated Databases. Nucleic Acids Res. 2018, 46, D1083–D1090. [Google Scholar] [CrossRef] [PubMed]
- Lindskog, S. Structure and Mechanism of Carbonic Anhydrase. Pharmacol. Ther. 1997, 74, 1–20. [Google Scholar] [CrossRef]
- Fujikawa-Adachi, K.; Nishimori, I.; Taguchi, T.; Onishi, S. Human Carbonic Anhydrase XIV (CA14): cDNA Cloning, mRNA Expression, and Mapping to Chromosome 1. Genomics 1999, 61, 74–81. [Google Scholar] [CrossRef] [PubMed]
- Parkkila, S.; Parkkila, A.K.; Rajaniemi, H.; Shah, G.N.; Grubb, J.H.; Waheed, A.; Sly, W.S. Expression of Membrane-Associated Carbonic Anhydrase XIV on Neurons and Axons in Mouse and Human Brain. Proc. Natl. Acad. Sci. USA 2001, 98, 1918–1923. [Google Scholar] [CrossRef] [Green Version]
- Kurihara-Shimomura, M.; Sasahira, T.; Nakamura, H.; Nakashima, C.; Kuniyasu, H.; Kirita, T. Zinc Finger AN1-Type Containing 4 Is a Novel Marker for Predicting Metastasis and Poor Prognosis in Oral Squamous Cell Carcinoma. J. Clin. Pathol. 2018, 71, 436–441. [Google Scholar] [CrossRef] [PubMed]
- Young, J.H.; Hunter Young, J.; Chang, Y.-P.C.; Kim, J.D.-O.; Chretien, J.-P.; Klag, M.J.; Levine, M.A.; Ruff, C.B.; Wang, N.-Y.; Chakravarti, A. Differential Susceptibility to Hypertension Is due to Selection during the out-of-Africa Expansion. PLoS Genet. 2005, 1, e82. [Google Scholar] [CrossRef]
- Neel, J.V. Diabetes Mellitus: A “Thrifty” Genotype Rendered Detrimental by “Progress”? Am. J. Hum. Genet. 1962, 14, 353–362. [Google Scholar]
- Guschanski, K.; Warnefors, M.; Kaessmann, H. The Evolution of Duplicate Gene Expression in Mammalian Organs. Genome Res. 2017, 27, 1461–1474. [Google Scholar] [CrossRef] [Green Version]
- Conant, G.C.; Wolfe, K.H. Turning a Hobby into a Job: How Duplicated Genes Find New Functions. Nat. Rev. Genet. 2008, 9, 938–950. [Google Scholar] [CrossRef]
- Pál, C.; Papp, B.; Hurst, L.D. Highly Expressed Genes in Yeast Evolve Slowly. Genetics 2001, 158, 927–931. [Google Scholar] [CrossRef]
- Zhang, J.; Yang, J.-R. Determinants of the Rate of Protein Sequence Evolution. Nat. Rev. Genet. 2015, 16, 409–420. [Google Scholar] [CrossRef] [Green Version]
- Drummond, D.A.; Bloom, J.D.; Adami, C.; Wilke, C.O.; Arnold, F.H. Why Highly Expressed Proteins Evolve Slowly. Proc. Natl. Acad. Sci. USA 2005, 102, 14338–14343. [Google Scholar] [CrossRef] [Green Version]
- Williams, G.C. Pleiotropy, Natural Selection, and the Evolution of Senescence. Evolution 1957, 11, 398–411. [Google Scholar] [CrossRef]
- Chen, H.; Li, C.; Zhou, Z.; Liang, H. Fast-Evolving Human-Specific Neural Enhancers Are Associated with Aging-Related Diseases. Cell Syst. 2018, 6, 604–611.e4. [Google Scholar] [CrossRef] [Green Version]


| Species (Latin Name) | Genome Version | Genome Size (bp) | Gene Annotation Source | No. of Protein-Coding Gene |
|---|---|---|---|---|
| Homo sapiens | GRCh38.p13 | 3,272,089,205 | NCBI Homo sapiens Updated Annotation Release 109.20210514 | 19,475 |
| Pan troglodytes | Clint_PTRv2 | 3,024,031,013 | NCBI Pan troglodytes Annotation Release 105 | 21,687 |
| Gorilla gorilla | Kamilah_GGO_v0 | 3,044,872,214 | NCBI Gorilla gorilla Annotation Release 102 | 20,784 |
| Pongo abelii | Susie_PABv2 | 3,065,035,716 | NCBI Pongo abelii Annotation Release 103 | 20,717 |
| Nomascus leucogenys | Asia_NLE_v1 | 2,843,982,884 | NCBI Nomascus leucogenys Annotation Release 103 | 20,656 |
| Macaca mulatta | Mmul_10 | 2,971,331,530 | NCBI Macaca mulatta Annotation Release 103 | 21,305 |
| Lineage | No. of PSG Candidates 1 | No. of PSG Candidates without Potential Alignment Error | No. of PSG Candidates without Low Quality Aligned Columns | No. of PSGs without Relaxed Selection |
|---|---|---|---|---|
| H. sapiens | 256 | 47 | 40 | 39 |
| P. troglodytes | 245 | 49 | 44 | 40 |
| Hominini | 65 | 11 | 11 | 11 |
| G. gorilla | 178 | 32 | 32 | 31 |
| Homininae | 80 | 40 | 39 | 39 |
| P. abelii | 181 | 38 | 35 | 34 |
| Hominidae | 55 | 22 | 20 | 20 |
| Lineage | No. of PSGs | No. of Positively Selected Sites | No. of PSGs with Fixed Positively Selected Sites | No. of Fixed Positively Selected Sites |
|---|---|---|---|---|
| H. sapiens | 39 | 111 | 31 | 91 |
| P. troglodytes | 40 | 133 | 40 | 129 |
| Hominini | 11 | 16 | 11 | 16 |
| G. gorilla | 31 | 76 | 31 | 75 |
| Homininae | 39 | 62 | 38 | 61 |
| P. abelii | 34 | 51 | 33 | 47 |
| Hominidae | 20 | 35 | 20 | 35 |
| Comparison | Human | Chimpanzee | |||
|---|---|---|---|---|---|
| This Study | Bakewell et al. [13] | This Study | Bakewell et al. [13] | ||
| Shared | 11 | 9 | |||
| Different Input | Different annotation versions | 3 | 58 | 1 | 75 |
| Different ortholog assignment 1 | - | 16 | - | 22 | |
| Problematic gene model in ortholog group 2 | - | 3 | - | 3 | |
| Different data processing method | No significance 3 | - | 25 | - | 96 |
| No significant sites 4 | - | 38 | - | 27 | |
| positively selected sites with low confidence 5 | - | 3 | - | 1 | |
| Newly detected PSGs in this study | 25 | - | 30 | - | |
| Total | 39 | 154 | 40 | 233 | |
| Gene Type | No. of PSGs | No. of Non-PSGs | Proportion of PSGs | p-Value 1 |
|---|---|---|---|---|
| H. sapiens | ||||
| No. of genes in multiple-gene family | 23 | 10,793 | 0.002 | 0.992 |
| No. of genes not in multiple-gene family | 16 | 3565 | 0.004 | |
| P. troglodytes | ||||
| No. of genes in multiple-gene family | 33 | 10,783 | 0.003 | 0.186 |
| No. of genes not in multiple-gene family | 7 | 3574 | 0.002 | |
| Hominini | ||||
| No. of genes in multiple-gene family | 10 | 10,806 | 0.001 | 0.200 |
| No. of genes not in multiple-gene family | 1 | 3580 | 0.000 | |
| G. gorilla | ||||
| No. of genes in multiple-gene family | 25 | 10,791 | 0.002 | 0.317 |
| No. of genes not in multiple-gene family | 6 | 3575 | 0.002 | |
| Homininae | ||||
| No. of genes in multiple-gene family | 28 | 10,788 | 0.003 | 0.753 |
| No. of genes not in multiple-gene family | 11 | 3570 | 0.003 | |
| P. abeli | ||||
| No. of genes in multiple-gene family | 23 | 10,793 | 0.002 | 0.885 |
| No. of genes not in multiple-gene family | 11 | 3570 | 0.003 | |
| Hominidae | ||||
| No. of genes in multiple-gene family | 14 | 10,802 | 0.001 | 0.790 |
| No. of genes not in multiple-gene family | 6 | 3575 | 0.002 | |
| Gene Type | No. of Disease Genes | No. of Non-Disease Genes | Proportion of Disease Genes | p-Value 1 |
|---|---|---|---|---|
| PSGs | 6 | 33 | 0.154 | 0.764 |
| Non-PSGs | 2690 | 11,668 | 0.187 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xie, D.; Chen, G.; Meng, X.; Wang, H.; Bi, X.; Fang, M.; Yang, C.; Zhou, Y.; Long, E.; Feng, S. Comparable Number of Genes Having Experienced Positive Selection among Great Ape Species. Animals 2021, 11, 3264. https://doi.org/10.3390/ani11113264
Xie D, Chen G, Meng X, Wang H, Bi X, Fang M, Yang C, Zhou Y, Long E, Feng S. Comparable Number of Genes Having Experienced Positive Selection among Great Ape Species. Animals. 2021; 11(11):3264. https://doi.org/10.3390/ani11113264
Chicago/Turabian StyleXie, Duo, Guangji Chen, Xiaoyu Meng, Haotian Wang, Xupeng Bi, Miaoquan Fang, Chentao Yang, Yang Zhou, Erping Long, and Shaohong Feng. 2021. "Comparable Number of Genes Having Experienced Positive Selection among Great Ape Species" Animals 11, no. 11: 3264. https://doi.org/10.3390/ani11113264
APA StyleXie, D., Chen, G., Meng, X., Wang, H., Bi, X., Fang, M., Yang, C., Zhou, Y., Long, E., & Feng, S. (2021). Comparable Number of Genes Having Experienced Positive Selection among Great Ape Species. Animals, 11(11), 3264. https://doi.org/10.3390/ani11113264





