Insights into the Karyotype Evolution of Charinidae, the Early-Diverging Clade of Whip Spiders (Arachnida: Amblypygi)
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Individuals
2.2. Chromosome Preparations and Evaluation of Karyotypes
2.3. Chromosome Banding Techniques
2.4. Telomeric FISH
2.5. Detection of 18S rDNA by FISH
2.6. Microscopy and Image Analysis
3. Results
3.1. Karyotype
3.2. Distribution and Composition of Constitutive Heterochromatin
3.3. Chromosomal Distribution of Telomeric Repeats
3.4. Distribution of 18S rDNA/NOR
4. Discussion
4.1. Patterns of Karyotype Differentiation
4.2. Distribution of rDNA and Telomeric Sequences
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Weygoldt, P. Whip Spiders (Chelicerata: Amblypygi). Their Biology, Morphology and Systematics; Apollo Books: Stenstrup, Denmark, 2000; 163p. [Google Scholar]
- Colla, A.; Legittimo, C.M.; Castelucci, F.; Simeon, E.; Miranda, G.S. First record of Amblypygi from Italy: Charinus ioanniticus (Charinidae). Arachnology 2020, 18, 642–648. [Google Scholar] [CrossRef]
- Harvey, M.S. Catalogue of the Smaller Arachnid Orders of the World. Amblypygi, Uropygi, Schizomida, Palpigradi, Ricinulei and Solifugae; CSIRO Publishing: Collingwood, Australia, 2003; 385p. [Google Scholar] [CrossRef] [Green Version]
- Garwood, R.J.; Dunlop, J.A.; Knecht, B.J.; Hegna, T.A. The phylogeny of fossil whip spiders. BMC Evol. Biol. 2017, 17, 105–118. [Google Scholar] [CrossRef] [Green Version]
- Esposito, A.L.; Bloom, T.; Caicedo-Quiroga, L.; Alicea-Serrano, A.M.; Sánchez-Ruiz, J.A.; May-Collado, L.J.; Blinford, G.J.; Agnarsson, I. Islands within islands: Diversification of tailless whip spiders (Amblypygi, Phrynus) in Caribbean caves. Mol. Phylogenet. Evol. 2015, 93, 107–117. [Google Scholar] [CrossRef] [PubMed]
- Reveillion, F.; Wattier, R.; Montuire, S.; Carvalho, L.S.; Bollache, L. Cryptic diversity within three South American whip spider species (Arachnida, Amblypygi). Zool. Res. 2020, 41, 595–598. [Google Scholar] [CrossRef]
- Seiter, M.; Reyes Lerma, A.C.; Král, J.; Sember, A.; Divišová, K.; Palacios, J.G.V.; Colmenares, P.A.; Loria, S.F.; Prendini, L. Cryptic diversity in the whip spider genus Paraphrynus (Amblypygi: Phrynidae): Integrating morphology, karyotype and DNA. Arthropod Syst. Phylogeny 2020, 78, 265–285. [Google Scholar] [CrossRef]
- Harvey, M.S. The neglected cousins: What do we know about the smaller arachnid orders? J. Arachnol. 2002, 30, 357–372. [Google Scholar] [CrossRef]
- Roskov, Y.; Ower, G.; Orrell, T.; Nicolson, D.; Bailly, N.; Kirk, P.M.; Bourgoin, T.; DeWalt, R.E.; Decock, W.; Van Nieukerken, E.; et al. Species 2000 & ITIS Catalogue of Life, 2019 Annual Checklist; Species 2000: Leiden, The Netherlands, 2019; ISSN 2405-884X; Available online: www.catalogueoflife.org/annual-checklist/2019. (accessed on 30 September 2021).
- Miranda, G.S.; Giupponi, A.P.L.; Scharff, N.; Prendini, L. Phylogeny and biogeography of the pantropical whip spider family Charinidae (Arachnida: Amblypygi). Zool. J. Linn. Soc. 2021, zlaa101. [Google Scholar] [CrossRef]
- Miranda, G.S.; Giupponi, A.P.; Prendini, L.; Scharff, N. Systematic revision of the pantropical whip spider family Charinidae Quintero, 1986 (Arachnida, Amblypygi). Eur. J. Taxon. 2021, 772, 1–409. [Google Scholar] [CrossRef]
- World Arachnida Catalog. World Amblypygi Catalog. Natural History Museum Bern. 2021. Available online: http://wac.nmbe.ch (accessed on 12 October 2021).
- Sharma, P.P.; Kaluziak, S.T.; Pérez-Porro, A.R.; González, V.L.; Hormiga, G.; Wheeler, W.C.; Giribet, G. Phylogenomic interrogation of Arachnida reveals systemic conflicts in phylogenetic signal. Mol. Biol. Evol. 2014, 31, 2963–2984. [Google Scholar] [CrossRef] [Green Version]
- Ballesteros, J.A.; Sharma, P.P. A critical appraisal of the placement of Xiphosura (Chelicerata) with account of known sources of phylogenetic error. Syst. Biol. 2019, 68, 896–917. [Google Scholar] [CrossRef]
- Lozano-Fernandez, J.; Tanner, A.R.; Giacomelli, M.; Carton, R.; Vinther, J.; Edgecombe, G.D.; Pisani, D. Increasing species sampling in chelicerate genomic-scale datasets provides support for monophyly of Acari and Arachnida. Nat. Commun. 2019, 10, 4534. [Google Scholar] [CrossRef] [Green Version]
- Millot, J.; Tuzet, O. La spermatogenèse chez les Pédipalpes. Bull. Biol. Fr. Belg. 1934, 68, 77–83. [Google Scholar]
- Vítková, M.; Král, J.; Traut, W.; Zrzavý, J.; Marec, F. The evolutionary origin of insect telomeric repeats, (TTAGG)n. Chromosome Res. 2005, 13, 145–156. [Google Scholar] [CrossRef] [PubMed]
- Paula-Neto, E.; Araujo, D.; Carvalho, L.S.; Cella, D.M.; Schneider, M.C. Chromosomal characteristics of a Brazilian whip spider (Amblypygi) and evolutionary relationships with other arachnid orders. Genet. Mol. Res. 2013, 12, 3726–3734. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miranda, G.S.; Giupponi, A.P.L.; Prendini, L.; Scharff, N. Weygoldtia, a new genus of Charinidae Quintero, 1986 (Arachnida, Amblypygi) with a reappraisal of the genera in the family. Zool. Anz. 2018, 273, 23–32. [Google Scholar] [CrossRef]
- Seiter, M.; Schramm, F.D.; Schwaha, T. Description of a new Charinus species (Amblypygi: Charinidae) from the Monseñor Nouel province, Dominican Republic. Zootaxa 2018, 4438, 349–361. [Google Scholar] [CrossRef]
- Miranda, G.S.; Reboleira, P.A.S. Amblypygids of Timor-Leste: First record of the order from the country with the description of a remarkable new species of Sarax (Arachnida, Amblypygi, Charinidae). Zookeys 2019, 820, 1–12. [Google Scholar] [CrossRef]
- Weygoldt, P. Evolutionary morphology of whip spiders: Towards a phylogenetic system (Chelicerata: Arachnida: Amblypygi). J. Zool. Syst. Evol. Res. 1996, 34, 185–202. [Google Scholar] [CrossRef]
- Traut, W. Pachytene mapping in the female silkworm, Bombyx mori L. (Lepidoptera). Chromosoma 1976, 108, 173–180. [Google Scholar] [CrossRef] [PubMed]
- Dolejš, P.; Kořínková, T.; Musilová, J.; Opatová, V.; Kubcová, L.; Buchar, J.; Král, J. Karyotypes of central European spiders of the genera Arctosa, Tricca and Xerolycosa (Aranea: Lycosidae). Eur. J. Entomol. 2011, 108, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Levan, A.K.; Fredga, K.; Sandberg, A.A. Nomenclature for centromeric position on chromosomes. Hereditas 1964, 52, 201–220. [Google Scholar] [CrossRef]
- Matthey, R. L’évolution de la formule chromosomiale chez les Vertébrés. Experientia 1945, I, 78–86. [Google Scholar] [CrossRef]
- Sumner, A.T. A simple technique for demonstrating centromeric heterochromatin. Exp. Cell Res. 1972, 75, 304–306. [Google Scholar] [CrossRef]
- Plíšková, J.; Kovařík, F.; Košulič, O.; Šťáhlavský, F. Description of a new species of Heterometrus Ehrenberg, 1828 (Scorpiones: Scorpionidae) from Thailand with remarks about the utilization of cytogenetic data in taxonomy of the genus. Annal. Zool. 2016, 66, 467–476. [Google Scholar] [CrossRef]
- Adilardi, R.S.; Ojanguren-Affilastro, A.A.; Martí, D.A.; Mola, L.M. Chromosome puzzle in the southernmost populations of the medically important scorpion Tityus bahiensis (Perty 1833) (Buthidae), a polymorphic species with striking structural rearrangements. Zool. Anz. 2020, 288, 139–150. [Google Scholar] [CrossRef]
- Sola, L.; Rossi, A.R.; Iaselli, V.; Rasch, E.M.; Monaco, P.J. Cytogenetics of bisexual/unisexual species of Poecilia. II. Analysis of heterochromatin and nucleolar organizer regions in Poecilia mexicana mexicana by C-banding and DAPI, quinacrine, chromomycin A3, and silver staining. Cytogenet. Cell Genet. 1992, 60, 229–235. [Google Scholar] [CrossRef]
- Mayr, B.; Ráb, P.; Kalat, M. Localisation of NORs and counterstain-enhanced fluorescence studies in Perca fluviatilis (Pisces, Percidae). Genetica 1985, 67, 51–56. [Google Scholar] [CrossRef]
- Miller, D.A.; Dev, V.G.; Tantravahi, R.; Miller, O.J. Suppression of human nucleolus organizer activity in mouse-human somatic hybrid cells. Exp. Cell Res. 1976, 101, 235–243. [Google Scholar] [CrossRef]
- Jiménez, R.; Burgos, M.; de la Guardia, R.D. A study of the Ag-staining significance in mitosis NORs. Heredity 1988, 60, 125–127. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ijdo, J.W.; Wells, R.A.; Baldini, A.; Readers, S.T. Improved telomere detection using a telomere repeat probe (TTAGGG)n generated by PCR. Nucleic Acids Res. 1991, 19, 4780. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sahara, K.; Marec, F.; Traut, W. TTAGG telomeric repeats in chromosomes of some insects and other arthropods. Chromosome Res. 1999, 7, 449–460. [Google Scholar] [CrossRef] [Green Version]
- Forman, M.; Nguyen, P.; Hula, V.; Král, J. Sex chromosome pairing and extensive NOR polymorphism in Wadicosa fidelis (Araneae: Lycosidae). Cytogenet. Genome Res. 2013, 141, 43–49. [Google Scholar] [CrossRef] [PubMed]
- Fuková, I.; Nguyen, P.; Marec, F. Codling moth cytogenetics: Karyotype, chromosomal location of rDNA, and molecular differentiation of sex chromosomes. Genome 2005, 48, 1083–1092. [Google Scholar] [CrossRef] [PubMed]
- Traut, W.; Sahara, K.; Otto, T.D.; Marec, F. Molecular differentiation of sex chromosomes probed by comparative genomic hybridization. Chromosoma 1999, 108, 173–180. [Google Scholar] [CrossRef] [PubMed]
- Schwager, E.E.; Sharma, P.P.; Clark, T.; Leite, D.J.; Wierschin, T.; Pechmann, M.; Akiyama-Oda, Y.; Esposito, L.; Bechsgaard, J.; Bilde, T.; et al. The house spider genome reveals an ancient whole-genome duplication during arachnid evolution. BMC Biol. 2017, 15, 62. [Google Scholar] [CrossRef] [PubMed]
- Gainett, G.; Sharma, P.P. Genomic resources and toolkits for developmental study of whip spiders (Amblypygi) provide insights into arachnid genome evolution and antenniform leg patterning. EvoDevo 2020, 11, 1–18. [Google Scholar] [CrossRef]
- Fan, Z.; Yuan, T.; Liu, P.; Wang, L.-Y.; Jin, J.-F.; Zhang, F.; Zhang, Z.S. A chromosome-level genome of the spider Trichonephila antipodiana reveals the genetic basis of its polyphagy and evidence of an ancient whole-genome duplication event. GigaScience 2021, 10, giab016. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, S. Cytological studies in spiders. III. Studies on the chromosomes of fifty-seven species of spiders belonging to seventeen families with general considerations on chromosomal evolution. J. Sci. Hiroshima Univ. B 1954, 15, 23–136. [Google Scholar]
- Deon, G.A.; Glugoski, L.; Vicari, M.R.; Nogaroto, V.; Sassi, F.M.C.; Cioffi, M.B.; Liehr, T.; Bertollo, L.A.C.; Moreira-Filho, O. Highly rearranged karyotypes and multiple sex chromosome systems in armored catfishes from the genus Harttia (Teleostei, Siluriformes). Genes 2020, 11, 1366. [Google Scholar] [CrossRef]
- Araujo, D.; Cella, D.M.; Brescovit, A.D. Cytogenetic analysis of the Neotropical spider Nephilengys cruentata (Araneomorphae, Tetragnathidae): Standard staining, NORs, C-bands and base-specific fluorochromes. Braz. J. Biol. 2005, 65, 193–202. [Google Scholar] [CrossRef] [Green Version]
- Almeida, B.R.R.; Milhomem-Paixão, S.S.R.; Noronha, R.C.R.; Nagamachi, C.Y.; Costa, M.J.R.; Oliveira-Pardal, P.P.; Coelho, J.S.; Pieczarka, J.C. Karyotype diversity and chromosomal organization of repetitive DNA in Tityus obscurus (Scorpiones, Buthidae). BMC Genet. 2017, 18, 35. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rincão, M.P.; Brescovit, A.D.; Dias, A.L. Insights on repetitive DNA behavior in two species of Ctenus Walckenaer, 1805 and Guasuctenus Polotow and Brescovit, 2019 (Araneae, Ctenidae): Evolutionary profile of H3 histone, 18S rRNA genes and heterochromatin distribution. PLoS ONE 2020, 15, e0231324. [Google Scholar] [CrossRef]
- Rocchi, M.; Archidiacono, N.; Schempp, W.; Capozzi, O.; Stanyon, R. Centromere repositioning in mammals. Heredity 2012, 108, 59–67. [Google Scholar] [CrossRef] [PubMed]
- Král, J.; Kořínková, T.; Krkavcová, L.; Musilová, J.; Forman, M.; Ávila Herrera, I.M.; Haddad, C.R.; Vítková, M.; Henriques, S.; Palacios Vargas, J.G.; et al. Evolution of karyotype, sex chromosomes, and meiosis in mygalomorph spiders (Araneae: Mygalomorphae). Biol. J. Linn. Soc. 2013, 109, 377–408. [Google Scholar] [CrossRef] [Green Version]
- Řezáč, M.; Arnedo, M.A.; Opatova, V.; Musilová, J.; Řezáčová, V.; Král, J. Taxonomic revision and insights into the speciation mode of the spider Dysdera erythrina species-complex (Araneae: Dysderidae): Sibling species with sympatric distributions. Invertebr. Syst. 2018, 32, 10–54. [Google Scholar] [CrossRef]
- Král, J.; Forman, M.; Kořínková, T.; Reyes Lerma, A.C.; Haddad, C.; Musilová, J.; Řezáč, M.; Ávila Herrera, I.M.; Thakur, S.; Dippenaar-Schoeman, A.S.; et al. Insights into the karyotype and genome evolution of haplogyne spiders indicate a polyploid origin of lineage with holokinetic chromosomes. Sci. Rep. 2019, 9, 3001. [Google Scholar] [CrossRef]
- Schneider, M.C.; Zacaro, A.A.; Pinto-da-Rocha, R.; Candido, D.M.; Cella, D.M. Complex meiotic configuration of the holocentric chromosomes: The intriguing case of the scorpion Tityus bahiensis. Chromosome Res. 2009, 17, 883–898. [Google Scholar] [CrossRef]
- Kovařík, F.; Lowe, G.; Šťáhlavský, F. Three new Chaerilus from Malaysia (Tioman Island) and Thailand (Scorpiones: Chaerilidae), with a review of C. cimrmani, C. sejnai, and C. tichyi. Euscorpius 2018, 268, 1–27. [Google Scholar] [CrossRef]
- Tsurusaki, N. Geographic variation of chromosomes in Sabacon makinoi Suzuki (Arachnida, Opiliones, Sabaconidae). Bull. Biogeogr. Soc. Jpn. 1989, 44, 111–116. [Google Scholar]
- Oliveira, R.M.; Zacaro, A.A.; Gnaspini, P.; Cella, D.M. Cytogenetics of three Brazilian Goniosoma species: A new record for diploid number in Laniatores (Opiliones, Gonyleptidae, Goniosomatinae). J. Arachnol. 2006, 34, 435–443. [Google Scholar] [CrossRef]
- Šťáhlavský, F.; Král, J.; Harvey, M.S.; Haddad, C.R. A karyotype study on the pseudoscorpion families Geogarypidae, Garypinidae and Olpiidae (Arachnida: Pseudoscorpiones). Eur. J. Entomol. 2006, 103, 277–289. [Google Scholar] [CrossRef] [Green Version]
- Šťáhlavský, F.; Král, J.; Harvey, M.S.; Haddad, C.R. The first cytogenetic characterisation of atemnids: Pseudoscorpions with the highest chromosome numbers (Arachnida: Pseudoscorpiones). Cytogenet. Genome Res. 2012, 137, 22–30. [Google Scholar] [CrossRef]
- Peng, J.C.; Karpen, G.H. Epigenetic regulation of heterochromatic DNA stability. Curr. Opin. Genet. Dev. 2008, 18, 204–211. [Google Scholar] [CrossRef] [Green Version]
- Brown, J.D.; O’Neill, R.J. Chromosomes, conflict, and epigenetics: Chromosomal speciation revisited. Annu. Rev. Genomics Hum. Genet. 2010, 11, 291–316. [Google Scholar] [CrossRef]
- King, M. Species Evolution: The Role of Chromosome Change, 1st ed.; Cambridge University Press: Cambridge, UK, 1993. [Google Scholar]
- Kandul, N.P.; Lukhtanov, V.A.; Pierce, N.E. Karyotypic diversity and speciation in Agrodiaetus butterflies. Evolution 2007, 61, 546–559. [Google Scholar] [CrossRef] [PubMed]
- Faria, R.; Navarro, A. Chromosomal speciation revisited: Rearranging theory with pieces of evidence. Trends Ecol. Evol. 2010, 25, 660–669. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.; Sun, X.; Cormack, B.P.; Boeke, J.D. Karyotype engineering by chromosome fusion leads to reproductive isolation in yeast. Nature 2018, 560, 392–396. [Google Scholar] [CrossRef] [PubMed]
- Damas, J.; Corbo, M.; Lewin, H.A. Vertebrate chromosome evolution. Annu. Rev. Anim. Biosci. 2021, 9, 1–27. [Google Scholar] [CrossRef] [PubMed]
- Guerrero, R.F.; Kirkpatrick, M. Local adaptation and the evolution of chromosome fusions. Evolution 2014, 68, 2747–2756. [Google Scholar] [CrossRef] [PubMed]
- Ortiz-Barrientos, D.; Engelstädter, J.; Rieseberg, L.H. Recombination rate evolution and the origin of species. Trends Ecol. Evol. 2016, 31, 226–236. [Google Scholar] [CrossRef] [Green Version]
- Wellenreuther, M.; Bernatchez, L. Eco-evolutionary genomics of chromosomal inversions. Trends Ecol. Evol. 2018, 33, 427–440. [Google Scholar] [CrossRef]
- Araujo, D.; Schneider, M.C.; Paula-Neto, E.; Cella, D.M. Sex chromosomes and meiosis in spiders: A review. In Meiosis—Molecular Mechanisms and Cytogenetic Diversity; Swan, A., Ed.; InTechOpen: Rieka, Croatia, 2012; pp. 87–108. [Google Scholar]
- Kořínková, T.; Král, J. Karyotypes, sex chromosomes, and meiotic division in spiders. In Spider Ecophysiology, 1st ed.; Nentwig, W., Ed.; Springer: Berlin, Germany, 2013; pp. 159–169. [Google Scholar] [CrossRef]
- Sember, A.; Pappová, M.; Forman, M.; Nguyen, P.; Marec, F.; Dalíková, M.; Divišová, K.; Doležálková-Kaštánková, M.; Zrzavá, M.; Sadílek, D.; et al. Patterns of sex chromosome differentiation in spiders: Insights from comparative genomic hybridisation. Genes 2020, 11, 849. [Google Scholar] [CrossRef]
- Kasturi Bai, A.R.; Parthasarathy, M.D. The chromosomes of Thelyphonus indicus Stoliczka. Proc. Indian Acad. Sci. B 1957, 24, 9–22. [Google Scholar]
- Král, J. Evolution of multiple sex chromosomes in the spider genus Malthonica (Araneae: Agelenidae) indicates unique structure of the spider sex chromosome systems. Chromosome Res. 2007, 15, 863–879. [Google Scholar] [CrossRef] [PubMed]
- Sochorová, J.; García, S.; Gálvez, F.; Symonová, R.; Kovařík, A. Evolutionary trends in animal ribosomal DNA loci: Introduction to a new online database. Chromosoma 2018, 127, 141–150. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cabral-de-Mello, D.C.; Marec, F. Universal fluorescence in situ hybridization (FISH) protocol for mapping repetitive DNAs in insects and other arthropods. Mol. Genet. Genom. 2021, 296, 513–552. [Google Scholar] [CrossRef] [PubMed]
- Provazníková, I.; Hejníčková, M.; Visser, S.; Dalíková, M.; Carabajal Paladino, L.Z.; Zrzavá, M.; Voleníková, A.; Marec, F.; Nguyen, P. Large-scale comparative analysis of cytogenetic markers across Lepidoptera. Sci. Rep. 2021, 11, 12214. [Google Scholar] [CrossRef]
- Rincão, M.P.; Chavari, J.L.; Brescovit, A.D.; Dias, A.L. Cytogenetic analysis of five Ctenidae species (Araneae): Detection of heterochromatin and 18S rDNA sites. Comp. Cytogenet. 2017, 11, 627–639. [Google Scholar] [CrossRef] [Green Version]
- Šťáhlavský, F.; Forman, M.; Just, P.; Denič, F.; Haddad, C.R.; Opatova, V. Cytogenetics of entelegyne spiders (Arachnida, Araneae) from southern Africa. Comp. Cytogenet. 2020, 14, 107–138. [Google Scholar] [CrossRef]
- Ávila Herrera, I.M.; Král, J.; Pastuchová, M.; Forman, M.; Musilová, J.; Kořínková, T.; Šťáhlavský, F.; Zrzavá, M.; Nguyen, P.; Just, P.; et al. Evolutionary pattern of karyotypes and meiosis in pholcid spiders (Araneae: Pholcidae): Implications for reconstructing chromosome evolution of araneomorph spiders. BMC Ecol. Evol. 2021, 21, 75. [Google Scholar] [CrossRef]
- Šťáhlavský, F.; Štundlová, J.; Lowe, G.; Stockmann, M.; Kovařík, F. Application of cytogenetic markers in the taxonomy of flat rock scorpions (Scorpiones: Hormuridae), with the description of Hadogenes weygoldti sp. n. Zool. Anz. 2018, 273, 173–182. [Google Scholar] [CrossRef]
- Ubinski, C.V.; Carvalho, L.S.; Schneider, M.C. Mechanisms of karyotype evolution in the Brazilian scorpions of the subfamily Centruroidinae (Buthidae). Genetica 2018, 146, 475–486. [Google Scholar] [CrossRef]
- Štundlová, J.; Šmíd, J.; Nguyen, P.; Šťáhlavský, F. Cryptic diversity and dynamic chromosome evolution in Alpine scorpions (Euscorpiidae: Euscorpius). Mol. Phylogenet. Evol. 2019, 134, 152–163. [Google Scholar] [CrossRef] [PubMed]
- Šťáhlavský, F.; Nguyen, P.; Sadílek, D.; Štundlová, J.; Just, P.; Haddad, C.R.; Koc, H.; Ranawana, K.B.; Stockmann, M.; Yagmur, E.A.; et al. Evolutionary dynamics of rDNA clusters on chromosomes of buthid scorpions (Chelicerata: Arachnida). Biol. J. Linn. Soc. 2020, 131, 547–565. [Google Scholar] [CrossRef]
- Šťáhlavský, F.; Kovařík, F.; Stockmann, M.; Opatova, V. Karyotype evolution and preliminary molecular assessment of genera in the family Scorpiopidae (Arachnida: Scorpiones). Zoology 2021, 144, 125882. [Google Scholar] [CrossRef]
- Červená, M.; Šťáhlavský, F.; Papáč, V.; Kováč, Ľ.; Christophoryová, J. Morphological and cytogenetic characteristics of Neobisium (Blothrus) slovacum Gulička, 1977 (Pseudoscorpiones, Neobisiidae), the northernmost troglobitic species of the subgenus Blothrus in Europe. Zookeys 2019, 817, 113–130. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Svojanovská, H.; Nguyen, P.; Hiřman, M.; Tuf, I.H.; Wahab, R.A.; Haddad, C.R.; Šťáhlavský, F. Karyotype evolution in harvestmen of the suborder Cyphophthalmi (Opiliones). Cytogenet. Genome Res. 2016, 148, 227–236. [Google Scholar] [CrossRef] [PubMed]
- Šťáhlavský, F.; Opatova, V.; Just, P.; Lotz, L.N.; Haddad, C.R. Molecular technique reveals high variability of 18S rDNA distribution in harvestmen (Opiliones, Phalangiidae) from South Africa. Comp. Cytogenet. 2018, 12, 41–59. [Google Scholar] [CrossRef] [Green Version]
- Jindrová, H.; Hiřman, M.; Sadílek, D.; Bezděčka, P.; Šťáhlavský, F. Distribution of 18S rDNA clusters in Central European harvestmen of the suborder Eupnoi (Arachnida: Opiliones). Eur. J. Entomol. 2020, 117, 282–288. [Google Scholar] [CrossRef]
- Hiřman, M.; Mohagan, A.; Šťáhlavský, F. Unexpectedly high number of 18S rRNA gene clusters in Miopsalis dillyi (Opiliones: Cyphophthalmi: Stylocellidae) from Mindanao, Philippines. J. Arachnol. 2021, 48, 322–328. [Google Scholar] [CrossRef]
- Hill, C.A.; Guerrero, F.D.; Van Zee, J.P.; Geraci, N.S.; Walling, J.G.; Stuart, J.J. The position of repetitive DNA sequence in the southern cattle tick genome permits chromosome identification. Chromosome Res. 2009, 17, 77–89. [Google Scholar] [CrossRef] [PubMed]
- Meyer, J.M.; Kurtti, T.J.; Van Zee, J.P.; Hill, C.A. Genome organization of major tandem repeats in the hard tick, Ixodes scapularis. Chromosome Res. 2010, 18, 357–370. [Google Scholar] [CrossRef]
- Král, J.; Musilová, J.; Šťáhlavský, F.; Řezáč, M.; Akan, Z.; Edwards, R.L.; Coyle, F.A.; Almerje, C.R. Evolution of the karyotype and sex chromosome systems in basal clades of araneomorph spiders (Araneae: Araneomorphae). Chromosome Res. 2006, 14, 859–880. [Google Scholar] [CrossRef]
- Gulia-Nuss, M.; Nuss, A.B.; Meyer, J.M.; Sonenshine, D.E.; Roe, R.M.; Waterhouse, R.M.; Sattelle, D.B.; de la Fuente, J.; Ribeiro, J.M.; Megy, K.; et al. Genomic insights into the Ixodes scapularis tick vector of Lyme disease. Nat. Commun. 2016, 7, 10507. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frydrychová, R.; Grossmann, P.; Trubač, P.; Vítková, M.; Marec, F. Phylogenetic distribution of TTAGG telomeric repeats in insects. Genome 2004, 47, 163–178. [Google Scholar] [CrossRef] [PubMed]
- Procházková Schrumpfová, P.; Schořová, Š.; Fajkus, J. Telomere- and telomerase-associated proteins and their functions in the plant cell. Front. Plant Sci. 2016, 7, 1–19. [Google Scholar] [CrossRef] [Green Version]
- Grozeva, S.; Anokhin, B.A.; Simov, N.; Kuznetsova, V.G. New evidence for the presence of the telomere motif (TTAGG)n in the family Reduviidae and its absence in the families Nabidae and Miridae (Hemiptera, Cimicomorpha). Comp. Cytogenet. 2019, 13, 283–295. [Google Scholar] [CrossRef]
- Kuznetsova, V.; Grozeva, S.; Gokhman, V. Telomere structure in insects: A review. J. Zool. Syst. Evol. Res. 2020, 58, 127–158. [Google Scholar] [CrossRef]
- Chirino, M.G.; Dalíková, M.; Marec, F.R.; Bressa, M.J. Chromosomal distribution of interstitial telomeric sequences as signs of evolution through chromosome fusion in six species of the giant water bugs (Hemiptera, Belostoma). Ecol. Evol. 2017, 7, 5227–5235. [Google Scholar] [CrossRef]
- Dutra, D.D.; Souza, L.H.B.; Cordeiro, L.M.; Araujo, D. The highest chromosome number and first chromosome fluorescent in situ hybridization in the velvet worms of the family Peripatidae. Zool. Stud. 2020, 59, e5. [Google Scholar] [CrossRef]
- Ruiz-Herrera, A.; Nergadze, S.G.; Santagostino, M.; Giulotto, E. Telomeric repeats far from the ends: Mechanisms of origin and role in evolution. Cytogenet. Genome Res. 2008, 122, 219–228. [Google Scholar] [CrossRef] [PubMed]
- Bolzán, A.D. Interstitial telomeric sequences in vertebrate chromosomes: Origin, function, instability and evolution. Mutat. Res. 2017, 773, 51–65. [Google Scholar] [CrossRef] [PubMed]
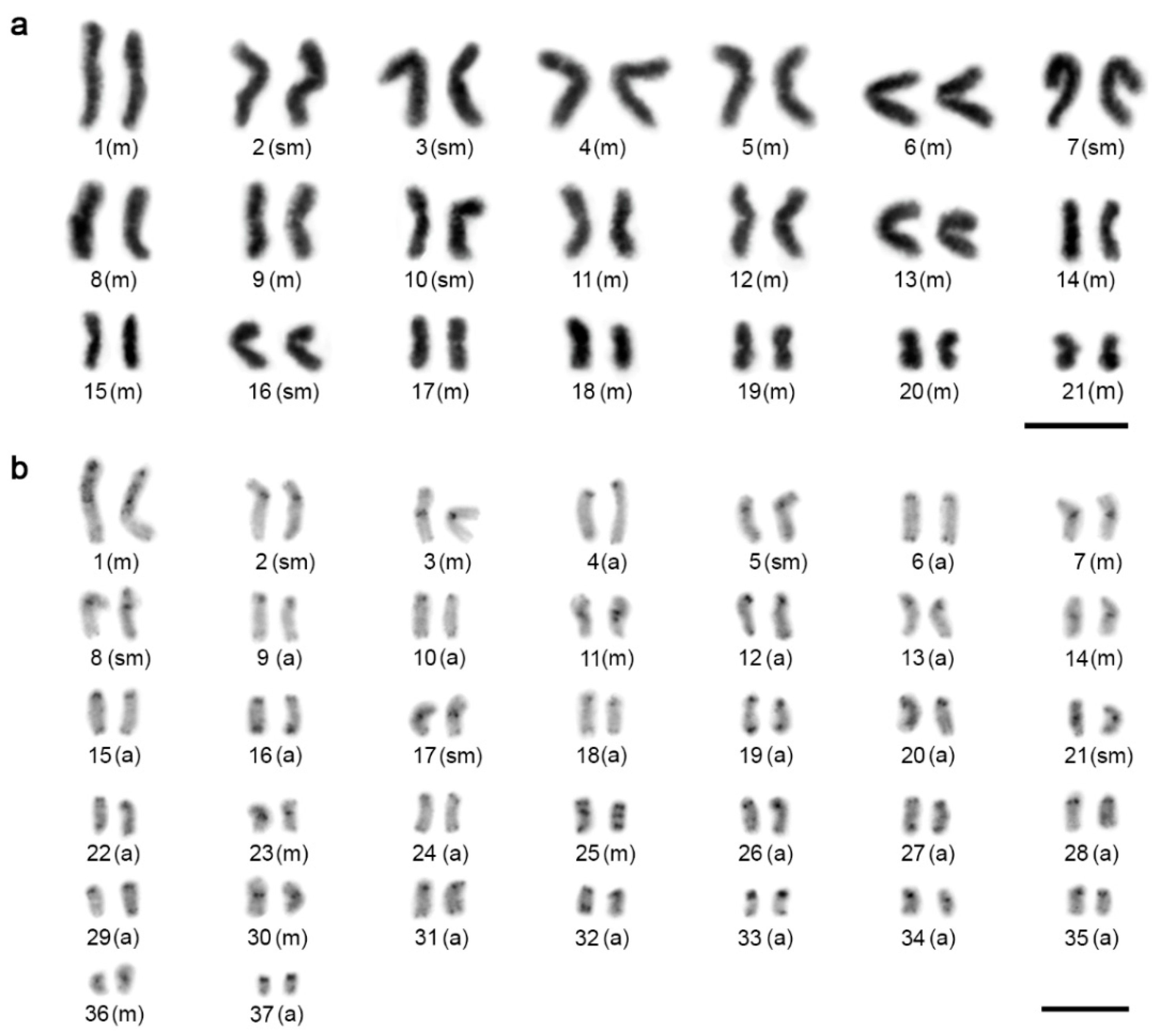



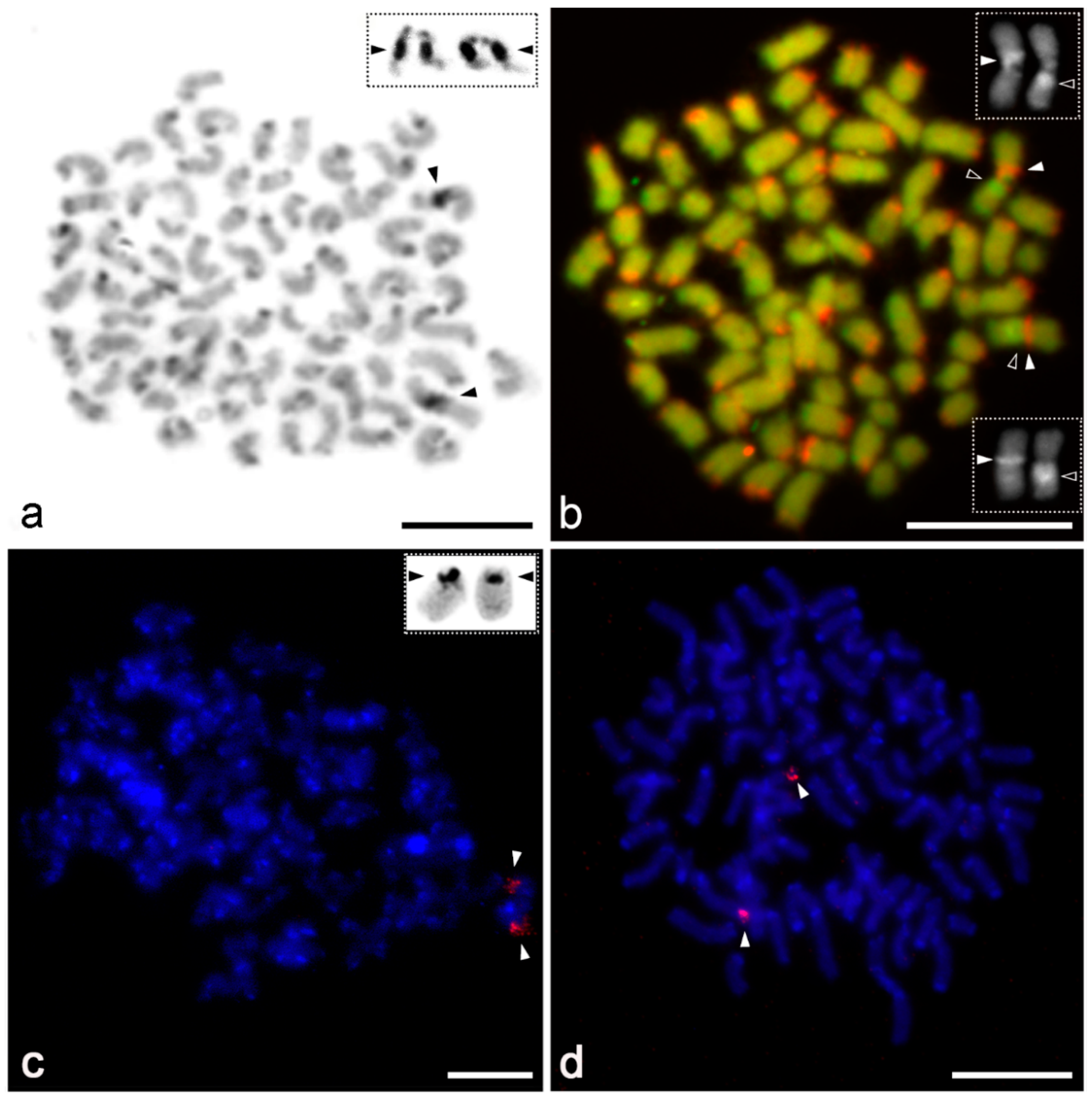
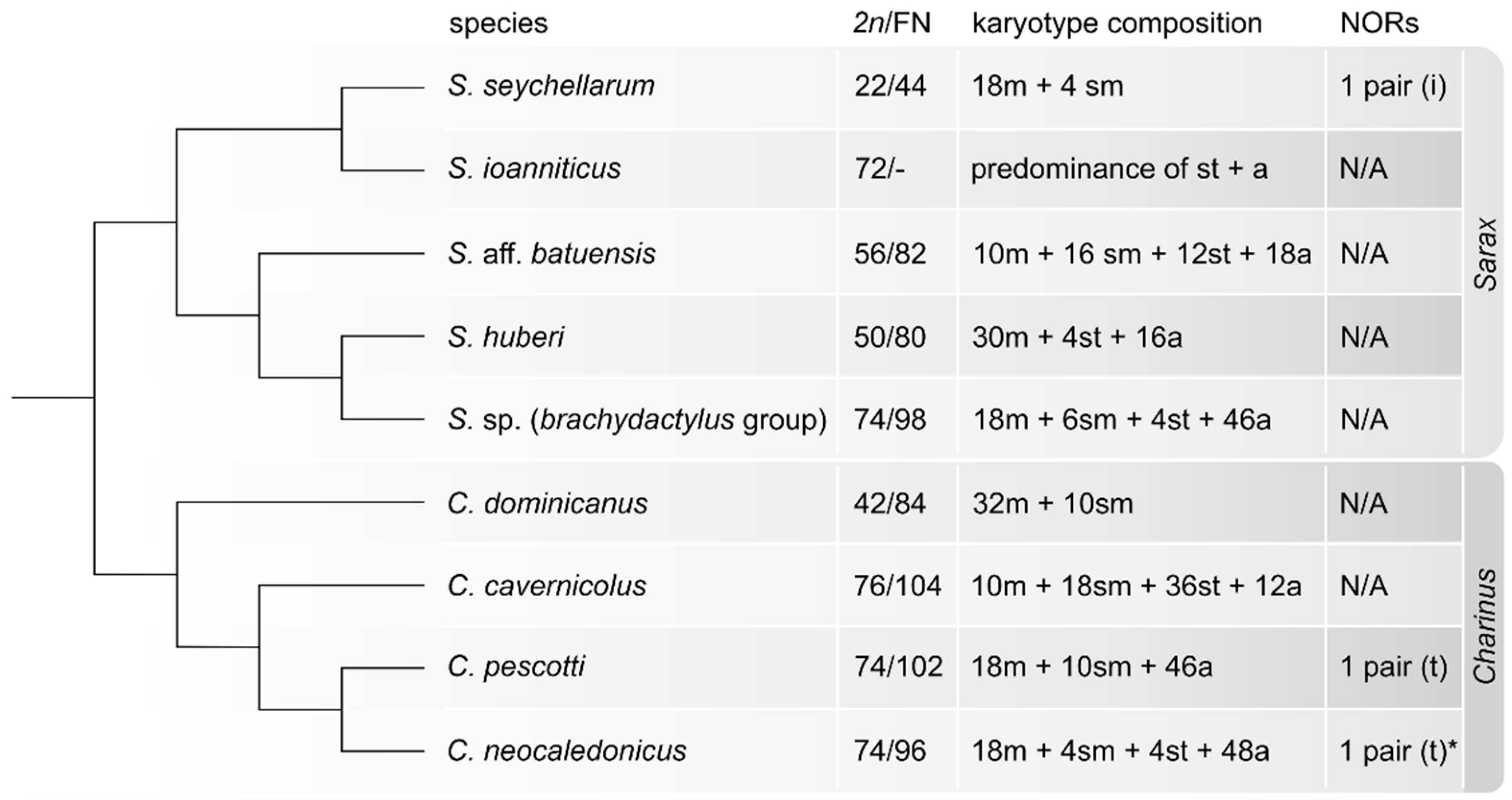
| Species | N | Source or Locality | Methods |
|---|---|---|---|
| Charinus cavernicolus Weygoldt, 2006 | 2♂, 1 juv | Grottes de Koumac, Koumac, New Caledonia | G, K |
| C. dominicanus Armas & Gonzáles Perez, 2001 | 1♂ | Los Charcos, San Rafael, La Ciénaga Municipality, Barahona Province, Dominican Republic | G, K |
| C. neocaledonicus Simon in Kraepelin, 1895 | 5♂ | Mount Coghis, close to Noumea, New Caledonia | G, K, C, F, Ag, rDNA |
| C. pescotti Dunn, 1949 | 3♂ | Mossman river Queensland, Australia | G, K, C, rDNA |
| Sarax aff. batuensis Roewer, 1962 | 1♂ | Malaysia breeding | G, K |
| S. huberi Seiter et al., 2015 | 1♂ | surroundings of the Busay cave, Moalboal, Cebu Island, Philippines | G, K |
| S. ioanniticus Kritscher, 1959 | 3♀ | casemates of the fortress, Rhodos, Rhodos Island, Greece | G |
| S. seychellarum Kraepelin, 1898 | 1♂, 2♀ | Mahé Island, Republic of Seychelles | G, K, rDNA, T |
| Sarax sp. * | 1♂ | Malaysia breeding | G, K |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Reyes Lerma, A.C.; Šťáhlavský, F.; Seiter, M.; Carabajal Paladino, L.Z.; Divišová, K.; Forman, M.; Sember, A.; Král, J. Insights into the Karyotype Evolution of Charinidae, the Early-Diverging Clade of Whip Spiders (Arachnida: Amblypygi). Animals 2021, 11, 3233. https://doi.org/10.3390/ani11113233
Reyes Lerma AC, Šťáhlavský F, Seiter M, Carabajal Paladino LZ, Divišová K, Forman M, Sember A, Král J. Insights into the Karyotype Evolution of Charinidae, the Early-Diverging Clade of Whip Spiders (Arachnida: Amblypygi). Animals. 2021; 11(11):3233. https://doi.org/10.3390/ani11113233
Chicago/Turabian StyleReyes Lerma, Azucena Claudia, František Šťáhlavský, Michael Seiter, Leonela Zusel Carabajal Paladino, Klára Divišová, Martin Forman, Alexandr Sember, and Jiří Král. 2021. "Insights into the Karyotype Evolution of Charinidae, the Early-Diverging Clade of Whip Spiders (Arachnida: Amblypygi)" Animals 11, no. 11: 3233. https://doi.org/10.3390/ani11113233







