Integration of Microfluidics, Photonic Integrated Circuits and Data Acquisition and Analysis Methods in a Single Platform for the Detection of Swine Viral Diseases
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Samples
2.2. Conventional PCR Assays
2.3. Quantitative PCR (qPCR) Assays
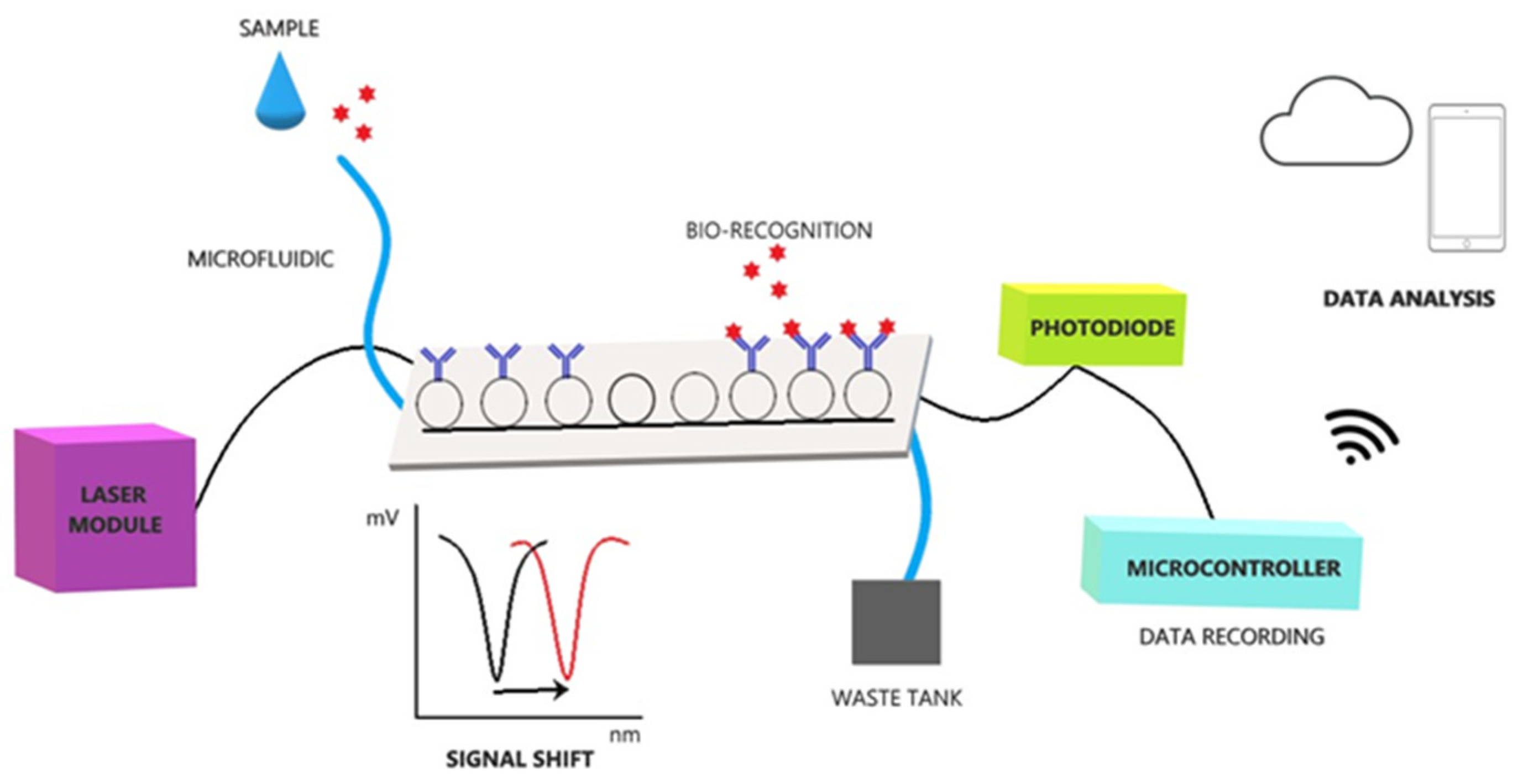
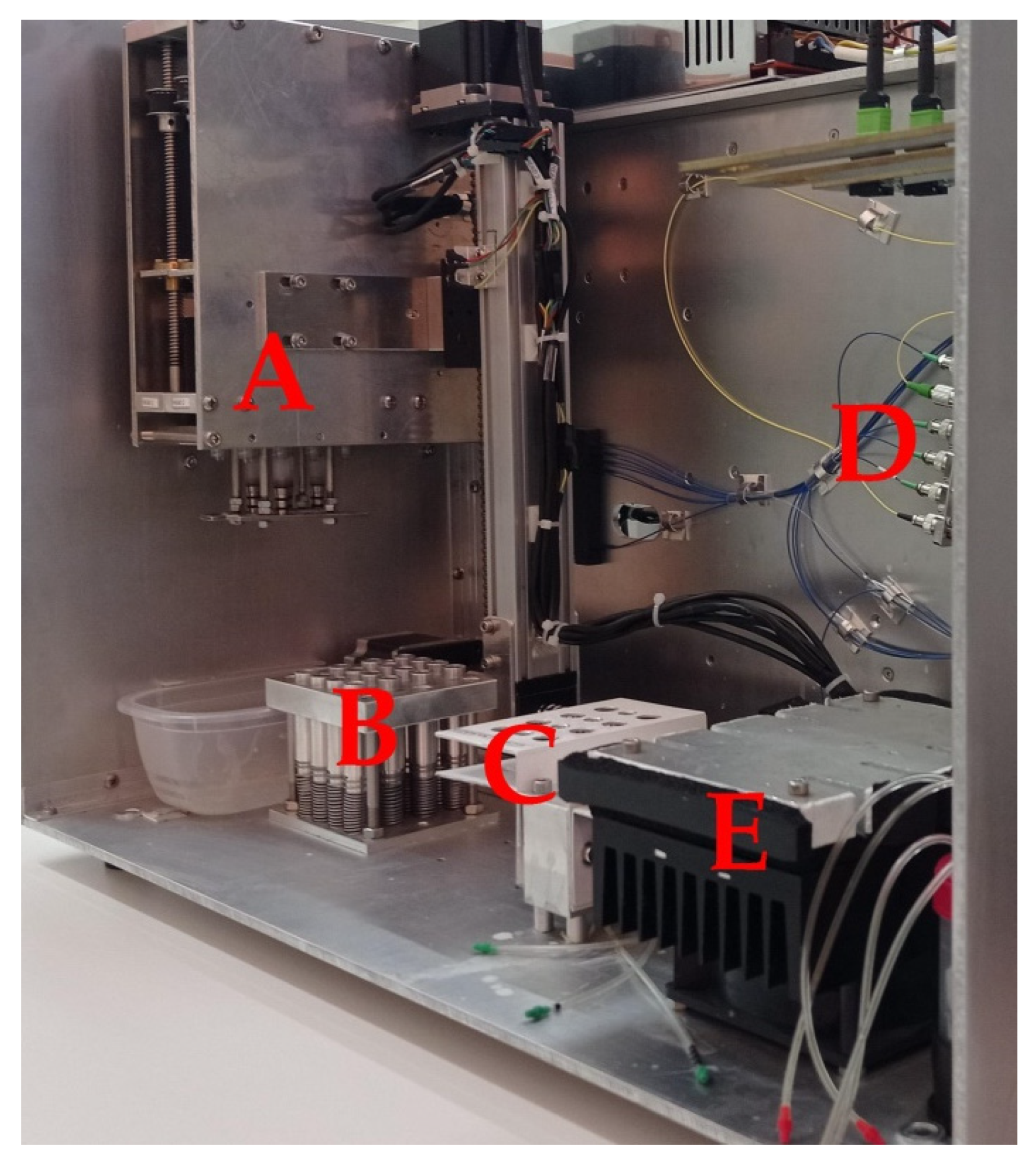
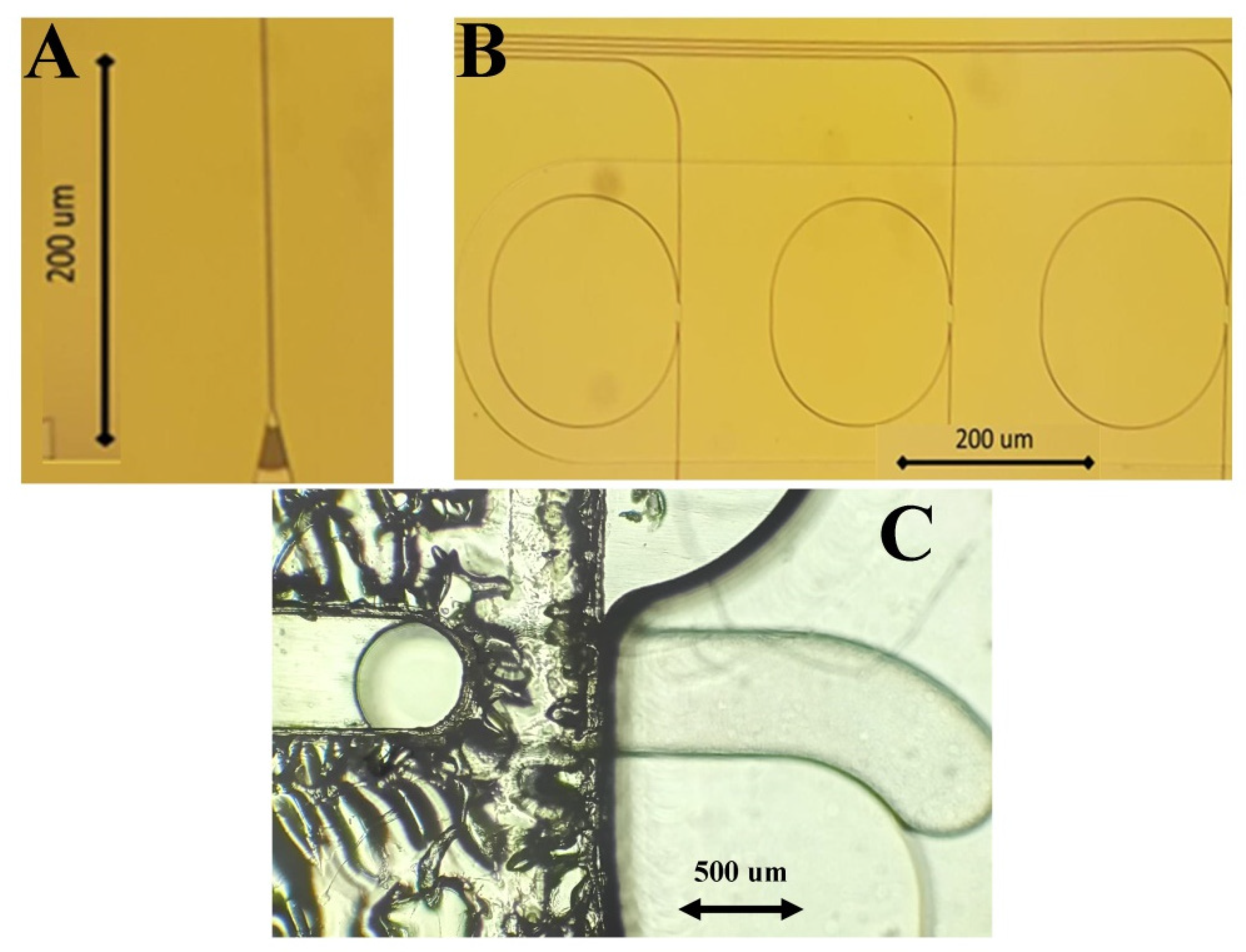
2.4. POC Device
2.5. Sensors, Antibodies, and Biofunctionalization
2.6. Analysis Protocol
- The buffer step: The buffer used was PBS + 0.05% v/v Tween 20 + 1% w/v BSA, pH = 7.4, which flowed for 15 min at a flow rate of 30 μL/min. During this step, a signal was stabilized to establish the signal baseline.
- The sample step: The sample was diluted at a ratio of 1:1 with PBS + 0.05% v/v Tween 20 + 1% w/v BSA, pH = 7.4. It flowed for 10 min at a flow rate of 30 μL/min. Binding of the analytes on the functionalized PIC surfaces occurred during this step.
- The washing step: The buffer used was PBS + 0.05% v/v Tween 20 + 1% w/v BSA, pH = 7.4, which flowed for 15 min at a flow rate of 30 μL/min. Unbound viral particles and sample residues that could affect photonic measurements were removed at this step.
- The PIC surface regeneration step: The buffer used was 50 mM Glycine + 50% v/v Ethylene Glycol, pH = 3, which flowed for 5 min at a flow rate of 30 μL/min. During this step, PIC surfaces were regenerated by releasing the captured antigens from the antibodies.
- The final washing step: The buffer used was PBS + 0.05% v/v Tween 20, pH = 7.4, which flowed for 5 min at a flow rate of 30 μL/min. Removal of the regeneration buffer was critical for the preparation of PIC surfaces for the next experiments. In this step, BSA was excluded from the washing buffer to prevent protein accumulation in the microfluidics.
2.7. Data Fitting
2.8. Data Analysis and Shift Calculation
2.9. Validation of the Device
2.9.1. Limit of Detection—LOD
2.9.2. System Performance
2.10. Statistical Analysis
3. Results
3.1. PCR Results
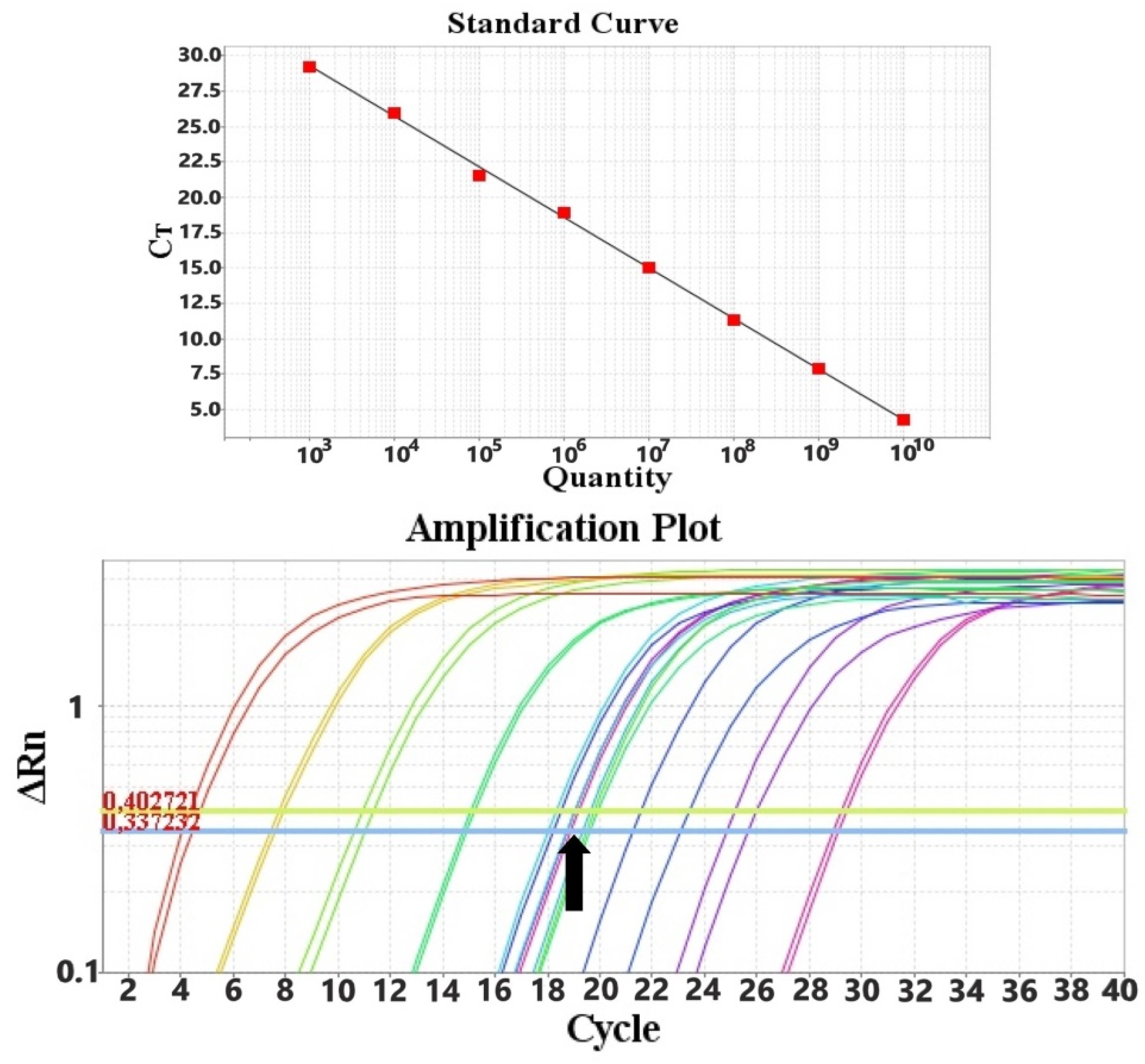
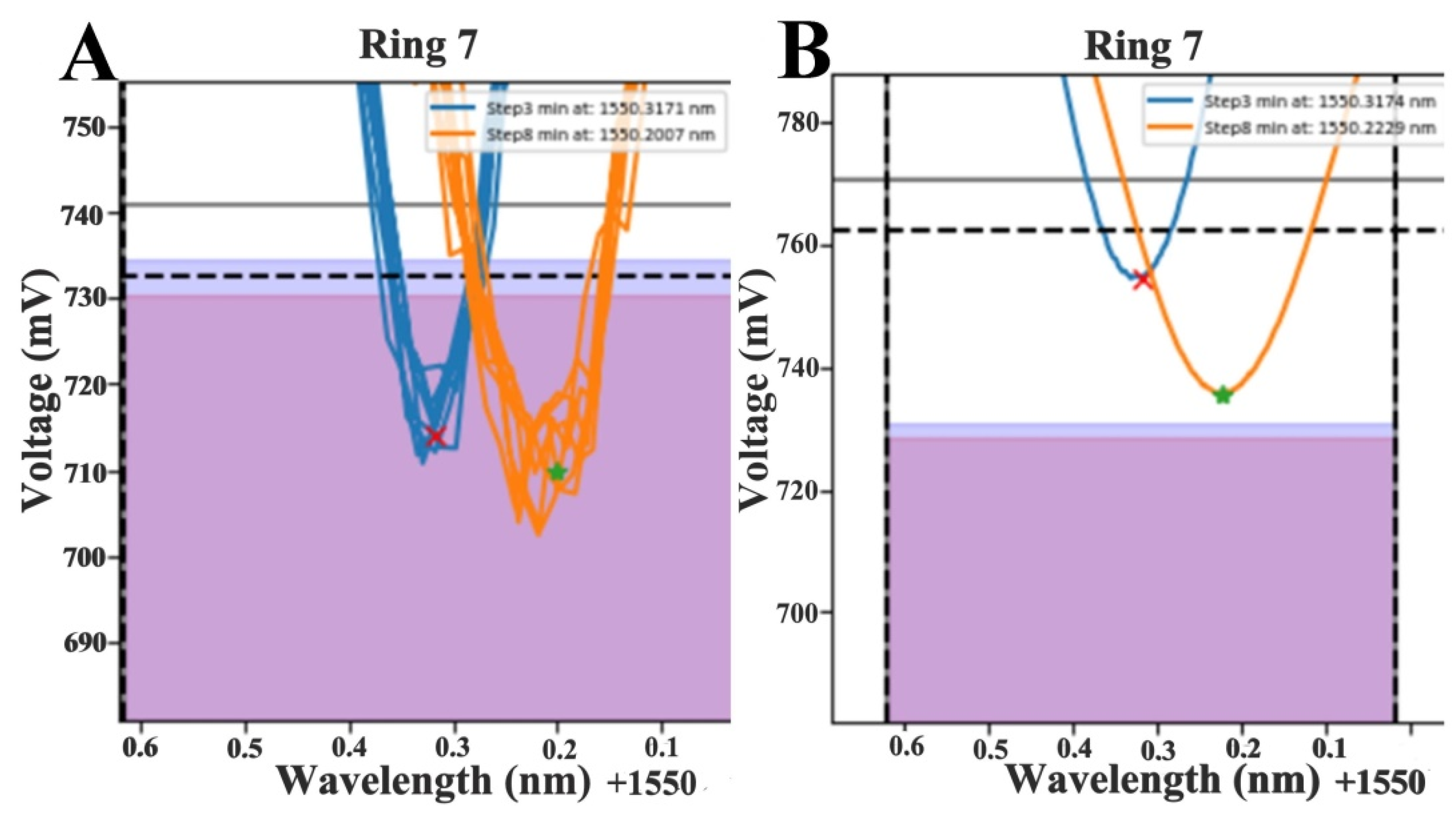
3.2. Data Fitting
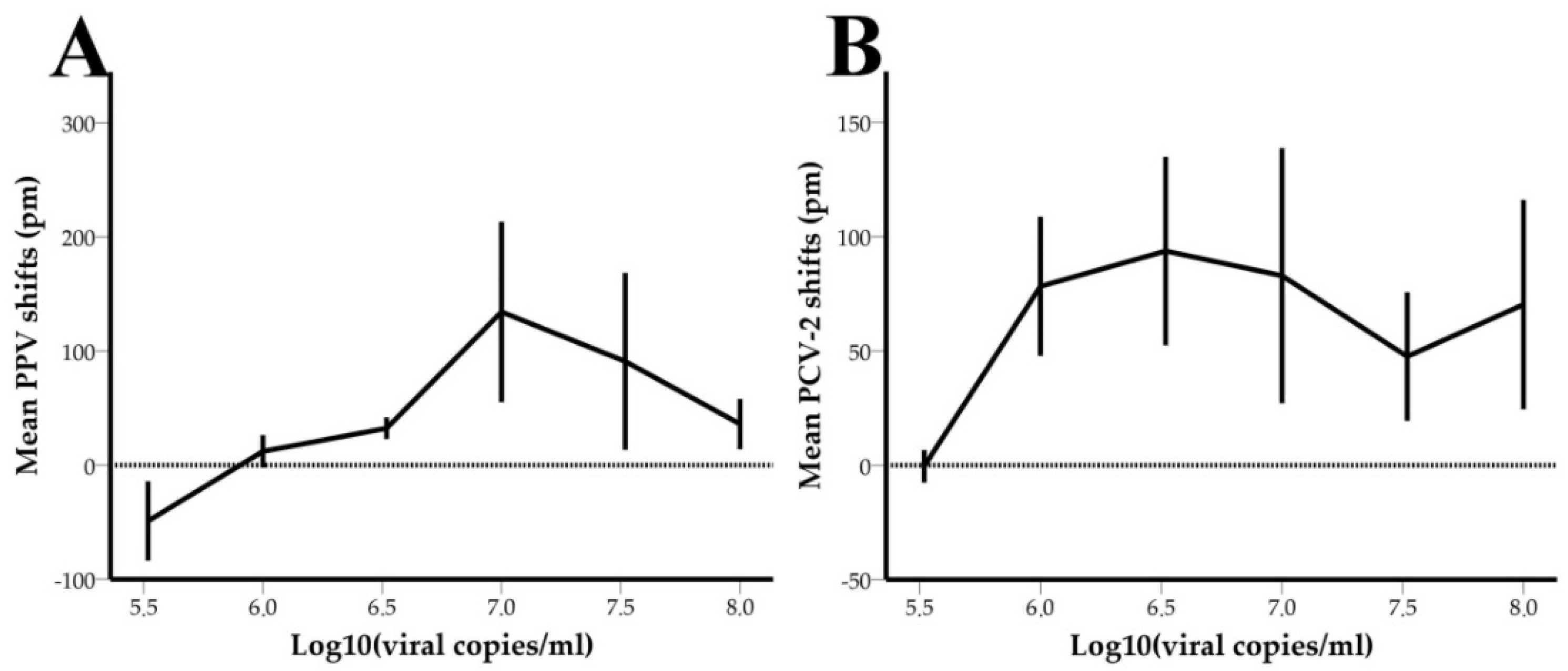
3.3. Limit of Detection-LOD
3.4. Performance of the System
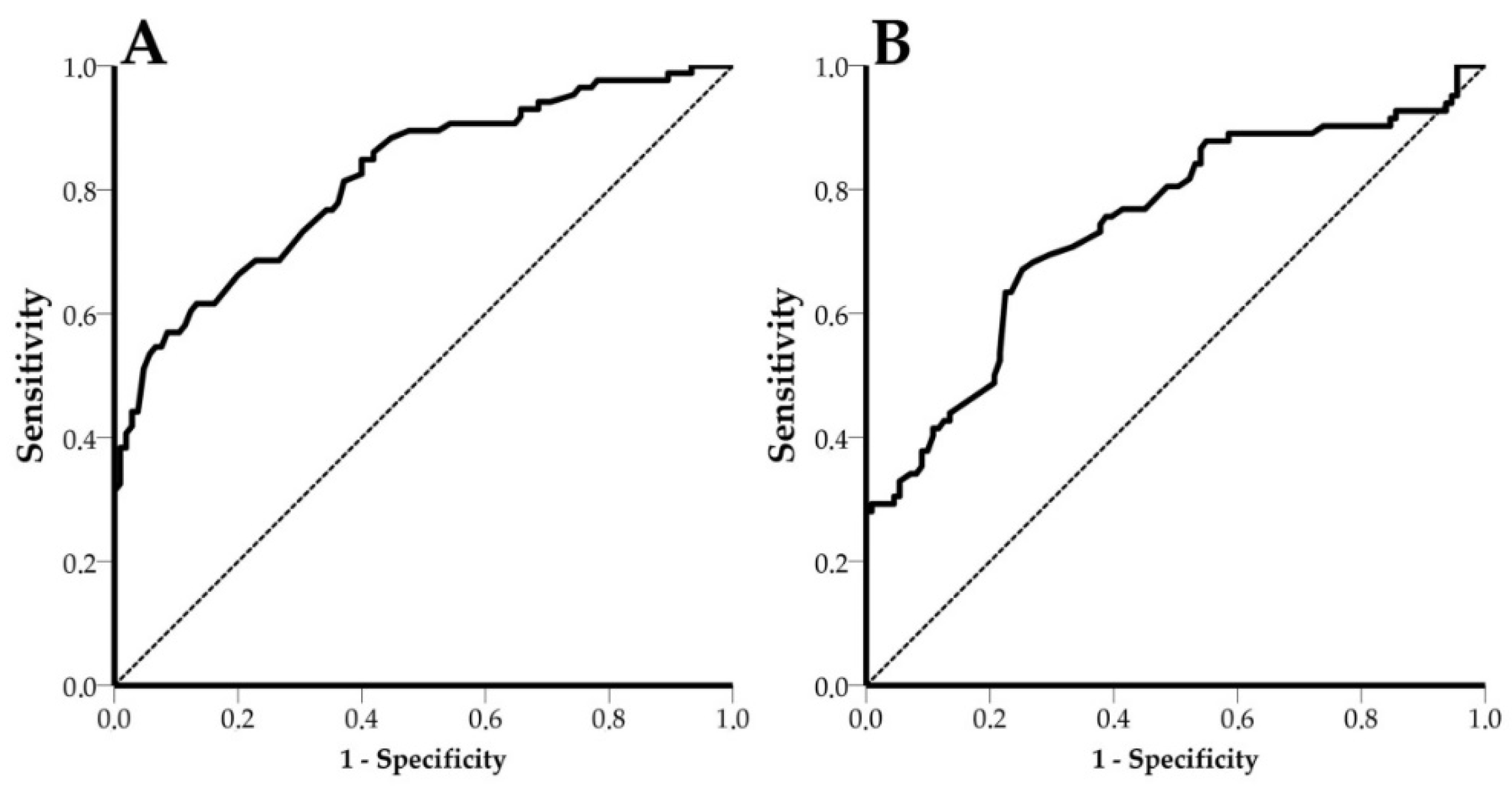
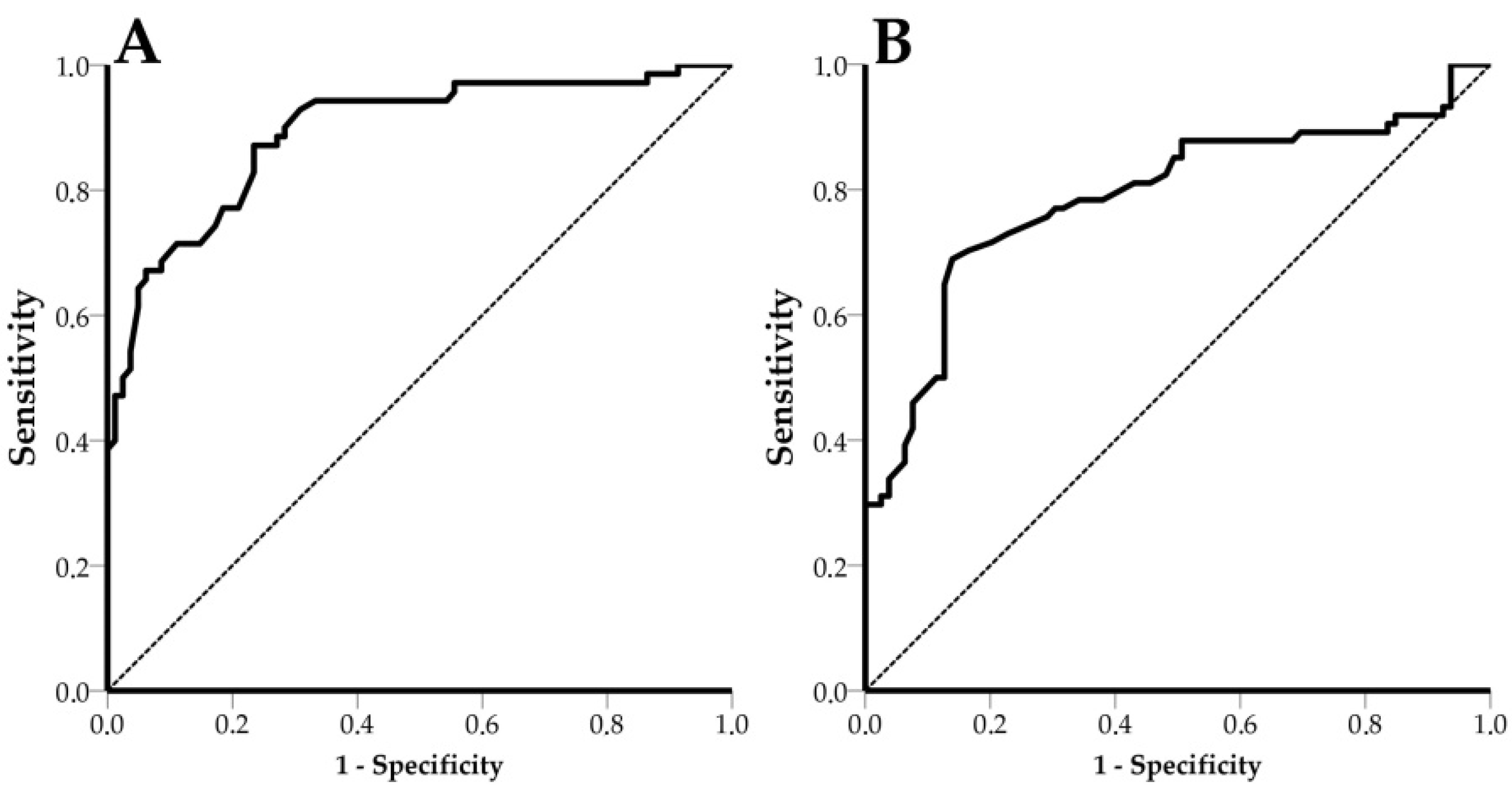
3.5. Receiver Operating Characteristic (ROC) Curves
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Acknowledgments
Conflicts of Interest
References
- Maes, D.G.D.; Dewulf, J.; Piñeiro, C.; Edwards, S.; Kyriazakis, I. A critical reflection on intensive pork production with an emphasis on animal health and welfare. J. Anim. Sci. 2020, 98, S15–S26. [Google Scholar] [CrossRef]
- Wang, H.; Long, W.; Chadwick, D.; Velthof, G.L.; Oenema, O.; Ma, W.; Wang, J.; Qin, W.; Hou, Y.; Zhang, F. Can dietary manipulations improve the productivity of pigs with lower environmental and economic cost? A global meta-analysis. Agric. Ecosyst. Environ. 2020, 289, 106748. [Google Scholar] [CrossRef]
- VanderWaal, K.; Deen, J. Global trends in infectious diseases of swine. Proc. Natl. Acad. Sci. USA 2018, 115, 11495–11500. [Google Scholar] [CrossRef]
- Morgan, N.; Prakash, A. International livestock markets and the impact of animal disease. OIE Rev. Sci. Tech. 2006, 25, 517–528. [Google Scholar] [CrossRef]
- Perry, B.D.; Grace, D.; Sones, K. Current drivers and future directions of global livestock disease dynamics. Proc. Natl. Acad. Sci. USA 2013, 110, 20871–20877. [Google Scholar] [CrossRef] [PubMed]
- Streck, A.F.; Canal, C.W.; Truyen, U. Molecular epidemiology evolution of porcine parvoviruses. Infect. Genet. Evol. 2015, 36, 300–306. [Google Scholar] [CrossRef] [PubMed]
- Nawagitgul, P.; Morozov, I.; Bolin, S.R.; Harms, P.A.; Sorden, S.D.; Paul, P.S. Open reading frame 2 of porcine circovirus type 2 encodes a major capsid protein. J. Gen. Virol. 2000, 81, 2281–2287. [Google Scholar] [CrossRef] [PubMed]
- Chae, C. Postweaning multisystemic wasting syndrome: A review of aetiology, diagnosis and pathology. Vet. J. 2004, 168, 41–49. [Google Scholar] [CrossRef]
- Krakowka, S.; Ellis, J.A.; McNeilly, F.; Ringler, S.; Rings, D.M.; Allan, G. Activation of the immune system is the pivotal event in the production of wasting disease in pigs infected with porcine circovirus-2 (PCV 2). Vet. Pathol. 2001, 38, 31–42. [Google Scholar] [CrossRef] [PubMed]
- Rovira, A.; Balasch, M.; Segalés, J.; García, L.; Plana-Durán, J.; Rosell, C.; Ellerbrok, H.; Mankertz, A.; Domingo, M. Experimental Inoculation of Conventional Pigs with Porcine Reproductive and Respiratory Syndrome Virus and Porcine Circovirus 2. J. Virol. 2002, 76, 3232–3239. [Google Scholar] [CrossRef] [PubMed]
- Manessis, G.; Gelasakis, A.; Bossis, I. The challenge of introducing Point of Care Diagnostics in Farm Animal Health Management. Biomed. J. Sci. Tech. Res. 2019, 14, 10–13. [Google Scholar] [CrossRef]
- Foudeh, A.M.; Fatanat Didar, T.; Veres, T.; Tabrizian, M. Microfluidic designs and techniques using lab-on-a-chip devices for pathogen detection for point-of-care diagnostics. Lab Chip 2012, 12, 3249–3266. [Google Scholar] [CrossRef] [PubMed]
- Cummins, B.M.; Ligler, F.S.; Walker, G.M. Point-of-care diagnostics for niche applications. Biotechnol. Adv. 2016, 34, 161–176. [Google Scholar] [CrossRef] [PubMed]
- Busin, V.; Wells, B.; Kersaudy-Kerhoas, M.; Shu, W.; Burgess, S.T.G. Opportunities and challenges for the application of microfluidic technologies in point-of-care veterinary diagnostics. Mol. Cell. Probes 2016, 30, 331–341. [Google Scholar] [CrossRef] [PubMed]
- Luppa, P.B.; Müller, C.; Schlichtiger, A.; Schlebusch, H. Point-of-care testing (POCT): Current techniques and future perspectives. TrAC Trends Anal. Chem. 2011, 30, 887–898. [Google Scholar] [CrossRef]
- Vashist, S.K.; Luppa, P.B.; Yeo, L.Y.; Ozcan, A.; Luong, J.H.T. Emerging Technologies for Next-Generation Point-of-Care Testing. Trends Biotechnol. 2015, 33, 692–705. [Google Scholar] [CrossRef]
- Jiang, Y.; Jiang, S.; Wu, Y.; Zhou, B.; Wang, K.; Jiang, L.; Long, Y.; Chen, G.; Zeng, D. Multiplex and on-site PCR detection of swine diseases based on the microfluidic chip system. BMC Vet. Res. 2021, 17, 1–8. [Google Scholar] [CrossRef]
- Zhou, L.; Chen, Y.; Fang, X.; Liu, Y.; Du, M.; Lu, X.; Li, Q.; Sun, Y.; Ma, J.; Lan, T. Microfluidic-RT-LAMP chip for the point-of-care detection of emerging and re-emerging enteric coronaviruses in swine. Anal. Chim. Acta 2020, 1125, 57–65. [Google Scholar] [CrossRef]
- Fu, Y.; Li, W.; Dai, B.; Zheng, L.; Zhang, Z.; Qi, D.; Cheng, X.; Zhang, D.; Zhuang, S. Diagnosis of mixed infections with swine viruses using an integrated microfluidic platform. Sens. Actuators B Chem. 2020, 312, 128005. [Google Scholar] [CrossRef]
- Montagnese, C.; Barattini, P.; Giusti, A.; Balka, G.; Bruno, U.; Bossis, I.; Gelasakis, A.; Bonasso, M.; Philmis, P.; Dénes, L.; et al. A diagnostic device for in-situ detection of swine viral diseases: The SWINOSTICS project. Sensors 2019, 19, 407. [Google Scholar] [CrossRef]
- Hänsel, A.; Heck, M.J.R. Opportunities for photonic integrated circuits in optical gas sensors. J. Phys. Photonics 2020, 2, 012002. [Google Scholar] [CrossRef]
- Wang, S.; Lifson, M.A.; Inci, F.; Liang, L.G.; Sheng, Y.F.; Demirci, U. Advances in addressing technical challenges of point-of-care diagnostics in resource-limited settings. Expert Rev. Mol. Diagn. 2016, 16, 449–459. [Google Scholar] [CrossRef] [PubMed]
- Chandrasekar, R.; Lapin, Z.J.; Nichols, A.S.; Braun, R.M.; Fountain, A.W. Photonic integrated circuits for Department of Defense-relevant chemical and biological sensing applications: State-of-the-art and future outlooks. Opt. Eng. 2019, 58, 1. [Google Scholar] [CrossRef]
- Soares, R.M.; Durigon, E.L.; Bersano, J.G.; Richtzenhain, L.J. Detection of porcine parvovirus DNA by the polymerase chain reaction assay using primers to the highly conserved nonstructural protein gene, NS-1. J. Virol. Methods 1999, 78, 191–198. [Google Scholar] [CrossRef]
- Huang, C.; Hung, J.J.; Wu, C.Y.; Chien, M.S. Multiplex PCR for rapid detection of pseudorabies virus, porcine parvovirus and porcine circoviruses. Vet. Microbiol. 2004, 101, 209–214. [Google Scholar] [CrossRef]
- Larochelle, R.; Bielanski, A.; Müller, P.; Magar, R. PCR detection and evidence of shedding of porcine circovirus type 2 in boar semen. J. Clin. Microbiol. 2000, 38, 4629–4632. [Google Scholar] [CrossRef]
- Yang, K.; Jiao, Z.; Zhou, D.; Guo, R.; Duan, Z.; Tian, Y. Development of a multiplex PCR to detect and discriminate porcine circoviruses in clinical specimens. BMC Infect. Dis. 2019, 19, 778. [Google Scholar] [CrossRef]
- Rincón Monroy, M.A.; Ramirez-Nieto, G.C.; Vera, V.J.; Correa, J.J.; Mogollón-Galvis, J.D. Detection and molecular characterization of porcine circovirus type 2 from piglets with Porcine Circovirus Associated Diseases in Colombia. Virol. J. 2014, 11, 1–11. [Google Scholar] [CrossRef][Green Version]
- Mudumba, S.; de Alba, S.; Romero, R.; Cherwien, C.; Wu, A.; Wang, J.; Gleeson, M.A.; Iqbal, M.; Burlingame, R.W. Photonic ring resonance is a versatile platform for performing multiplex immunoassays in real time. J. Immunol. Methods 2017, 448, 34–43. [Google Scholar] [CrossRef]
- Griol, A.; Peransi, S.; Rodrigo, M.; Hurtado, J.; Bellieres, L.; Ivanova, T.; Zurita, D.; Carles, S.; Recuero, S.; Hern, A.; et al. Design and Development of Photonic Biosensors for Swine Viral Diseases Detection. Sensors 2019, 19, 3985. [Google Scholar] [CrossRef]
- Cleveland, W.S. Robust locally weighted regression and smoothing scatterplots. J. Am. Stat. Assoc. 1979, 74, 829–836. [Google Scholar] [CrossRef]
- Rabenau, H.F.; Kessler, H.H.; Kortenbusch, M.; Steinhorst, A.; Raggam, R.B.; Berger, A. Verification and validation of diagnostic laboratory tests in clinical virology. J. Clin. Virol. 2007, 40, 93–98. [Google Scholar] [CrossRef] [PubMed]
- Belák, S. Molecular diagnosis of viral diseases, present trends and future aspects. A view from the OIE Collaborating Centre for the Application of Polymerase Chain Reaction Methods for Diagnosis of Viral Diseases in Veterinary Medicine. Vaccine 2007, 25, 5444–5452. [Google Scholar] [CrossRef] [PubMed]
- Hobbs, E.C.; Colling, A.; Gurung, R.B.; Allen, J. The potential of diagnostic point-of-care tests (POCTs) for infectious and zoonotic animal diseases in developing countries: Technical, regulatory and sociocultural considerations. Transbound. Emerg. Dis. 2021, 68, 1835–1849. [Google Scholar] [CrossRef]
- Nannucci, L.; Barattini, P.; Bossis, I.; Wozniakowski, G.; Balka, G.; Pugliese, C. Point-of-service diagnostic technology for detection of swine viral diseases. J. Vet. Res. 2020, 64, 15–23. [Google Scholar] [CrossRef]
- Ni, J.; Qiao, C.; Han, X.; Han, T.; Kang, W.; Zi, Z.; Cao, Z.; Zhai, X.; Cai, X. Identification and genomic characterization of a novel porcine parvovirus (PPV6) in china. Virol. J. 2014, 11, 203. [Google Scholar] [CrossRef]
- Bøtner, A.; Belsham, G.J. Virus survival in slurry: Analysis of the stability of foot-and-mouth disease, classical swine fever, bovine viral diarrhoea and swine influenza viruses. Vet. Microbiol. 2012, 157, 41–49. [Google Scholar] [CrossRef]
- Mészáros, I.; Olasz, F.; Cságola, A.; Tijssen, P.; Zádori, Z. Biology of porcine parvovirus (Ungulate parvovirus 1). Viruses 2017, 9, 393. [Google Scholar] [CrossRef]
- Miłek, D.; Woźniak, A.; Guzowska, M.; Stadejek, T. Detection patterns of porcine parvovirus (PPV) and novel porcine parvoviruses 2 through 6 (PPV2–PPV6) in Polish swine farms. Viruses 2019, 11, 474. [Google Scholar] [CrossRef]
- Segalés, J.; Allan, G.M.; Domingo, M. Porcine circovirus diseases. Anim. Health Res. Rev. 2005, 6, 119–142. [Google Scholar] [CrossRef]
- Karuppannan, A.K.; Opriessnig, T. Porcine circovirus type 2 (PCV2) vaccines in the context of current molecular epidemiology. Viruses 2017, 9, 99. [Google Scholar] [CrossRef] [PubMed]
- Miao, L.F.; Zhang, C.F.; Chen, C.M.; Cui, S.J. Real-time PCR to detect and analyze virulent PPV loads in artificially challenged sows and their fetuses. Vet. Microbiol. 2009, 138, 145–149. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Ullman, K.; Chowdry, V.; Reining, M.; Benyeda, Z.; Baule, C.; Juremalm, M.; Wallgren, P.; Schwarz, L.; Zhou, E.; et al. Molecular investigations on the prevalence and viral load of enteric viruses in pigs from five European countries. Vet. Microbiol. 2016, 182, 75–81. [Google Scholar] [CrossRef] [PubMed]
- Olvera, A.; Sibila, M.; Calsamiglia, M.; Segalés, J.; Domingo, M. Comparison of porcine circovirus type 2 load in serum quantified by a real time PCR in postweaning multisystemic wasting syndrome and porcine dermatitis and nephropathy syndrome naturally affected pigs. J. Virol. Methods 2004, 117, 75–80. [Google Scholar] [CrossRef] [PubMed]
- Henao-Diaz, A.; Giménez-Lirola, L.; Baum, D.H.; Zimmerman, J. Guidelines for oral fluid-based surveillance of viral pathogens in swine. Porc. Health Manag. 2020, 6, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Zimmerman, J.; Bjustrom-Kraft, J.; Christopher-Hennings, J.; Daly, R.; Main, R.; Torrison, J.; Thurn, M.; Zimmerman DVM, J. The use of oral fluid diagnostics in swine medicine. J. Swine Health Prod. 2018, 26, 262–269. [Google Scholar]
- Ramirez, A.; Wang, C.; Prickett, J.R.; Pogranichniy, R.; Yoon, K.J.; Main, R.; Johnson, J.K.; Rademacher, C.; Hoogland, M.; Hoffmann, P.; et al. Efficient surveillance of pig populations using oral fluids. Prev. Vet. Med. 2012, 104, 292–300. [Google Scholar] [CrossRef] [PubMed]
- Latkowski, S.; Pustakhod, D.; Yao, W.; Leijtens, X.; Williams, K. Test Methods and Processes in Manufacturing Chain of Photonic Integrated Circuits. Int. Conf. Transparent Opt. Netw. 2018, 2018, 1–4. [Google Scholar] [CrossRef]

| Primer Set | Target | Primer Sequence (5′-3′) | Amplicon Length (bp) | Literature |
|---|---|---|---|---|
| PPV_Set_1 | NS1 gene | Forward: TTGGTAATGTTGGTTGCTACAATGC Reverse: ACCTGAACATATGGCTTTGAATTGG | 127 | [24] |
| PPV_Set_2 | NS1 gene | Forward: AGCCAAAAATGCAAACCCCAATA Reverse: CTCCACGGCTCCAAGGCTAAAG | 142 | [25] |
| PPV_Set_3 | NS1 gene | Forward: ATACAATTCTATTTCATGGGCCAGC Reverse: TATGTTCTGGTCTTTCCTCGCATC | 330 | [24] |
| PCV2_Set_1 | PCV-2 Capsid protein gene | Forward: TAGGTTAGGGCTGTGGCCTT Reverse: CCGCACCTTCGGATATACTG | 263 | [26] |
| PCV2_Set_2 | PCV-2 Rep gene | Forward: CACATCGAGAAAGCGAAAGGAAC Reverse: TGCGGGCCAAAAAAGGTACAGTT | 505 | [27] |
| PCV2_Set_3 | PCV-2 ORF1 | Forward: GCCAGTTCGTCACCCTTTC Reverse: CTCCCGCACCTTCGGATAT | 657 | [28] |
| Primer Set | Pre-Denaturation at 95 °C | Cycles | Denaturation at 94 °C | Annealing for 30 s at | Extension at 72 °C | Final Extension at 72 °C |
|---|---|---|---|---|---|---|
| PPV_Set_1 | 2 min | 32 | 20 s | 62 °C | 30 s | 1 min |
| PPV_Set_2 | 2 min | 32 | 20 s | 59 °C | 30 s | 1 min |
| PPV_Set_3 | 2 min | 32 | 20 s | 62 °C | 30 s | 1 min |
| PCV2_Set_1 | 2 min | 32 | 20 s | 60 °C | 30 s | 1 min |
| PCV2_Set_2 | 2 min | 32 | 20 s | 62 °C | 40 s | 1 min |
| PCV2_Set_3 | 2 min | 32 | 20 s | 59 °C | 40 s | 1 min |
| Positive and Negative Calibrators | Low Positive Calibrators | |||||
|---|---|---|---|---|---|---|
| Exp. Number | Sample Type | Negative/Positive | Viral Copies/mL | Sample Matrix | Virus | Viral Copies/mL |
| 1st | Oral fluids | Negative for PCV2 & PPV | 0 | Oral fluids | PPV, PCV-2 | 7 × 105 9 × 105 |
| 2nd | Oral fluids | Positive for PCV2 & PPV | 108 | Oral fluids | PPV, PCV-2 | 9 × 105 3 × 106 |
| 3rd | Oral fluids | Negative for PCV2 & PPV | 0 | Oral fluids | PPV, PCV-2 | 1.1 × 106 4 × 106 |
| 4th | Oral fluids | Positive for PPV | 108 | Feces | PCV-2 | 6 × 106 |
| 5th | Oral fluids | Negative for PCV2 & PPV | 0 | Feces | PPV, PCV-2 | 9 × 105 4 × 106 |
| 6th | Oral fluids | Positive for PCV2 | 108 | |||
| Status of Samples According to PCR–PPV | ||||
|---|---|---|---|---|
| Positives | Negatives | Total | ||
| Screening results | Positives | 59 (TP) | 24 (FP) | 83 |
| Negatives | 27 (FN) | 81 (TN) | 108 | |
| Total | 86 | 105 | 191 | |
| Status of Samples According to PCR–PCV-2 | ||||
|---|---|---|---|---|
| Positives | Negatives | Total | ||
| Screening results | Positives | 57 (TP) | 33 (FP) | 90 |
| Negatives | 25 (FN) | 78 (TN) | 103 | |
| Total | 82 | 111 | 193 | |
| Metrics | PPV % | PCV-2 % |
|---|---|---|
| Sensitivity | 68.6 | 69.5 |
| Specificity | 77.1 | 70.3 |
| Accuracy | 73.3 | 69.9 |
| Precision | 71.1 | 63.3 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Manessis, G.; Mourouzis, C.; Griol, A.; Zurita-Herranz, D.; Peransi, S.; Sanchez, C.; Giusti, A.; Gelasakis, A.I.; Bossis, I. Integration of Microfluidics, Photonic Integrated Circuits and Data Acquisition and Analysis Methods in a Single Platform for the Detection of Swine Viral Diseases. Animals 2021, 11, 3193. https://doi.org/10.3390/ani11113193
Manessis G, Mourouzis C, Griol A, Zurita-Herranz D, Peransi S, Sanchez C, Giusti A, Gelasakis AI, Bossis I. Integration of Microfluidics, Photonic Integrated Circuits and Data Acquisition and Analysis Methods in a Single Platform for the Detection of Swine Viral Diseases. Animals. 2021; 11(11):3193. https://doi.org/10.3390/ani11113193
Chicago/Turabian StyleManessis, Georgios, Christos Mourouzis, Amadeu Griol, David Zurita-Herranz, Sergio Peransi, Carlos Sanchez, Alessandro Giusti, Athanasios I. Gelasakis, and Ioannis Bossis. 2021. "Integration of Microfluidics, Photonic Integrated Circuits and Data Acquisition and Analysis Methods in a Single Platform for the Detection of Swine Viral Diseases" Animals 11, no. 11: 3193. https://doi.org/10.3390/ani11113193
APA StyleManessis, G., Mourouzis, C., Griol, A., Zurita-Herranz, D., Peransi, S., Sanchez, C., Giusti, A., Gelasakis, A. I., & Bossis, I. (2021). Integration of Microfluidics, Photonic Integrated Circuits and Data Acquisition and Analysis Methods in a Single Platform for the Detection of Swine Viral Diseases. Animals, 11(11), 3193. https://doi.org/10.3390/ani11113193








