Genome-Wide Expression Profiling of mRNAs, lncRNAs and circRNAs in Skeletal Muscle of Two Different Pig Breeds
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals and Tissue Samples
2.2. RNA Isolation and Library Construction
2.3. Illumina Sequencing and Quantification
2.4. Identification of Novel lncRNAs and Targets Prediction
2.5. CircRNA Prediction and Biological Analysis
2.6. Gene Ontology (GO) Term and KEGG Pathway Enrichment Analysis
3. Results
3.1. Overview of Sequencing Data
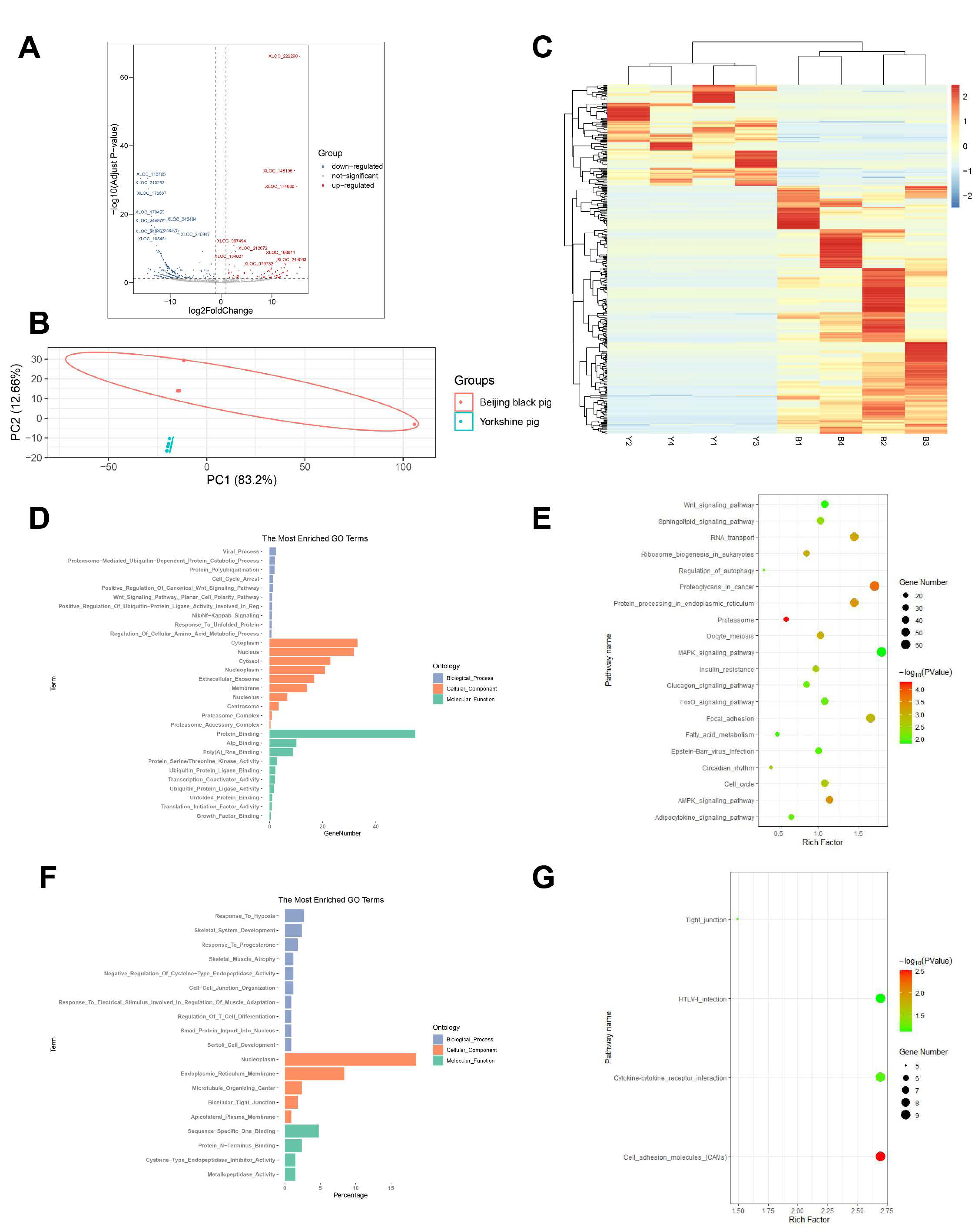
3.2. Identification of lncRNAs in Porcine Skeletal Muscle
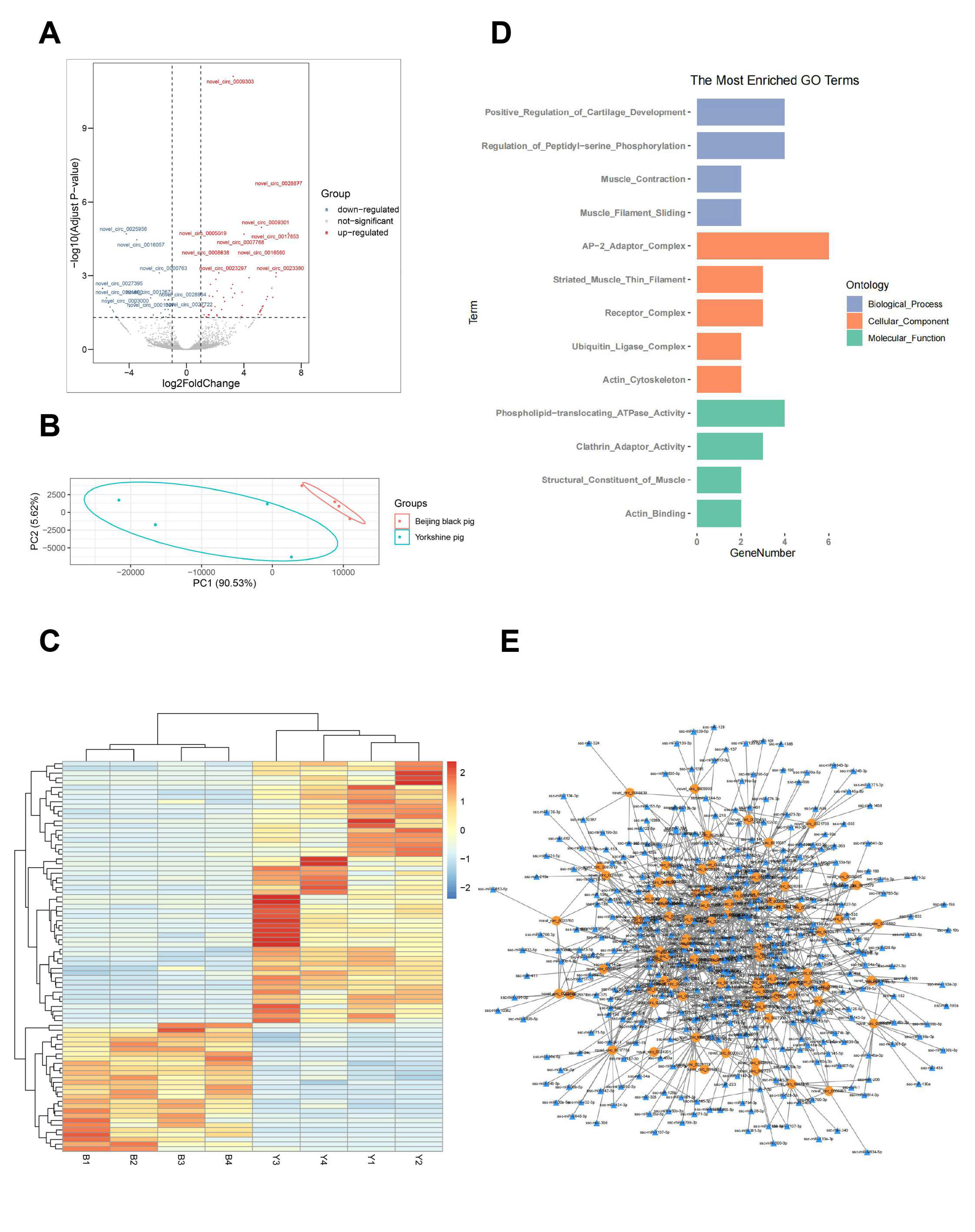
3.3. CircRNA Detection in Porcine Skeletal Muscle
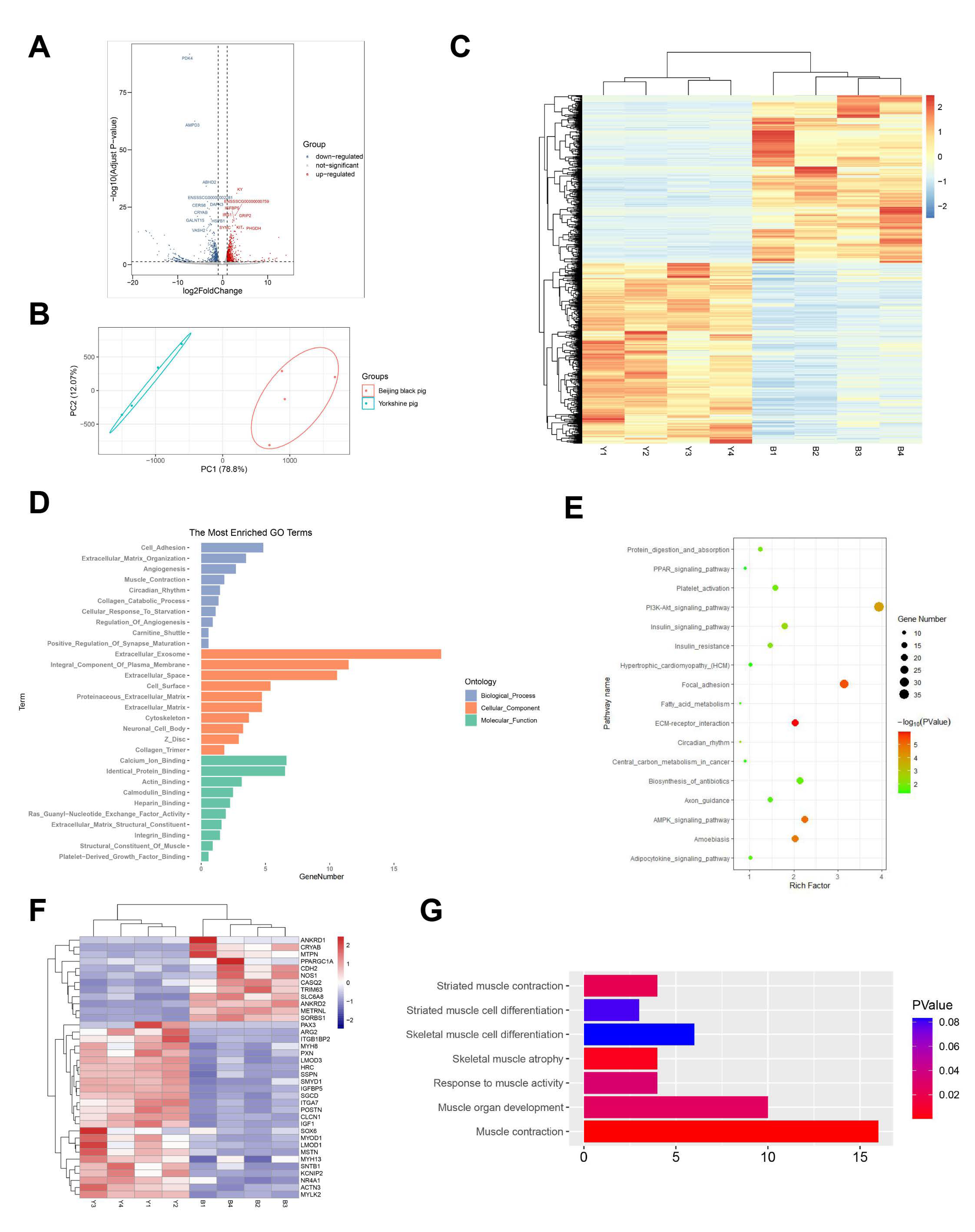
3.4. Differential Expression Analysis
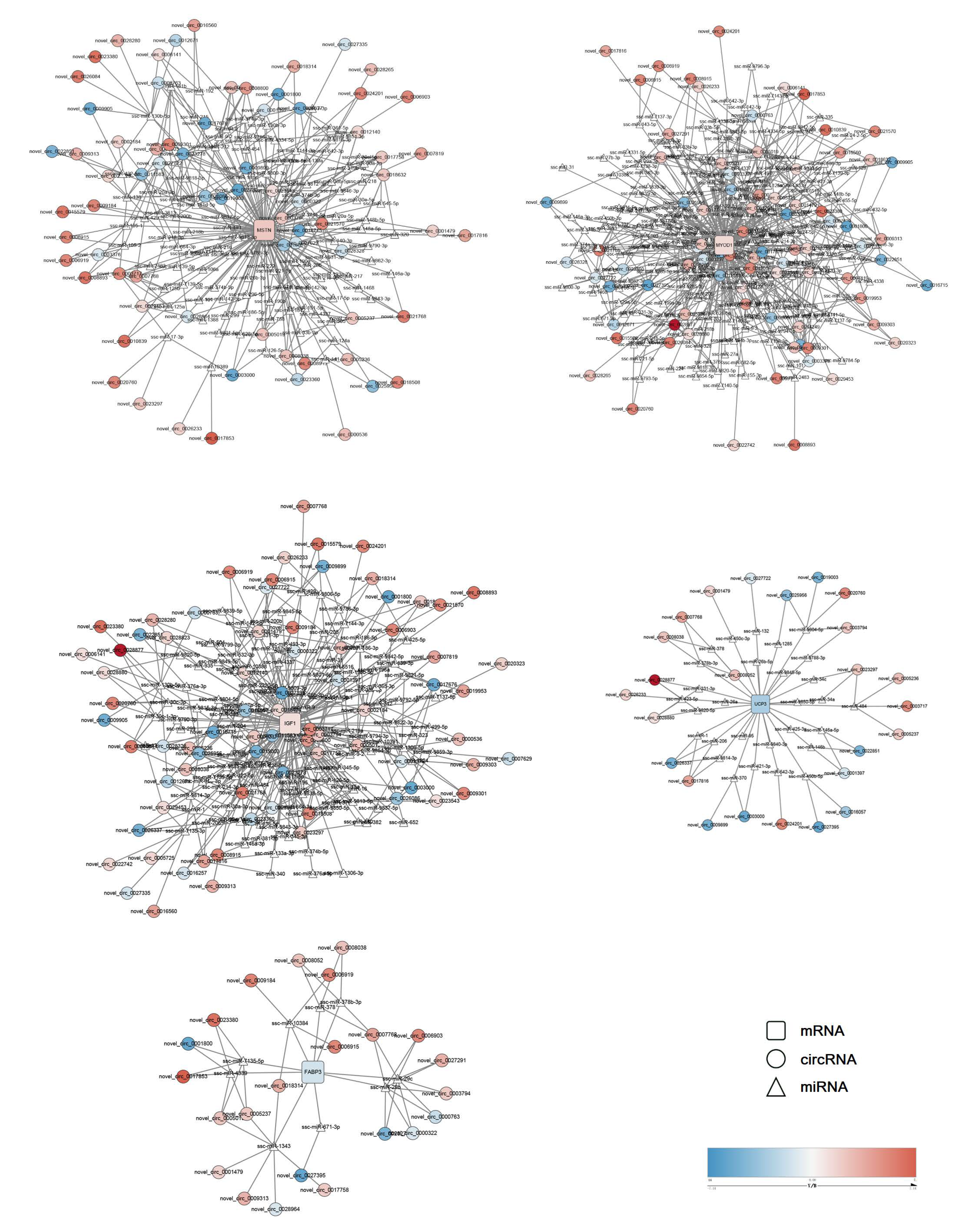
3.5. Functional Analysis of Transcriptome Data
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Lefaucheur, L.; Milan, D.; Ecolan, P.; Le Callennec, C. Myosin heavy chain composition of different skeletal muscles in Large White and Meishan pigs. J. Anim. Sci. 2004, 82, 1931–1941. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.R.; Yu, B.; Mao, X.B.; Zheng, P.; He, J.; Yu, J.; He, Y.; Reecy, J.M.; Chen, D.W. Lean and obese pig breeds exhibit differences in prenatal gene expression profiles of muscle development. Anim. Int. J. Anim. Biosci. 2015, 9, 28–34. [Google Scholar] [CrossRef] [PubMed]
- Gao, P.F.; Guo, X.H.; Du, M.; Cao, G.Q.; Yang, Q.C.; Pu, Z.D.; Wang, Z.Y.; Zhang, Q.; Li, M.; Jin, Y.S.; et al. LncRNA profiling of skeletal muscles in Large White pigs and Mashen pigs during development. J. Anim. Sci. 2017, 95, 4239–4250. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Mo, D.; Li, A.; Gong, W.; Xiao, S.; Zhang, Y.; Qin, L.; Niu, Y.; Guo, Y.; Liu, X.; et al. Comparative analyses by sequencing of transcriptomes during skeletal muscle development between pig breeds differing in muscle growth rate and fatness. PLoS ONE 2011, 6, e19774. [Google Scholar] [CrossRef]
- Wu, T.; Zhang, Z.; Yuan, Z.; Lo, L.J.; Chen, J.; Wang, Y.; Peng, J. Distinctive genes determine different intramuscular fat and muscle fiber ratios of the longissimus dorsi muscles in Jinhua and landrace pigs. PLoS ONE 2013, 8, e53181. [Google Scholar] [CrossRef]
- Yu, K.; Shu, G.; Yuan, F.; Zhu, X.; Gao, P.; Wang, S.; Wang, L.; Xi, Q.; Zhang, S.; Zhang, Y.; et al. Fatty acid and transcriptome profiling of longissimus dorsi muscles between pig breeds differing in meat quality. Int. J. Biol. Sci. 2013, 9, 108–118. [Google Scholar] [CrossRef]
- Kim, K.; Kim, D.; Min, Y.; Jeong, D.; Son, Y.O.; Do, K. Myogenic regulatory factors are key players in determining muscle mass and meat quality in Jeju native and Berkshire pigs. Vet. Med. Sci. 2021, 7, 735–745. [Google Scholar] [CrossRef]
- Xu, J.; Wang, C.; Jin, E.; Gu, Y.; Li, S.; Li, Q. Identification of differentially expressed genes in longissimus dorsi muscle between Wei and Yorkshire pigs using RNA sequencing. Genes Genom. 2018, 40, 413–421. [Google Scholar] [CrossRef]
- Xu, Y.J.; Jin, M.L.; Wang, L.J.; Zhang, A.D.; Zuo, B.; Xu, D.Q.; Ren, Z.Q.; Lei, M.G.; Mo, X.Y.; Li, F.E.; et al. Differential proteome analysis of porcine skeletal muscles between Meishan and Large White. J. Anim. Sci. 2009, 87, 2519–2527. [Google Scholar] [CrossRef]
- Wang, J.; Ren, Q.; Hua, L.; Chen, J.; Zhang, J.; Bai, H.; Li, H.; Xu, B.; Shi, Z.; Cao, H.; et al. Comprehensive Analysis of Differentially Expressed mRNA, lncRNA and circRNA and Their ceRNA Networks in the Longissimus Dorsi Muscle of Two Different Pig Breeds. Int. J. Mol. Sci. 2019, 20, 1107. [Google Scholar] [CrossRef]
- Li, X.J.; Zhou, J.; Liu, L.Q.; Qian, K.; Wang, C.L. Identification of genes in longissimus dorsi muscle differentially expressed between Wannanhua and Yorkshire pigs using RNA-sequencing. Anim. Genet. 2016, 47, 324–333. [Google Scholar] [CrossRef]
- Gao, Y.; Wang, J.; Zhao, F. CIRI: An efficient and unbiased algorithm for de novo circular RNA identification. Genome Biol. 2015, 16, 4. [Google Scholar] [CrossRef]
- Memczak, S.; Jens, M.; Elefsinioti, A.; Torti, F.; Krueger, J.; Rybak, A.; Maier, L.; Mackowiak, S.D.; Gregersen, L.H.; Munschauer, M.; et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013, 495, 333–338. [Google Scholar] [CrossRef]
- Xia, J.; Xin, L.; Zhu, W.; Li, L.; Li, C.; Wang, Y.; Mu, Y.; Yang, S.; Li, K. Characterization of long non-coding RNA transcriptome in high-energy diet induced nonalcoholic steatohepatitis minipigs. Sci. Rep. 2016, 6, 30709. [Google Scholar] [CrossRef]
- Ruszkowska, M.; Nynca, A.; Paukszto, L.; Sadowska, A.; Swigonska, S.; Orlowska, K.; Molcan, T.; Jastrzebski, J.P.; Ciereszko, R.E. Identification and characterization of long non-coding RNAs in porcine granulosa cells exposed to 2,3,7,8-tetrachlorodibenzo-p-dioxin. J. Anim. Sci. Biotechnol. 2018, 9, 72. [Google Scholar] [CrossRef]
- Hansen, T.B.; Jensen, T.I.; Clausen, B.H.; Bramsen, J.B.; Finsen, B.; Damgaard, C.K.; Kjems, J. Natural RNA circles function as efficient microRNA sponges. Nature 2013, 495, 384–388. [Google Scholar] [CrossRef]
- Thomas, M.; Langley, B.; Berry, C.; Sharma, M.; Kirk, S.; Bass, J.; Kambadur, R. Myostatin, a negative regulator of muscle growth, functions by inhibiting myoblast proliferation. J. Biol. Chem. 2000, 275, 40235–40243. [Google Scholar] [CrossRef]
- Florini, J.R.; Ewton, D.Z.; Coolican, S.A. Growth hormone and the insulin-like growth factor system in myogenesis. Endocr. Rev. 1996, 17, 481–517. [Google Scholar] [CrossRef]
- Hasty, P.; Bradley, A.; Morris, J.H.; Edmondson, D.G.; Venuti, J.M.; Olson, E.N.; Klein, W.H. Muscle deficiency and neonatal death in mice with a targeted mutation in the myogenin gene. Nature 1993, 364, 501–506. [Google Scholar] [CrossRef]
- Glatz, J.F.; Schaap, F.G.; Binas, B.; Bonen, A.; van der Vusse, G.J.; Luiken, J.J. Cytoplasmic fatty acid-binding protein facilitates fatty acid utilization by skeletal muscle. Acta Physiol. Scand. 2003, 178, 367–371. [Google Scholar] [CrossRef]
- Wang, S.; Subramaniam, A.; Cawthorne, M.A.; Clapham, J.C. Increased fatty acid oxidation in transgenic mice overexpressing UCP3 in skeletal muscle. Diabetes Obes. Metab. 2003, 5, 295–301. [Google Scholar] [CrossRef]
- Spiegler, V.; Hensel, A.; Seggewiß, J.; Lubisch, M.; Liebau, E. Transcriptome analysis reveals molecular anthelmintic effects of procyanidins in C. elegans. PLoS ONE 2017, 12, e0184656. [Google Scholar] [CrossRef]
- Talbott, H.; Hou, X.; Qiu, F.; Zhang, P.; Guda, C.; Yu, F.; Cushman, R.A.; Wood, J.R.; Wang, C.; Cupp, A.S.; et al. Transcriptomic and bioinformatics analysis of the early time-course of the response to prostaglandin F2 alpha in the bovine corpus luteum. Data Brief 2017, 14, 695–706. [Google Scholar] [CrossRef]
- Wang, J.; Koganti, P.P.; Yao, J.; Wei, S.; Cleveland, B. Comprehensive analysis of lncRNAs and mRNAs in skeletal muscle of rainbow trout (Oncorhynchus mykiss) exposed to estradiol. Sci. Rep. 2017, 7, 11780. [Google Scholar] [CrossRef]
- Picard, B.; Lefaucheur, L.; Berri, C.; Duclos, M.J. Muscle fibre ontogenesis in farm animal species. Reprod. Nutr. Dev. 2002, 42, 415–431. [Google Scholar] [CrossRef]
- Rehfeldt, C.; Fiedler, I.; Dietl, G.; Ender, K. Myogenesis and postnatal skeletal muscle cell growth as influenced by selection. Livest. Prod. Sci. 2000, 66, 177–188. [Google Scholar] [CrossRef]
- Picard, B.; Berri, C.; Lefaucheur, L.; Molette, C.; Sayd, T.; Terlouw, C. Skeletal muscle proteomics in livestock production. Brief. Funct. Genom. 2010, 9, 259–278. [Google Scholar] [CrossRef]
- Ashmore, C.R.; Addis, P.B.; Doerr, L. Development of muscle fibers in the fetal pig. J. Anim. Sci. 1973, 36, 1088–1093. [Google Scholar] [CrossRef]
- Liu, H.; Xi, Y.; Liu, G.; Zhao, Y.; Li, J.; Lei, M. Comparative transcriptomic analysis of skeletal muscle tissue during prenatal stages in Tongcheng and Yorkshire pig using RNA-seq. Funct. Integr. Genom. 2018, 18, 195–209. [Google Scholar] [CrossRef]
- Cagnazzo, M.; te Pas, M.F.; Priem, J.; de Wit, A.A.; Pool, M.H.; Davoli, R.; Russo, V. Comparison of prenatal muscle tissue expression profiles of two pig breeds differing in muscle characteristics. J. Anim. Sci. 2006, 84, 1–10. [Google Scholar] [CrossRef]
- Lefaucheur, L.; Vigneron, P. Post-natal changes in some histochemical and enzymatic characteristics of three pig muscles. Meat Sci. 1986, 16, 199–216. [Google Scholar] [CrossRef]
- Rehfeldt, C.; Fiedler, I.; Weikard, R.; Kanitz, E.; Ender, K. It is possible to increase skeletal muscle fibre number in utero. Biosci. Rep. 1993, 13, 213–220. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Liu, Z.Y.; Liu, Q.X. Gene coexpression networks reveal key drivers of phenotypic divergence in porcine muscle. BMC Genom. 2015, 16, 50. [Google Scholar] [CrossRef] [PubMed]
- Cai, C.; Li, M.; Zhang, Y.; Meng, S.; Yang, Y.; Gao, P.; Guo, X.; Cao, G.; Li, B. Comparative Transcriptome Analyses of Longissimus thoracis Between Pig Breeds Differing in Muscle Characteristics. Front. Genet. 2020, 11, 526309. [Google Scholar] [CrossRef]
- Xu, X.; Mishra, B.; Qin, N.; Sun, X.; Zhang, S.; Yang, J.; Xu, R. Differential Transcriptome Analysis of Early Postnatal Developing Longissimus Dorsi Muscle from Two Pig Breeds Characterized in Divergent Myofiber Traits and Fatness. Anim. Biotechnol. 2019, 30, 63–74. [Google Scholar] [CrossRef]
- McPherron, A.C.; Lee, S.J. Double muscling in cattle due to mutations in the myostatin gene. Proc. Natl. Acad. Sci. USA 1997, 94, 12457–12461. [Google Scholar] [CrossRef]
- McPherron, A.C.; Lee, S.J. Suppression of body fat accumulation in myostatin-deficient mice. J. Clin. Investig. 2002, 109, 595–601. [Google Scholar] [CrossRef]
- McPherron, A.C.; Lawler, A.M.; Lee, S.J. Regulation of skeletal muscle mass in mice by a new TGF-beta superfamily member. Nature 1997, 387, 83–90. [Google Scholar] [CrossRef]
- Khandwala, H.M.; McCutcheon, I.E.; Flyvbjerg, A.; Friend, K.E. The effects of insulin-like growth factors on tumorigenesis and neoplastic growth. Endocr. Rev. 2000, 21, 215–244. [Google Scholar] [CrossRef]
- Sparkman, A.M.; Schwartz, T.S.; Madden, J.A.; Boyken, S.E.; Ford, N.B.; Serb, J.M.; Bronikowski, A.M. Rates of molecular evolution vary in vertebrates for insulin-like growth factor-1 (IGF-1), a pleiotropic locus that regulates life history traits. Gen. Comp. Endocrinol. 2012, 178, 164–173. [Google Scholar] [CrossRef]
- Powell-Braxton, L.; Hollingshead, P.; Warburton, C.; Dowd, M.; Pitts-Meek, S.; Dalton, D.; Gillett, N.; Stewart, T.A. IGF-I is required for normal embryonic growth in mice. Genes Dev. 1993, 7, 2609–2617. [Google Scholar] [CrossRef]
- Wang, L.; Lei, M.; Xiong, Y. Molecular characterization and different expression patterns of the muscle ankyrin repeat protein (MARP) family during porcine skeletal muscle development in vitro and in vivo. Anim. Biotechnol. 2011, 22, 87–99. [Google Scholar] [CrossRef]
- Bean, C.; Facchinello, N.; Faulkner, G.; Lanfranchi, G. The effects of Ankrd2 alteration indicate its involvement in cell cycle regulation during muscle differentiation. Biochim. Biophys. Acta 2008, 1783, 1023–1035. [Google Scholar] [CrossRef]
- Cenni, V.; Bavelloni, A.; Beretti, F.; Tagliavini, F.; Manzoli, L.; Lattanzi, G.; Maraldi, N.M.; Cocco, L.; Marmiroli, S. Ankrd2/ARPP is a novel Akt2 specific substrate and regulates myogenic differentiation upon cellular exposure to H2O2. Mol. Biol. Cell 2011, 22, 2946–2956. [Google Scholar] [CrossRef]
- Singh, B.N.; Rao, K.S.; Rao Ch, M. Ubiquitin-proteasome-mediated degradation and synthesis of MyoD is modulated by alphaB-crystallin, a small heat shock protein, during muscle differentiation. Biochim. Biophys. Acta 2010, 1803, 288–299. [Google Scholar] [CrossRef]
- Conley, C.A.; Fritz-Six, K.L.; Almenar-Queralt, A.; Fowler, V.M. Leiomodins: Larger members of the tropomodulin (Tmod) gene family. Genomics 2001, 73, 127–139. [Google Scholar] [CrossRef]
- Yuen, M.; Sandaradura, S.A.; Dowling, J.J.; Kostyukova, A.S.; Moroz, N.; Quinlan, K.G.; Lehtokari, V.L.; Ravenscroft, G.; Todd, E.J.; Ceyhan-Birsoy, O.; et al. Leiomodin-3 dysfunction results in thin filament disorganization and nemaline myopathy. J. Clin. Investig. 2014, 124, 4693–4708. [Google Scholar] [CrossRef]
- Lin, F.H.; Wang, A.; Dai, W.; Chen, S.; Ding, Y.; Sun, L.V. Lmod3 promotes myoblast differentiation and proliferation via the AKT and ERK pathways. Exp. Cell Res. 2020, 396, 112297. [Google Scholar] [CrossRef]
- Bodine, S.C.; Latres, E.; Baumhueter, S.; Lai, V.K.; Nunez, L.; Clarke, B.A.; Poueymirou, W.T.; Panaro, F.J.; Na, E.; Dharmarajan, K.; et al. Identification of ubiquitin ligases required for skeletal muscle atrophy. Science 2001, 294, 1704–1708. [Google Scholar] [CrossRef]
- Damon, M.; Wyszynska-Koko, J.; Vincent, A.; Hérault, F.; Lebret, B. Comparison of muscle transcriptome between pigs with divergent meat quality phenotypes identifies genes related to muscle metabolism and structure. PLoS ONE 2012, 7, e33763. [Google Scholar] [CrossRef]
- Sentandreu, M.A.; Coulis, G.; Ouali, A. Role of muscle endopeptidases and their inhibitors in meat tenderness. Trends Food Sci. Technol. 2002, 13, 400–421. [Google Scholar] [CrossRef]
- Thorsteinsdóttir, S.; Deries, M.; Cachaço, A.S.; Bajanca, F. The extracellular matrix dimension of skeletal muscle development. Dev. Biol. 2011, 354, 191–207. [Google Scholar] [CrossRef]
- Wheeler, T.L.; Shackelford, S.D.; Koohmaraie, M. Variation in proteolysis, sarcomere length, collagen content, and tenderness among major pork muscles. J. Anim. Sci. 2000, 78, 958–965. [Google Scholar] [CrossRef]
- Zhao, S.M.; Ren, L.J.; Chen, L.; Zhang, X.; Cheng, M.L.; Li, W.Z.; Zhang, Y.Y.; Gao, S.Z. Differential expression of lipid metabolism related genes in porcine muscle tissue leading to different intramuscular fat deposition. Lipids 2009, 44, 1029–1037. [Google Scholar] [CrossRef]
- Furuhashi, M.; Hotamisligil, G.S. Fatty acid-binding proteins: Role in metabolic diseases and potential as drug targets. Nat. Rev. Drug Discov. 2008, 7, 489–503. [Google Scholar] [CrossRef]
- Yi, B.; Wang, J.; Wang, S.; Yuan, D.; Sun, J.; Li, Z.; Mao, Y.; Hou, Q.; Liu, W. Overexpression of Banna mini-pig inbred line fatty acid binding protein 3 promotes adipogenesis in 3T3-L1 preadipocytes. Cell Biol. Int. 2014, 38, 918–923. [Google Scholar] [CrossRef]
- Li, X.; Kim, S.W.; Choi, J.S.; Lee, Y.M.; Lee, C.K.; Choi, B.H.; Kim, T.H.; Choi, Y.I.; Kim, J.J.; Kim, K.S. Investigation of porcine FABP3 and LEPR gene polymorphisms and mRNA expression for variation in intramuscular fat content. Mol. Biol. Rep. 2010, 37, 3931–3939. [Google Scholar] [CrossRef]
- Cho, K.H.; Kim, M.J.; Jeon, G.J.; Chung, H.Y. Association of genetic variants for FABP3 gene with back fat thickness and intramuscular fat content in pig. Mol. Biol. Rep. 2011, 38, 2161–2166. [Google Scholar] [CrossRef]
- Arnyasi, M.; Grindflek, E.; Jávor, A.; Lien, S. Investigation of two candidate genes for meat quality traits in a quantitative trait locus region on SSC6: The porcine short heterodimer partner and heart fatty acid binding protein genes. J. Anim. Breed. Genet. 2006, 123, 198–203. [Google Scholar] [CrossRef]
- Majer, M.; Popov, K.M.; Harris, R.A.; Bogardus, C.; Prochazka, M. Insulin downregulates pyruvate dehydrogenase kinase (PDK) mRNA: Potential mechanism contributing to increased lipid oxidation in insulin-resistant subjects. Mol. Genet. Metab. 1998, 65, 181–186. [Google Scholar] [CrossRef]
- Boulatnikov, I.; Popov, K.M. Formation of functional heterodimers by isozymes 1 and 2 of pyruvate dehydrogenase kinase. Biochim. Biophys. Acta 2003, 1645, 183–192. [Google Scholar] [CrossRef]
- Lan, J.; Lei, M.G.; Zhang, Y.B.; Wang, J.H.; Feng, X.T.; Xu, D.Q.; Gui, J.F.; Xiong, Y.Z. Characterization of the porcine differentially expressed PDK4 gene and association with meat quality. Mol. Biol. Rep. 2009, 36, 2003–2010. [Google Scholar] [CrossRef] [PubMed]
- Zhe, C.; Zhao, X.; Zhu, H.; Guo, X.; Xu, N. Association of porcine UCP3 gene polymorphisms with fatness traits in a Pietrain×Jinhua F2 population. Afr. J. Biotechnol. 2011, 10, 3296–3300. [Google Scholar] [CrossRef][Green Version]
- Chen, W.; Fang, G.-f.; Wang, S.-d.; Wang, H.; Zeng, Y.-q. Longissimus lumborum muscle transcriptome analysis of Laiwu and Yorkshire pigs differing in intramuscular fat content. Genes Genom. 2017, 39, 759–766. [Google Scholar] [CrossRef]
- Díaz-Rúa, R.; Palou, A.; Oliver, P. Cpt1a gene expression in peripheral blood mononuclear cells as an early biomarker of diet-related metabolic alterations. Food Nutr. Res. 2016, 60, 33554. [Google Scholar] [CrossRef]
- Perdomo, G.; Commerford, S.R.; Richard, A.M.; Adams, S.H.; Corkey, B.E.; O’Doherty, R.M.; Brown, N.F. Increased beta-oxidation in muscle cells enhances insulin-stimulated glucose metabolism and protects against fatty acid-induced insulin resistance despite intramyocellular lipid accumulation. J. Biol. Chem. 2004, 279, 27177–27186. [Google Scholar] [CrossRef]
- Miao, Z.; Wei, P.; Khan, M.A.; Zhang, J.; Guo, L.; Liu, D.; Zhang, X.; Bai, Y.; Wang, S. Transcriptome analysis reveals differential gene expression in intramuscular adipose tissues of Jinhua and Landrace pigs. J. Vet. Med Sci. 2018, 80, 953–959. [Google Scholar] [CrossRef]
- Luo, M.; Wang, L.; Yin, H.; Zhu, W.; Fu, J.; Dong, Z. Integrated analysis of long non-coding RNA and mRNA expression in different colored skin of koi carp. BMC Genom. 2019, 20, 515. [Google Scholar] [CrossRef]
- Guttman, M.; Donaghey, J.; Carey, B.W.; Garber, M.; Grenier, J.K.; Munson, G.; Young, G.; Lucas, A.B.; Ach, R.; Bruhn, L.; et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature 2011, 477, 295–300. [Google Scholar] [CrossRef]
- Yu, A.D.; Wang, Z.; Morris, K.V. Long noncoding RNAs: A potent source of regulation in immunity and disease. Immunol. Cell Biol. 2015, 93, 277–283. [Google Scholar] [CrossRef]
- Kornfeld, J.W.; Brüning, J.C. Regulation of metabolism by long, non-coding RNAs. Front. Genet. 2014, 5, 57. [Google Scholar] [CrossRef]
- Arber, S.; Halder, G.; Caroni, P. Muscle LIM protein, a novel essential regulator of myogenesis, promotes myogenic differentiation. Cell 1994, 79, 221–231. [Google Scholar] [CrossRef]
- Barash, I.A.; Mathew, L.; Lahey, M.; Greaser, M.L.; Lieber, R.L. Muscle LIM protein plays both structural and functional roles in skeletal muscle. Am. J. Physiol. Cell Physiol. 2005, 289, C1312–C1320. [Google Scholar] [CrossRef]
- Reis, E.P.; Paixão, D.M.; Brustolini, O.J.; Silva, F.F.; Silva, W.; Araújo, F.M.; Salim, A.C.; Oliveira, G.; Guimarães, S.E. Expression of myogenes in longissimus dorsi muscle during prenatal development in commercial and local Piau pigs. Genet. Mol. Biol. 2016, 39, 589–599. [Google Scholar] [CrossRef]
- Zhu, Z.; Miller, J.B. MRF4 can substitute for myogenin during early stages of myogenesis. Dev. Dyn. Off. Publ. Am. Assoc. Anat. 1997, 209, 233–241. [Google Scholar] [CrossRef]
- Kassar-Duchossoy, L.; Gayraud-Morel, B.; Gomès, D.; Rocancourt, D.; Buckingham, M.; Shinin, V.; Tajbakhsh, S. Mrf4 determines skeletal muscle identity in Myf5:Myod double-mutant mice. Nature 2004, 431, 466–471. [Google Scholar] [CrossRef]
- Green, B.N.; Jones, S.B.; Streck, R.D.; Wood, T.L.; Rotwein, P.; Pintar, J.E. Distinct expression patterns of insulin-like growth factor binding proteins 2 and 5 during fetal and postnatal development. Endocrinology 1994, 134, 954–962. [Google Scholar] [CrossRef]
- Pampusch, M.S.; Xi, G.; Kamanga-Sollo, E.; Loseth, K.J.; Hathaway, M.R.; Dayton, W.R.; White, M.E. Production of recombinant porcine IGF-binding protein-5 and its effect on proliferation of porcine embryonic myoblast cultures in the presence and absence of IGF-I and Long-R3-IGF-I. J. Endocrinol. 2005, 185, 197–206. [Google Scholar] [CrossRef]
- Li, X.; McFarland, D.C.; Velleman, S.G. Extracellular matrix proteoglycan decorin-mediated myogenic satellite cell responsiveness to transforming growth factor-beta1 during cell proliferation and differentiation Decorin and transforming growth factor-beta1 in satellite cells. Domest. Anim. Endocrinol. 2008, 35, 263–273. [Google Scholar] [CrossRef]
- Wu, Z.; Woodring, P.J.; Bhakta, K.S.; Tamura, K.; Wen, F.; Feramisco, J.R.; Karin, M.; Wang, J.Y.; Puri, P.L. p38 and extracellular signal-regulated kinases regulate the myogenic program at multiple steps. Mol. Cell. Biol. 2000, 20, 3951–3964. [Google Scholar] [CrossRef]
- Ulloa, F.; Itasaki, N.; Briscoe, J. Inhibitory Gli3 activity negatively regulates Wnt/beta-catenin signaling. Curr. Biol. CB 2007, 17, 545–550. [Google Scholar] [CrossRef]
- Yokoyama, T.; Takano, K.; Yoshida, A.; Katada, F.; Sun, P.; Takenawa, T.; Andoh, T.; Endo, T. DA-Raf1, a competent intrinsic dominant-negative antagonist of the Ras-ERK pathway, is required for myogenic differentiation. J. Cell Biol. 2007, 177, 781–793. [Google Scholar] [CrossRef]
- Hirata, A.; Masuda, S.; Tamura, T.; Kai, K.; Ojima, K.; Fukase, A.; Motoyoshi, K.; Kamakura, K.; Miyagoe-Suzuki, Y.; Takeda, S. Expression profiling of cytokines and related genes in regenerating skeletal muscle after cardiotoxin injection: A role for osteopontin. Am. J. Pathol. 2003, 163, 203–215. [Google Scholar] [CrossRef]
- Evans, R.M.; Barish, G.D.; Wang, Y.X. PPARs and the complex journey to obesity. Nat. Med. 2004, 10, 355–361. [Google Scholar] [CrossRef]
- Kim, D.J.; Bility, M.T.; Billin, A.N.; Willson, T.M.; Gonzalez, F.J.; Peters, J.M. PPARbeta/delta selectively induces differentiation and inhibits cell proliferation. Cell Death Differ. 2006, 13, 53–60. [Google Scholar] [CrossRef]
- Chen, L.; Yang, G. PPARs Integrate the Mammalian Clock and Energy Metabolism. PPAR Res. 2014, 2014, 653017. [Google Scholar] [CrossRef]
- Nakae, J.; Kitamura, T.; Kitamura, Y.; Biggs, W.H., 3rd; Arden, K.C.; Accili, D. The forkhead transcription factor Foxo1 regulates adipocyte differentiation. Dev. Cell 2003, 4, 119–129. [Google Scholar] [CrossRef]
- Zhan, T.; Poppelreuther, M.; Ehehalt, R.; Füllekrug, J. Overexpressed FATP1, ACSVL4/FATP4 and ACSL1 increase the cellular fatty acid uptake of 3T3-L1 adipocytes but are localized on intracellular membranes. PLoS ONE 2012, 7, e45087. [Google Scholar] [CrossRef]
- Phan, J.; Péterfy, M.; Reue, K. Lipin expression preceding peroxisome proliferator-activated receptor-gamma is critical for adipogenesis in vivo and in vitro. J. Biol. Chem. 2004, 279, 29558–29564. [Google Scholar] [CrossRef]
- Zhang, P.; Chao, Z.; Zhang, R.; Ding, R.; Wang, Y.; Wu, W.; Han, Q.; Li, C.; Xu, H.; Wang, L.; et al. Circular RNA Regulation of Myogenesis. Cells 2019, 8, 885. [Google Scholar] [CrossRef]
- Zhang, P.; Xu, H.; Li, R.; Wu, W.; Chao, Z.; Li, C.; Xia, W.; Wang, L.; Yang, J.; Xu, Y. Assessment of myoblast circular RNA dynamics and its correlation with miRNA during myogenic differentiation. Int. J. Biochem. Cell Biol. 2018, 99, 211–218. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Gui, W.; Lin, X.; Li, H. Knock-down of circular RNA H19 induces human adipose-derived stem cells adipogenic differentiation via a mechanism involving the polypyrimidine tract-binding protein 1. Exp. Cell Res. 2020, 387, 111753. [Google Scholar] [CrossRef] [PubMed]
- Yan, B.; Guo, J.-T.; Zhao, L.-H.; Zhao, J.-L. microRNA expression signature in skeletal muscle of Nile tilapia. Aquaculture 2012, 364–365, 240–246. [Google Scholar] [CrossRef]
- Yan, B.; Zhu, C.D.; Guo, J.T.; Zhao, L.H.; Zhao, J.L. miR-206 regulates the growth of the teleost tilapia (Oreochromis niloticus) through the modulation of IGF-1 gene expression. J. Exp. Biol. 2013, 216, 1265–1269. [Google Scholar] [CrossRef] [PubMed]
- Jia, L.; Li, Y.F.; Wu, G.F.; Song, Z.Y.; Lu, H.Z.; Song, C.C.; Zhang, Q.L.; Zhu, J.Y.; Yang, G.S.; Shi, X.E. MiRNA-199a-3p regulates C2C12 myoblast differentiation through IGF-1/AKT/mTOR signal pathway. Int. J. Mol. Sci. 2013, 15, 296–308. [Google Scholar] [CrossRef]
- Tan, S.B.; Li, J.; Chen, X.; Zhang, W.; Zhang, D.; Zhang, C.; Li, D.; Zhang, Y. Small molecule inhibitor of myogenic microRNAs leads to a discovery of miR-221/222-myoD-myomiRs regulatory pathway. Chem. Biol. 2014, 21, 1265–1270. [Google Scholar] [CrossRef][Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hou, X.; Wang, L.; Zhao, F.; Liu, X.; Gao, H.; Shi, L.; Yan, H.; Wang, L.; Zhang, L. Genome-Wide Expression Profiling of mRNAs, lncRNAs and circRNAs in Skeletal Muscle of Two Different Pig Breeds. Animals 2021, 11, 3169. https://doi.org/10.3390/ani11113169
Hou X, Wang L, Zhao F, Liu X, Gao H, Shi L, Yan H, Wang L, Zhang L. Genome-Wide Expression Profiling of mRNAs, lncRNAs and circRNAs in Skeletal Muscle of Two Different Pig Breeds. Animals. 2021; 11(11):3169. https://doi.org/10.3390/ani11113169
Chicago/Turabian StyleHou, Xinhua, Ligang Wang, Fuping Zhao, Xin Liu, Hongmei Gao, Lijun Shi, Hua Yan, Lixian Wang, and Longchao Zhang. 2021. "Genome-Wide Expression Profiling of mRNAs, lncRNAs and circRNAs in Skeletal Muscle of Two Different Pig Breeds" Animals 11, no. 11: 3169. https://doi.org/10.3390/ani11113169
APA StyleHou, X., Wang, L., Zhao, F., Liu, X., Gao, H., Shi, L., Yan, H., Wang, L., & Zhang, L. (2021). Genome-Wide Expression Profiling of mRNAs, lncRNAs and circRNAs in Skeletal Muscle of Two Different Pig Breeds. Animals, 11(11), 3169. https://doi.org/10.3390/ani11113169






