Composition of Fecal Microbiota in Grazing and Feedlot Angus Beef Cattle
Abstract
:Simple Summary
Abstract
1. Introduction
2. Material and Methods
2.1. Fecal Sample Collection
2.2. DNA Extraction and Sequencing
2.3. Bioinformatics Analysis
3. Results
3.1. Gene Sequencing Data Summary
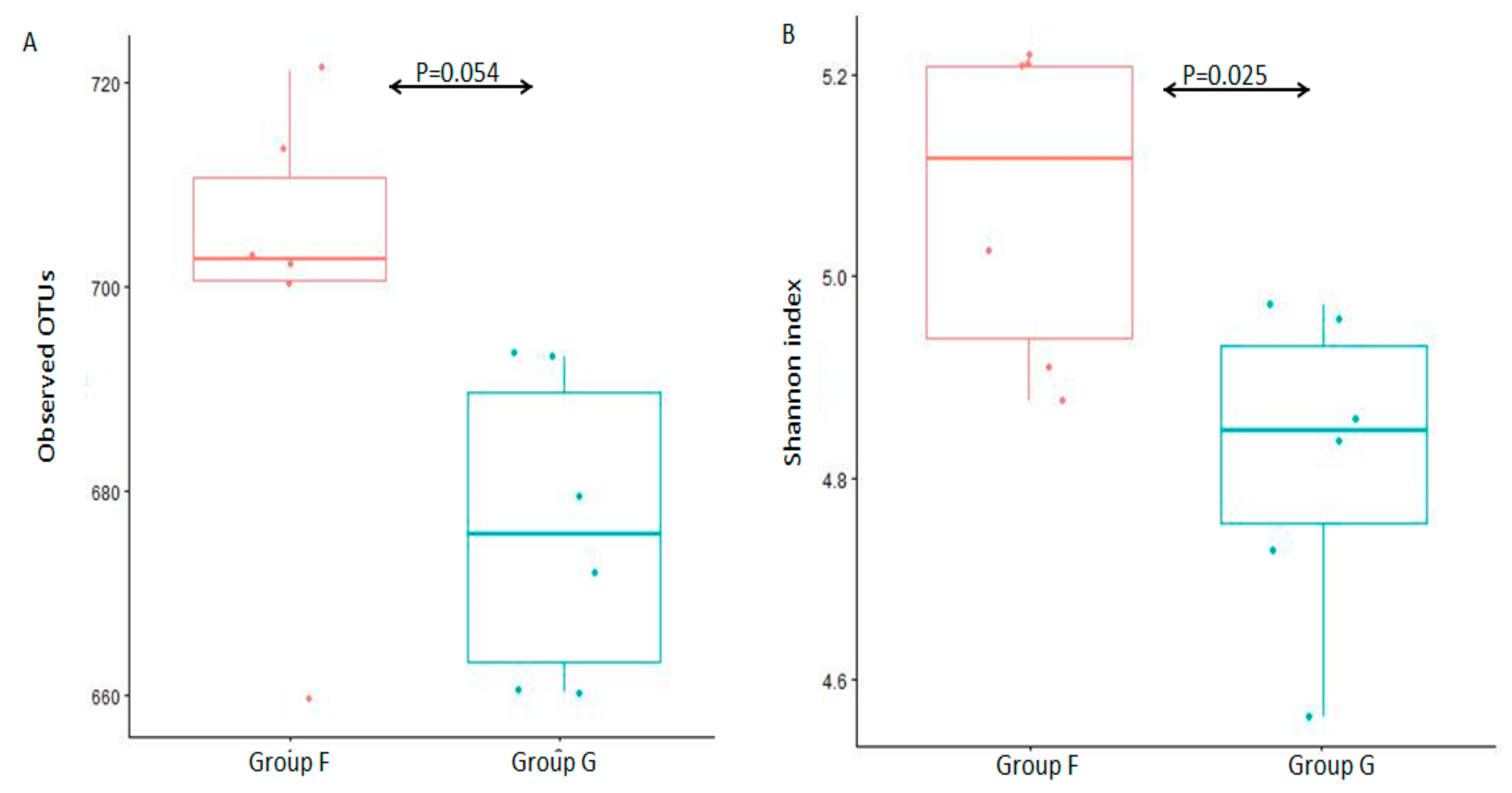
3.2. Analysis of Bacterial Diversity
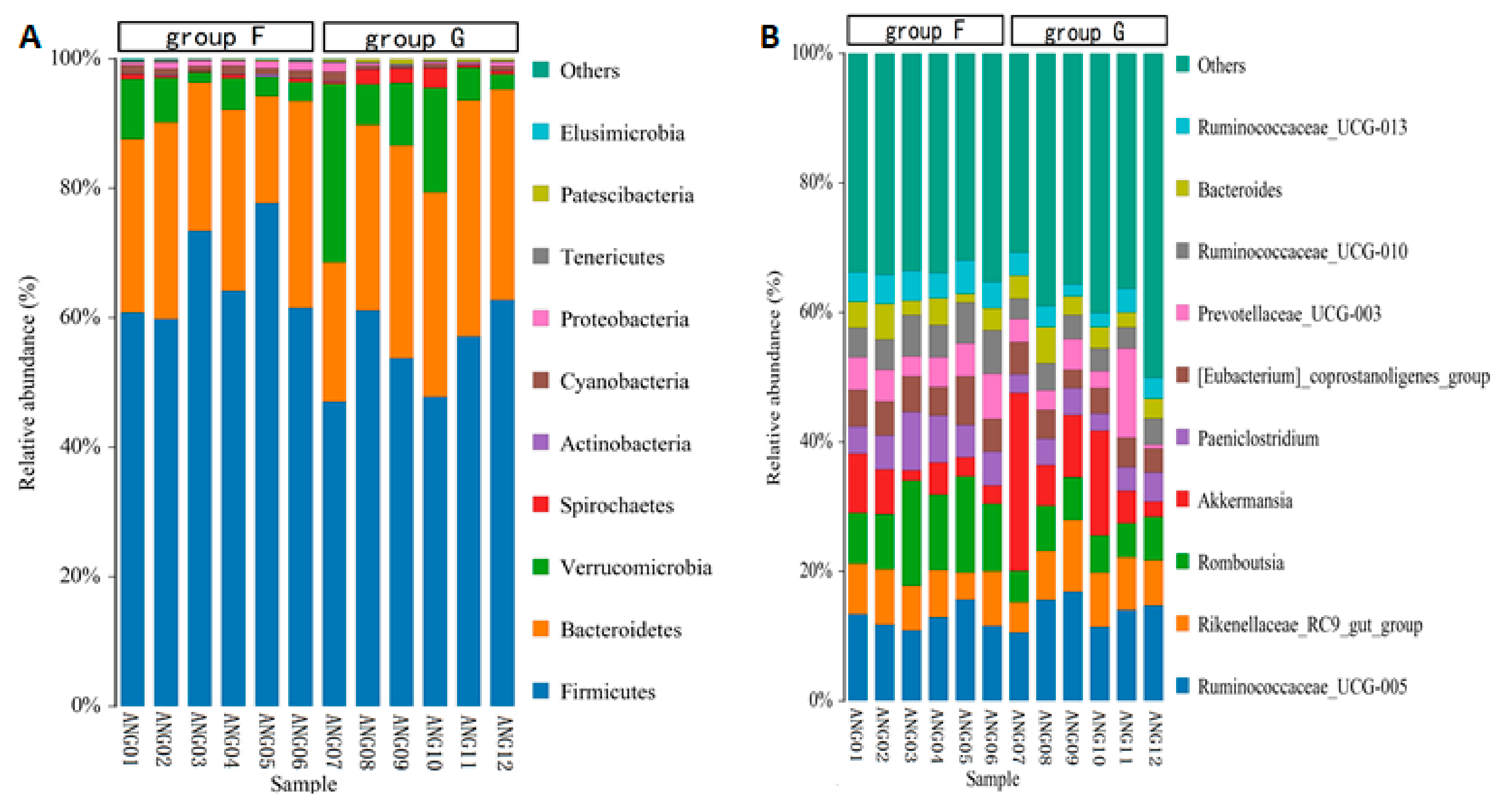
3.3. Analysis of Bacterial Taxonomy and Function
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Prache, S.; Martin, B.; Coppa, M. Review: Authentication of grass-fed meat and dairy products from cattle and sheep. Animal 2020, 14, 854–863. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Daley, C.A.; Abbott, A.; Doyle, P.S.; A Nader, G.; Larson, S. A review of fatty acid profiles and antioxidant content in grass-fed and grain-fed beef. Nutr. J. 2010, 9, 10. [Google Scholar] [CrossRef] [Green Version]
- Kim, M.; Wells, J.E. A Meta-analysis of Bacterial Diversity in the Feces of Cattle. Curr. Microbiol. 2015, 72, 145–151. [Google Scholar] [CrossRef]
- Kim, M.; Kim, J.; Kuehn, L.; Bono, J.L.; Berry, E.D.; Kalchayanand, N.; Freetly, H.C.; Benson, A.K.; Wells, J.E. Investigation of bacterial diversity in the feces of cattle fed different diets. J. Anim. Sci. 2014, 92, 683–694. [Google Scholar] [CrossRef] [Green Version]
- Rice, W.C.; Galyean, M.L.; Cox, S.B.; Dowd, S.E.; Cole, N.A. Influence of wet distillers grains diets on beef cattle fecal bacterial community structure. BMC Microbiol. 2012, 12, 25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ran, T.; Jiao, P.; AlZahal, O.; Xie, X.; A Beauchemin, K.; Niu, D.; Yang, W. Fecal bacterial community of finishing beef steers fed ruminally protected and non-protected active dried yeast. J. Anim. Sci. 2020, 98, skaa058. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.-L.; Xu, T.-W.; Wang, X.-G.; Geng, Y.-Y.; Liu, H.-J.; Hu, L.-Y.; Zhao, N.; Kang, S.-P.; Zhang, W.-M.; Xu, S.-X. The Effect of Transitioning between Feeding Methods on the Gut Microbiota Dynamics of Yaks on the Qinghai–Tibet Plateau. Animals 2020, 10, 1641. [Google Scholar] [CrossRef]
- Albonico, F.; Barelli, C.; Albanese, D.; Manica, M.; Partel, E.; Rosso, F.; Ripellino, S.; Pindo, M.; Donati, C.; Zecconi, A.; et al. Raw milk and fecal microbiota of commercial Alpine dairy cows varies with herd, fat content and diet. PLoS ONE 2020, 15, e0237262. [Google Scholar] [CrossRef]
- Magoč, T.; Salzberg, S. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [Green Version]
- Edgar, R.C.; Haas, B.J.; Clemente, J.C.; Quince, C.; Knight, R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 2011, 27, 2194–2200. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; I Gordon, J.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huson, D.H.; Mitra, S.; Ruscheweyh, H.-J.; Weber, N.; Schuster, S.C. Integrative analysis of environmental sequences using MEGAN4. Genome Res. 2011, 21, 1552–1560. [Google Scholar] [CrossRef] [Green Version]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing mothur: Open-Source, Platform-Independent, Community-Supported Software for Describing and Comparing Microbial Communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Langille, M.G.I.; Zaneveld, J.; Caporaso, J.G.; McDonald, D.; Knights, D.; Reyes, J.A.; Clemente, J.C.; Burkepile, D.E.; Thurber, R.L.V.; Knight, R.; et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 2013, 31, 814–821. [Google Scholar] [CrossRef]
- Mwangi, F.W.; Charmley, E.; Gardiner, C.P.; Malau-Aduli, B.S.; Kinobe, R.T.; Malau-Aduli, A.E.O. Diet and Genetics Influence Beef Cattle Performance and Meat Quality Characteristics. Foods 2019, 8, 648. [Google Scholar] [CrossRef] [Green Version]
- Vestergaard, M.; Oksbjerg, N.; Henckel, P. Influence of feeding intensity, grazing and finishing feeding on muscle fibre characteristics and meat colour of semitendinosus, longissimus dorsi and supraspinatus muscles of young bulls. Meat Sci. 1999, 54, 177–185. [Google Scholar] [CrossRef]
- Cassar-Malek, I.; Jurie, C.; Bernard, C.; Barnola, I.; Micol, D.; Hocquette, J.-F. Pasture-feeding of Charolais steers influences skeletal muscle metabolism and gene expression. J. Physiol. Pharmacol. Off. J. Pol. Physiol. Soc. 2009, 60, 83–90. [Google Scholar]
- Sun, J.; Wang, Y.; Li, N.; Zhong, H.; Xu, H.; Zhu, Q.; Liu, Y. Comparative Analysis of the Gut Microbial Composition and Meat Flavor of Two Chicken Breeds in Different Rearing Patterns. BioMed. Res. Int. 2018, 2018, 4343196. [Google Scholar] [CrossRef] [Green Version]
- Durso, L.M.; Harhay, G.; Smith, T.P.L.; Bono, J.L.; DeSantis, T.Z.; Harhay, D.M.; Andersen, G.; Keen, J.E.; Laegreid, W.W.; Clawson, M.L. Animal-to-Animal Variation in Fecal Microbial Diversity among Beef Cattle. Appl. Environ. Microbiol. 2010, 76, 4858–4862. [Google Scholar] [CrossRef] [Green Version]
- Tanes, C.; Bittinger, K.; Gao, Y.; Friedman, E.S.; Nessel, L.; Paladhi, U.R.; Chau, L.; Panfen, E.; Fischbach, M.A.; Braun, J.; et al. Role of dietary fiber in the recovery of the human gut microbiome and its metabolome. Cell Host Microbe 2021, 29, 394–407.e5. [Google Scholar] [CrossRef] [PubMed]
- Ley, R.; Bäckhed, F.; Turnbaugh, P.; Lozupone, C.A.; Knight, R.; Gordon, J.I. Obesity alters gut microbial ecology. Proc. Natl. Acad. Sci. USA 2005, 102, 11070–11075. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jie, Z.; Yu, X.; Liu, Y.; Sun, L.; Chen, P.; Ding, Q.; Gao, Y.; Zhang, X.; Yu, M.; Liu, Y.; et al. The Baseline Gut Microbiota Directs Dieting-Induced Weight Loss Trajectories. Gastroenterology 2021, 160, 2029–2042.e16. [Google Scholar] [CrossRef] [PubMed]
- Grigor’eva, I.N. Gallstone Disease, Obesity and the Firmicutes/Bacteroidetes Ratio as a Possible Biomarker of Gut Dysbiosis. J. Pers. Med. 2021, 11, 13. [Google Scholar]
- Ferrazzano, G.; Papa, C.; Pollio, A.; Ingenito, A.; Sangianantoni, G.; Cantile, T. Cyanobacteria and Microalgae as Sources of Functional Foods to Improve Human General and Oral Health. Molecules 2020, 25, 5164. [Google Scholar] [CrossRef]
- Shin, N.-R.; Whon, T.W.; Bae, J.-W. Proteobacteria: Microbial signature of dysbiosis in gut microbiota. Trends Biotechnol. 2015, 33, 496–503. [Google Scholar] [CrossRef]
- Doud, D.F.R.; Bowers, R.M.; Schulz, F.; De Raad, M.; Deng, K.; Tarver, A.; Glasgow, E.; Meulen, K.V.; Fox, B.; Deutsch, S.; et al. Function-driven single-cell genomics uncovers cellulose-degrading bacteria from the rare biosphere. ISME J. 2019, 14, 659–675. [Google Scholar] [CrossRef] [Green Version]
- Antushevich, H. Fecal microbiota transplantation in disease therapy. Clin. Chim. Acta 2020, 503, 90–98. [Google Scholar] [CrossRef]
- Kim, H.S.; Whon, T.W.; Sung, H.; Jeong, Y.-S.; Jung, E.S.; Shin, N.-R.; Hyun, D.-W.; Kim, P.S.; Lee, J.-Y.; Lee, C.H.; et al. Longitudinal evaluation of fecal microbiota transplantation for ameliorating calf diarrhea and improving growth performance. Nat. Commun. 2021, 12, 161. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Z.; Yang, L.; He, Y.; Luo, X.; Zhao, S.; Jia, X. Composition of Fecal Microbiota in Grazing and Feedlot Angus Beef Cattle. Animals 2021, 11, 3167. https://doi.org/10.3390/ani11113167
Zhang Z, Yang L, He Y, Luo X, Zhao S, Jia X. Composition of Fecal Microbiota in Grazing and Feedlot Angus Beef Cattle. Animals. 2021; 11(11):3167. https://doi.org/10.3390/ani11113167
Chicago/Turabian StyleZhang, Zhimin, Li Yang, Yang He, Xinmao Luo, Shaokang Zhao, and Xianbo Jia. 2021. "Composition of Fecal Microbiota in Grazing and Feedlot Angus Beef Cattle" Animals 11, no. 11: 3167. https://doi.org/10.3390/ani11113167





