1. Introduction
Myostatin protein was initially a subject of research interest, due the appearance of animals with significantly high muscle mass. This observation led to the coining of the term “muscular hypertrophy”. The first written account of bovine muscular hypertrophy (mh) was by a British farmer in his stock almanac [
1]. This was later described in some depth in 1888 by Kaiser [
2], who changed the perception of the meat characteristics of cattle, marking the beginning of a novel perspective on animal breeding with increased muscle mass. This phenotype has since become increasingly widespread among European beef cattle and has been termed “double-muscling” (DM). It has occurred at a high frequency in some breeds of cattle, such as Belgian Blue and Piedmontese [
3]. To differentiate between DM and the normal phenotype among animals, researchers have used different symbols. These include double-muscled or normal; DM or N; D or n; DM or dm; C or N; c or n; A or a; and mh or + [
4].
In addition, the influence of the myostatin gene on bone development has been established: newborn myostatin null mice (MSTN−/−) possess larger vertebral bone volume fractions (bone volume/total volume; BV/TV) due to increased BV, trabecular thickness (Tb, Th), and bone area (BA), as well as increased tissue mineral density (TMD) and BMD, as compared to newborn wildtype (Wt) mice [
5].
Interestingly, the molecular structure of the myostatin protein is conserved across vertebrates so that its C-terminal sequences are 100% identical in mice, rats, humans, pigs, and chickens [
6].
Myostatin, or growth and differentiation factor 8 (
GDF8), was initially identified as the factor causing a double-muscling phenotype due the presence of mutations inactivating gene, and, therefore, leading to the loss of the ability to stop muscle fiber growth [
6]. The genetic study of the myostatin gene (
MSTN) began during the last century [
7,
8].
The influence of
MSTN variability on the productivity traits of agriculturally useful animals has been investigated in different species. In particular, the polymorphisms of the promoter region of the
GDF8 gene influence meat color and final pH, affecting meat quality in various sheep breed groups [
9]. PCR with single-stranded conformational polymorphism (PCR-SSCP) analysis of the intron-1 of sheep (
Ovis aries) was used to identify five variants, designated
A–E (Gene Bank No. FJ858196-FJ858200) of the
MSTN gene [
10]. General linear mixed effect models revealed that the presence of
A allele in a lamb’s genotype was associated with decreased leg, loin, and total yield of lean meat, whereas the presence of
B allele was associated with increased loin yield and proportion loin yield (loin yield divided by total yield expressed as percentage). The effect of the number of allele copies present was investigated, and it was found that the absence of
A allele, or the presence of two copies of
B allele, was associated with increased mean leg yield, loin yield, and total yield. Two copies of
B allele were also associated with a decrease in the proportion of shoulder yield, whereas two copies of
A allele were associated with a decrease in the proportion of loin yield [
10].
The main reason for the appearance of mh is the gene mutations that result in a lack of
MSTN expression and continuous muscle growth [
11,
12]. This was confirmed when the mh locus was mapped 3.1 cM from microsatellite TGLA44 on the centromeric end of bovine chromosome 2. The positional candidate approach carried out by Grobet et al. in 1997 demonstrated that a mutation in bovine
MSTN, which encoded myostatin, a member of the TGFβ superfamily, is responsible for the double-muscled phenotype [
13]. This was also reported on in regard to 11 bp deletion in the coding sequence for the bioactive carboxy-terminal domain of the protein, causing muscular hypertrophy observed in Belgian Blue cattle [
14]. When the gene underlying the trait was identified as myostatin [
12], it was found to have a surprisingly high number of polymorphisms [
15].
Similar to other animal species, the myostatin gene in cattle is highly polymorphic [
12]. To date, reports on several mutations in all coding regions of myostatin (exons 1–3) have described them as silent and causing non-synonymous changes. The most examined mutations of the
MSTN gene mainly single nucleotide polymorphisms (SNP) appearing in the various cattle breeds are summarized in
Table 1.
Phylogenetic analysis has shown that there was positive selection pressure for non-synonymous mutations within the myostatin gene family around the time of the divergence of cattle, sheep, and goats, and these positive selective pressures on non-ancestral myostatin are relatively recent [
16]. Unfortunately, breed management problems exist for double-muscled cattle, such as birthing difficulties, which can be overcome through genetically controlled breeding programs [
17].
We are interested in the polymorphisms of the myostatin gene caused by
nt821(del11) and
F94L MSTN mutations. In our opinion, these considerably influence the productivity traits of the
nt821(del11) MSTN mutation associated with the double-muscled trait in cattle, which is characterized by an increase in muscle mass of approximately 20%, resulting in substantially higher meat yield, a higher proportion of expensive cuts of meat, and lean and very tender meat, for which a substantial premium is paid [
17]. This trait is autosomal recessive, and the locus is referred to as mh. It occurs at such a high frequency in Piedmontese and Belgian Blue cattle that it is characteristic of these breeds [
5]. However, it also occurs in other breeds (
Table 1). In addition to its obvious advantages, double-muscling also has one major drawback—a greatly increased incidence of calving difficulties to the extent that cesarian sections are required for deliveries within these breeds [
17] and so it was classified as a genetic defect (M1). However, due to its sufficient advantages, double-muscled cattle play a major role in animal agriculture in several countries, and they can be found in many regions of the world [
18,
19].
F94L MSTN is a single-nucleotide polymorphism (SNP) of g. 433C > A transversion, that causes amino acid substitution of phenylalanine via leucine at position 94 of the protein sequence. Previous studies have shown the additive effect of this mutation on cattle carcass traits and strong evidence that an
MSTN genotype can produce an intermediate, non-double-muscling phenotype, which is expected to be of significant value for beef cattle producers [
20,
21].
Currently, the most precise methods of
nt821(del11) and
F94L MSTN diagnostics are DNA analyses, which are based on different modifications of the polymerase chain reaction (PCR) technique, and the most modern methods of DNA analyses are based on sequencing or real-time PCR [
9,
10,
11,
12,
13,
14,
15,
16].
The aim of this study is to determine the haplotype frequencies of the F94L and nt821(del11) polymorphisms of the myostatin gene among the Russian cattle populations of Aberdeen Angus, Limousin, Simmental, and Belgian Blue breeds using DNA analysis.
2. Materials and Methods
The study was approved by the bioethical commission of L.K. Ernst Federal Research Center for Animal Husbandry (Protocol No. 5 from 22 March 2021). We used the DNA samples (
n = 930) from Aberdeen Angus, Simmental, Limousin, and Belgian Blue cattle (
Table 2). The animals used in this study were selected from breeding farms located in different regions of Russia. All animals of the herds were semi-sibs and were managed and fed in the same manner.
The DNA were extracted from the available biomaterial of the animals (skin, sperm, milk, and blood) by methods traditionally used in the lab of genetics and genomics of cattle at L.K. Ernst Federal Research Center for Animal Husbandry (Dubrovitsy, Moscow, Russia) [
22].
The genotyping methods were chosen based on the structures of the studied mutations. Due the presence of the small deletion for
nt821(del11) genotyping, we used the allele-specific PCR method (AS-PCR) [
22]. For
F94L polymorphism diagnostics, we chose the PCR-RFLP (PCR with the subsequent analysis of restriction fragments length polymorphism) method due the existence of a mutation area recognition site for the
TaqI restriction endonuclease site in the DNA.
For the development of the
nt821(del11) and
F94L MSTN genotyping tests, we chose the last version of the
Bos taurus myostatin gene DNA sequence from NCBI (Accession Number MK 214682.1). The conducted alignment of the sequences using BLAST 2.12.0+ (
https://blast.ncbi.nlm.nih.gov/Blast.cgi#1692837278, accessed on 20 August 2021) revealed high similarity between the chosen sequence and other
Bos taurus myostatin gene sequences (99.46–100%).
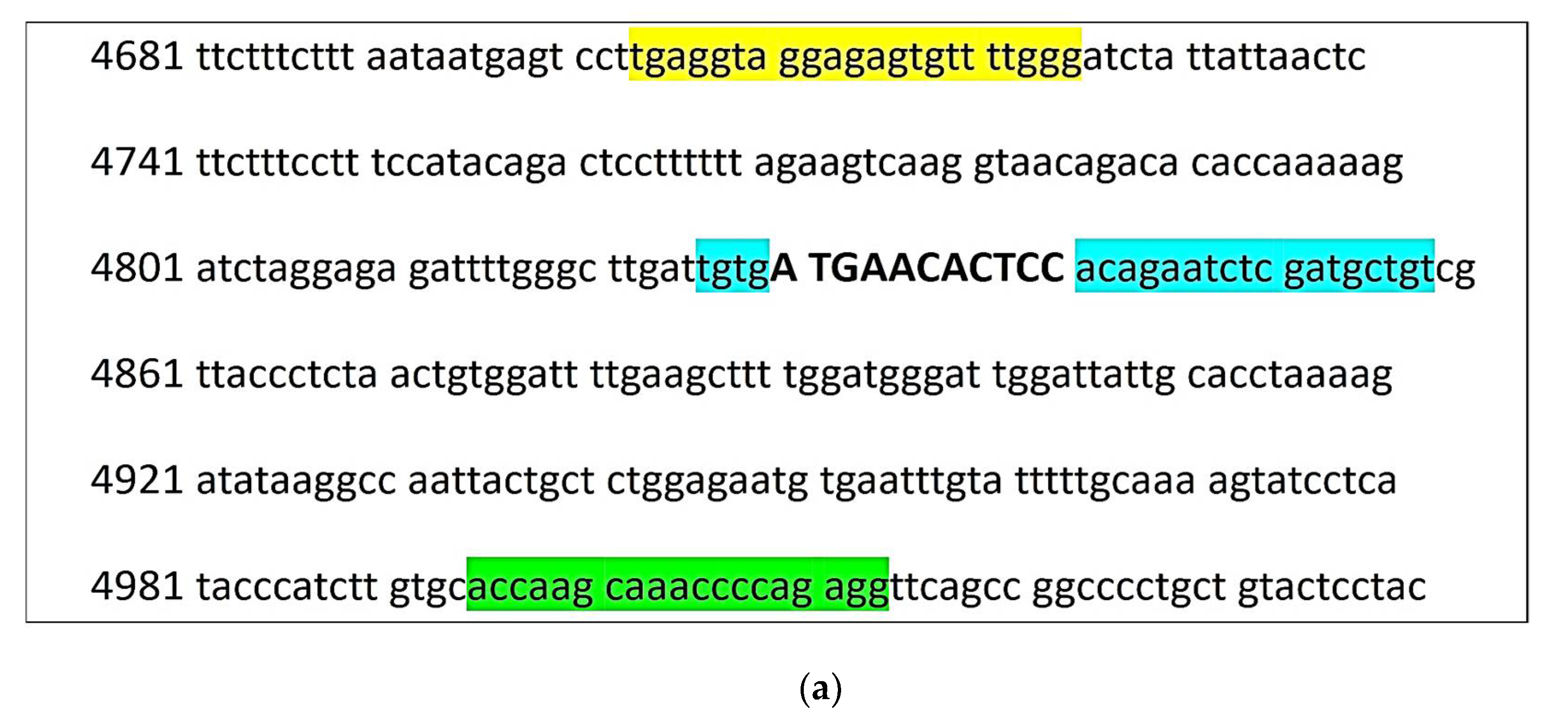
The schemes of the test systems for
nt821(del11) and
F94L MSTN genotyping are presented in
Figure 1.
Primer design was carried out using the Primer3Plus Software (
https://primer3plus.com, accessed on 29 June 2021) and the selection of the restriction endonuclease was made using NEBcutter V.2.0. (
https://nc2.neb.com/NEBcutter2/, accessed on 29 June 2021). The primer characteristics for the DNA amplification of the mutation area of
nt821(del11) and
F94L MSTN polymorphisms are presented in
Table 3.
The PCRs were carried out on a Biorad T100 thermocycler (Biorad, Singapore) in a mixture of 10 μL M1 and 15 μL F94L containing dNTP mix, 10x PCR buffer, 0.03 μL of each oligonucleotide primer at a 100 mM concentration, and 1 U of Taq-polymerase. The volume of the DNA was 1 μL with the content of genomic DNA at 50 ng. The temperature–time PCR conditions included the initial denaturation under 95 °C for 3 min; 35 cycles of denaturation under 95 °C for 45 s; annealing under 60 °C (for M1) or 55 °C (for F94L) for 30 s; elongation under 72 °C for 30 s; and the final extension under 72 °C for 4 min.
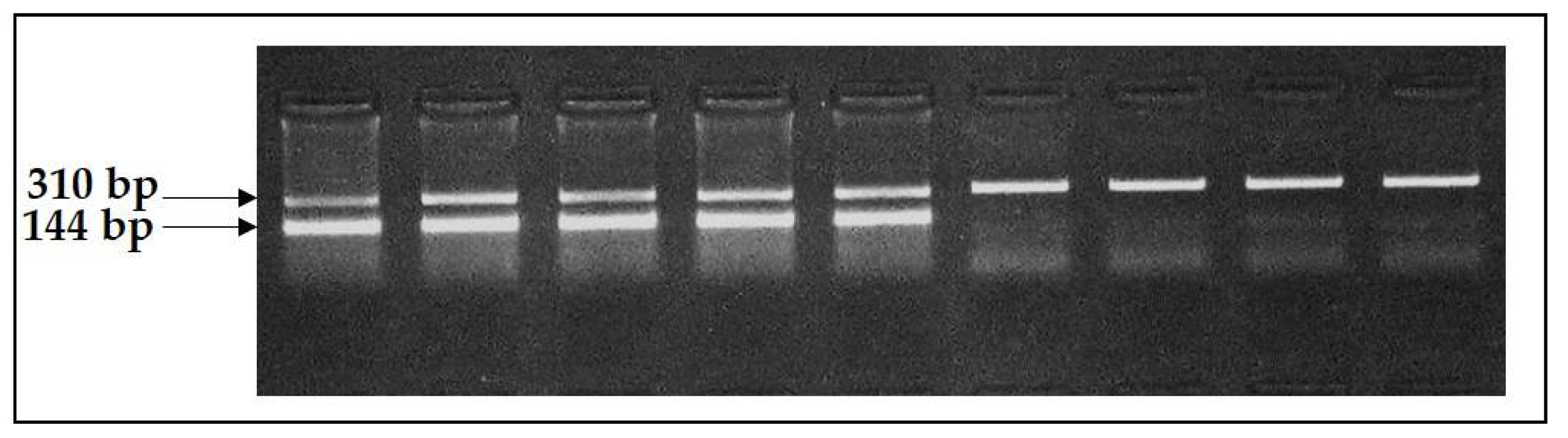
The PCR-RFLP method was used to test the F94L MSTN polymorphism. Due to the observation of the restriction site for TaqI endonuclease restriction (T↓CGA) in the wild type but not in the mutant allele, we used this enzyme for RFLP analysis (1 U on a probe with exposition per night). PCR product detection was conducted by the method of electrophoresis in agarose gel with agarose content of 3% under a voltage of 120 V for 30 min. The 10x TAE buffer was used as a conducting medium.
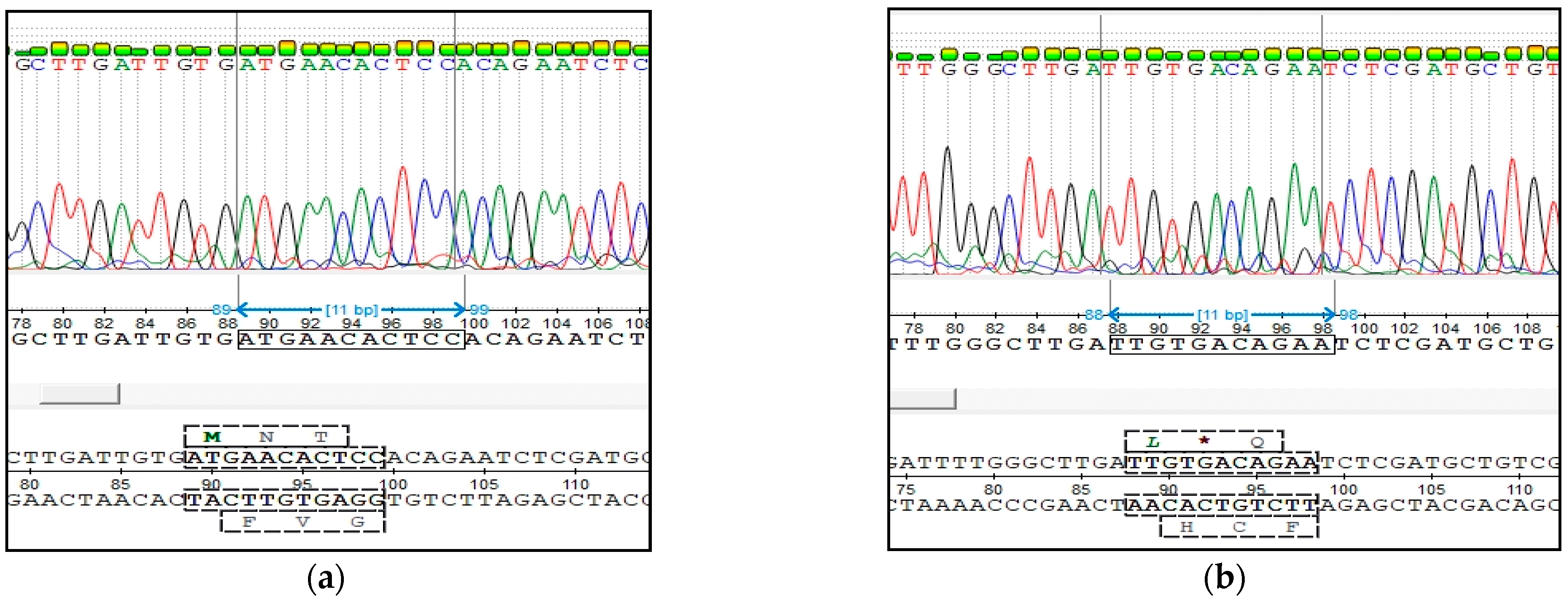
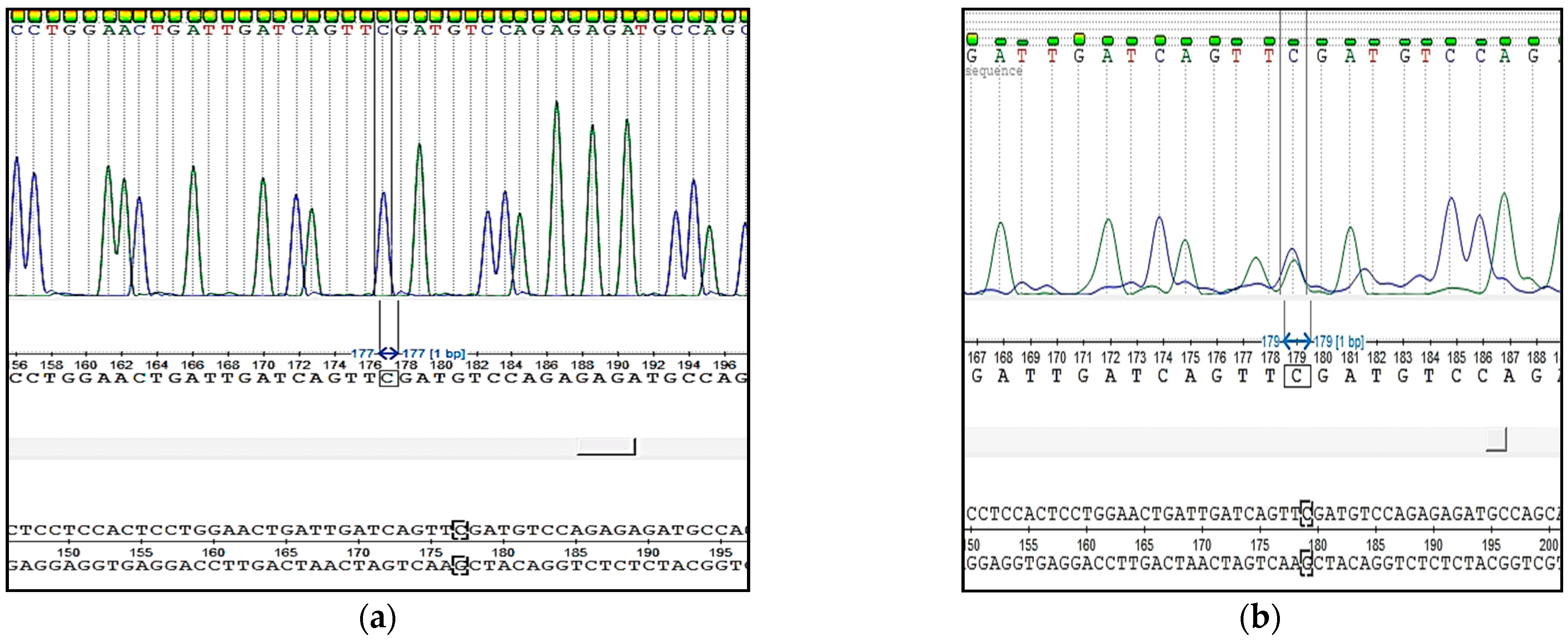
Verification of the correct work of our test systems was carried out using the Sanger sequencing method. Sequencing was performed by Eurogene (Moscow, Russia). The results were evaluated using UGENE Software 40.0.
The statistical evaluation of the data was conducted using traditional methods [
23,
24] and with the assistance of Microsoft Office Excel and online calculators’ software (
https://www.statskingdom.com, accessed on 28 June 2021). For the evaluation of the influence of breed factors on the frequencies of the desirable alleles of
nt821(del11) and
F94L MSTN mutations, we used two-way ANOVA (
https://www.statskingdom.com/two-way-anova-calculator.html, accessed on 20 August 2021).
4. Discussion
The conducted investigations showed the sufficient variability of the myostatin gene and revealed almost all genotypes of nt821(del11) and F94L MSTN mutations in the cattle populations of Aberdeen Angus, Limousin, Simmental, and Belgian Blue.
Previous investigations of
nt821(del11) MSTN in Belgian Blue cattle have shown that all heads of this breed were carriers of the mutant allele associated with double-muscling [
25]. The additional
nt821(del11) MSTN genotyping conducted in the frame of the current work has confirmed the earlier obtained data and conclusion of our colleagues overseas, that the mutant allele associated with the double-muscling genetic defect was fixed in the Belgian Blue cattle breed due the long-time selection in regard to increased muscle mass [
26]. On the one hand, this polymorphism has a positive influence on meat quality by increasing fat content in carcasses [
26], but, on the other hand,
nt821(del11) in the homozygous station is a reason behind the calving difficulties or dystocia ultimately leading to the wide use of the cesarian section in Belgian Blue cattle [
19,
27].
The
F94L MSTN polymorphism occurred due to the considerable interest in the single-nucleotide polymorphism c.282C > A (or g.433C > A) showed by breeders. In fact, this mutation has the same action as that of
nt821(del11) on fat content, but it does not have an influence on the ease of calving due the absence of influence on birth weight, thereby making this polymorphism a possible genetic marker of productivity traits [
28].
Previous investigations have shown the nonsignificant effect of the
A allele of
F94L MSTN on birth and growth traits, and intramuscular fat content in carcasses of Limousin backcross calves in Australian and New Zealand populations. However, a significant increase in total carcass fat weight (−16.5% and −8.1% trait means for Australian and New Zealand populations;
p < 0.001 and
p < 0.05, respectively), meat weight (7.3% and 5.9% trait means for Australian and New Zealand populations, respectively,
p < 0.001), and a reduction in fat depth (−18.7%,
p < 0.001 in the Australian population) have been established. Meat tenderness, pH, and cooking loss of
M. longissimus dorsi were not affected by the
F94L variant. The results offer strong evidence that this
F94L MSTN variant provides an intermediate and more useful phenotype than the more severe double-muscling phenotype caused by knockout mutations in the myostatin gene [
20]. In accordance with other data, the g.433A allele was found to be associated with a 5.5% increase in silverside percentage and eye muscle area (EMA), and a 2.3% increase in total meat percentage relative to the g.433C allele. The phenotypic effects of the g.433A allele were partially recessive, providing strong evidence that an
MSTN genotype can produce an intermediate, non-double-muscling phenotype, which is expected to be of significant value for beef cattle producers [
21].
A population analysis previously conducted on fifteen cattle breeds (
n = 1140) located in Australia demonstrated the presence of SNP 433C > A in the Simmental (0.8%), Piedmontese (2%), Droughtmaster (4%), and Limousin (94.2%) breeds, but not in the Salers, Aberdeen Angus, Hereford, Charole, Jersey, Holstein, Shorthorn, Gelbvieh, and Maine-Anjou breeds [
20].
In the current study, we obtained data that are consistent with the data previously obtained by other researchers [
28], according to which the most genetically preferable breed in terms of
F94L MSTN polymorphism is the Limousin breed. The data on the Simmental breed are also in accordance with those of an Australian study—0.04%
AA animals in the Russian population and 0.08% in the Australian one. Regarding the Aberdeen Angus breed, unlike in the Australian populations, in our herds, SNP 433C > A was found in
CA animals with frequencies of 6.90% and 5.60% in Aberdeen Angus populations one and two, respectively. It is also noteworthy that if among the populations of the Aberdeen Angus breed, the animals that are carrying the A allele are completely heterozygous (
CA genotype), then the animals of the Simmental and Belgian Blue breed populations that have the
A allele in their genotype are homozygous for this allele.
In general, the populations we studied are characterized by a low proportion of heterozygosity for both of the studied MSTN polymorphisms. F94L MSTN was observed in one of the populations of the Limousin breed as well as in the populations of the Simmental and Belgian breeds, but this was not the case for animals with the CA genotype. Regarding nt821(del11) MSTN polymorphism, the heterozygous animals (M1C or MSTN+/MSTN−) were absent from almost all investigated populations, excluding the Belgian Blue breed. These facts are probably related both to the purebred breeding practiced in the farms that owned the studied populations, and to the historical origin of the studied cattle breeds. Currently, among these indicators, we can observe a fairly high stability of the genome of the Russian cattle of the Aberdeen Angus, Simmental, Limousin, and Belgian Blue breeds in relation to the studied mutations of the myostatin gene.
The obtained results also allow for the proposal of the high genetic potential of the Limousin and Belgian Blue cattle breeds. In our opinion, the Belgian Blue cattle could be used in selection programs for the purpose of productivity improvement, but with caution due to the high risk of the appearance of reproductive problems. The Limousin breed is extremely promising, due to the high frequency of the polymorphism linked with the increase in muscle weight but without calving difficulties. The high frequencies of A allele F94L MSTN polymorphism may be the result of a directed, long-term selection of animals with a high muscle mass combined with the simultaneous culling of animals with the appearance of calving difficulties.
Despite the low frequency of AA genotype animals in the populations of the Aberdeen Angus and Simmental cattle breeds, the presence of animals carrying the A allele genotype represents the possibility of cattle selection with consideration of F94L MSTN polymorphism. In addition, we cannot discount the good acclimatization abilities and endurance of these animals as a result of this—they are already spread widely over the territory of Russia and form the breeding core of beef cattle breeding.
The results of the current study show the high genetic potential of Russian Aberdeen Angus and Limousin livestock.
In our opinion, the issue of polymorphisms of the myostatin gene has not yet been thoroughly examined. In 2012–2013, four double-muscled German Gelbvieh cattle were born with an abnormally high birth weight of more than 60 kg, and specialists proposed that this effect could be explained by either the homozygous c.821del11 mutation or compound heterozygosity of the c.191T > C and c.821del11 mutations [
29].
In addition, we should not forget the prospective programs of genome selection, which allow one to select the most productive animals based on the knowledge of information regarding polymorphism of all genomes. In 2013, the reference population of 3060 beef cattle, including pure-bred Limousin, Limousin cross-bred with Angus, and pure-bred Angus, were genotyped using a BovineSNP50 BeadChip and directly for the
MSTN-
F94L variant. The study demonstrated that the
MSTN-
F94L variant was the most strongly associated with five traits (birth weight, easy and direct calving, milk, weaning weight, and yield grade) among the thirteen measured traits in the Limousin and mixed (Angus x Limousin) populations. Fitting the
MSTN-
F94L variant as a random effect, the genomic prediction accuracies for birth weight increased by 2.7% in purebred Limousin, by 2.2% in the mixed populations, and by 3.2% in the multibreed population. Prediction accuracies for five traits increased in the multibreed analysis. Fitting
MSTN-F94L as a fixed effect in purebred Limousin, Angus x Limousin, and multibreed analyses resulted in increased prediction accuracy in purebred Limousin for eight traits. Prediction accuracies can be improved by including a causal variant in genomic evaluation rather than simply using whole-genome SNP markers [
30].
It should be noted that recent research findings in humans and other mammalian and non-mammalian species support the potent regulatory role of myostatin in the morphology and function of muscle, as well as cellular differentiation and metabolism, with real-life implications in agricultural meat production and human disease. Myostatin-null mice (
MSTN−/−) exhibit skeletal muscle fiber hyperplasia and hypertrophy, whereas myostatin deficiency in larger mammals, such as sheep and pigs, can result in muscle fiber hyperplasia. Myostatin’s impact extends beyond muscles, with alterations in myostatin present in the pathophysiology of myocardial infarctions, inflammation, insulin resistance, diabetes, aging, cancer cachexia, and musculoskeletal disease [
31].
5. Conclusions
The results of the investigations conducted in this study allow us to conclude that the myostatin gene is extremely variable and not unambiguous because in this gene, there are polymorphisms that have a positive effect on meat productivity traits as well as those that have negative effects on their reproductive traits, for example, calving difficulties (dystocia). The genotyping results of Russian populations of Aberdeen Angus, Limousin, Simmental, and Belgian Blue cattle show that the domestic herds of these breeds could be considered as beneficial for further breeding. The most investigated populations had low frequencies of animal carriers of the double-muscling genetic defect caused by nt821(del11) mutation of the myostatin gene. The exception was the Belgian Blue cattle population, which consisted of 100% animal carriers of the mutant allele generated due the 11 bp deletion and associated with the M1 genetic defect (double-muscling).
The study of F94L MSTN polymorphism revealed the high frequency of the desirable A allele in terms of productivity in the Limousin populations, making this cattle breed attractive in terms of an improvement in the meat productivity without negative consequences related to animal health.
The Aberdeen Angus cattle populations showed the presence of A allele F94L and the mutant allele of nt821(del11) MSTN (although at low frequencies), which suggests that it may be possible to develop their meat qualities, facilitating the production of beef with a larger weight and lower fat depth in carcasses.
Thus, the studied populations of cattle of the Aberdeen Angus, Limousin, Simmental, and Belgian Blue breeds showed a sufficiently high genetic potential, suggesting the possibility of increasing the productivity of herds while preserving their health.
Currently, searches are being carried out to identify a method of obtaining animals with knock-out myostatin using CRISPR/CAS9 technology [
32]. However, in reality, the selection of animals based on genotyping information seems to be the most affordable way to increase the productivity of beef breeds of cattle.