Advantage of including Genomic Information to Predict Breeding Values for Lactation Yields of Milk, Fat, and Protein or Somatic Cell Score in a New Zealand Dairy Goat Herd
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Data
2.2. Genotyping
2.3. Methods
2.3.1. Pedigree-Based BLUP Evaluation
2.3.2. Single-Step BayesC Genomic Evaluation
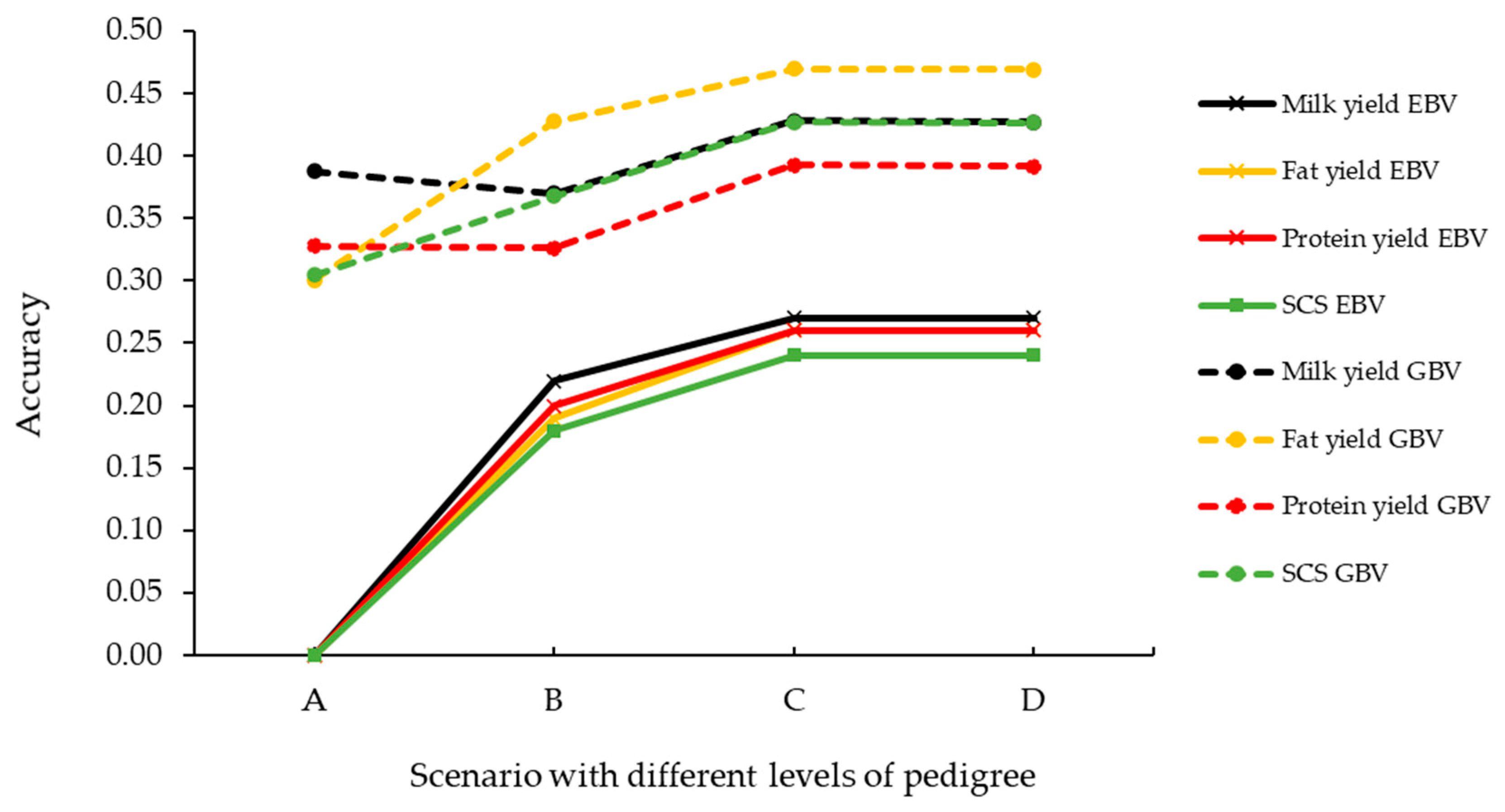
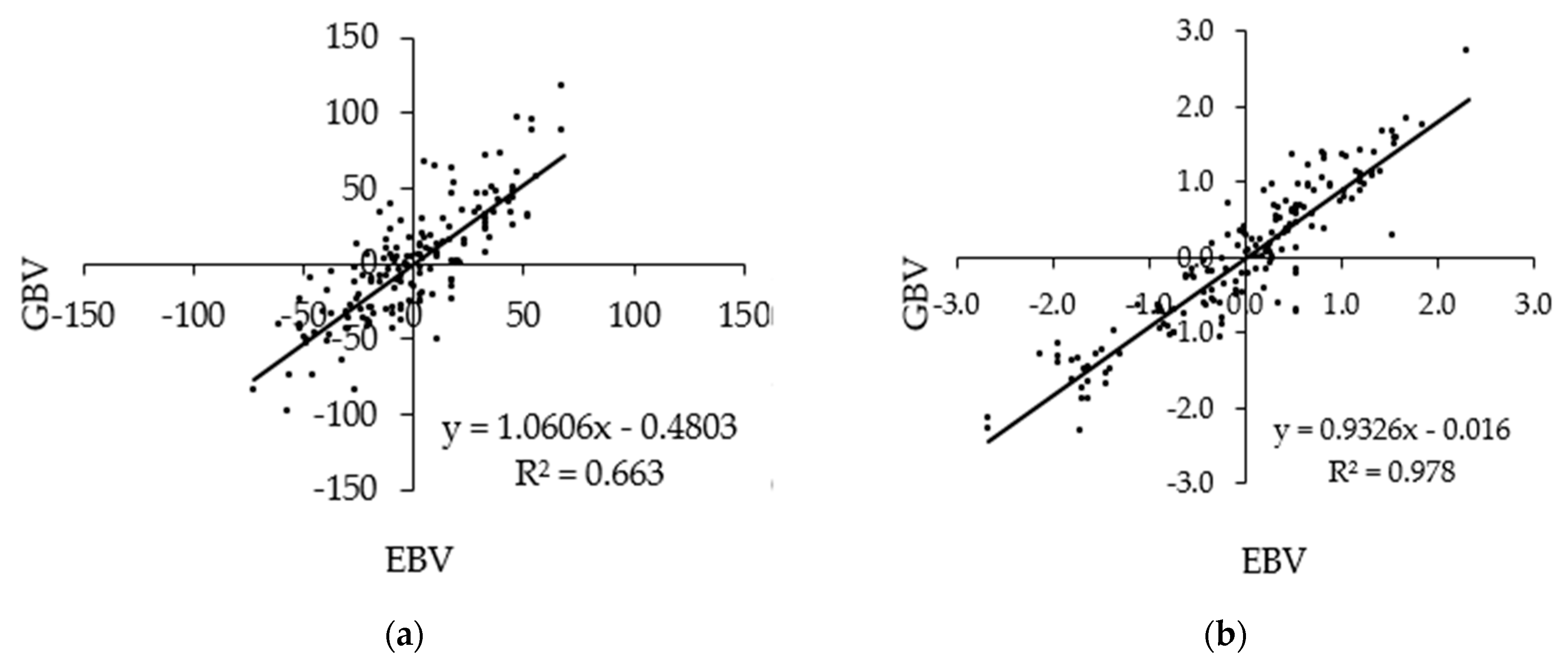
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Cole, J.; VanRaden, P. Symposium review: Possibilities in an age of genomics: The future of selection indices. J. Dairy Sci. 2018, 101, 3686–3701. [Google Scholar] [CrossRef] [PubMed]
- Garrick, D.; Fernando, R. Genomic prediction and genome-wide association studies in beef and dairy cattle. In The Genetics of Cattle; CABI: Wallingford, UK, 2014; pp. 474–501. [Google Scholar]
- Wright, S. Coefficients of inbreeding and relationship. Am. Nat. 1922, 56, 330–338. [Google Scholar] [CrossRef]
- Legarra, A.; Aguilar, I.; Misztal, I. A relationship matrix including full pedigree and genomic information. J. Dairy Sci. 2009, 92, 4656–4663. [Google Scholar] [CrossRef] [PubMed]
- Henderson, C. Selection index and expected genetic advance. Stat. Genet. Plant Breed. 1963, 982, 141–163. [Google Scholar]
- Chen, C.; Misztal, I.; Aguilar, I.; Legarra, A.; Muir, W. Effect of different genomic relationship matrices on accuracy and scale. J. Anim. Sci. 2011, 89, 2673–2679. [Google Scholar] [CrossRef]
- Misztal, I.; Legarra, A.; Aguilar, I. Using recursion to compute the inverse of the genomic relationship matrix. J. Dairy Sci. 2014, 97, 3943–3952. [Google Scholar] [CrossRef]
- Fernando, R.; Dekkers, J.; Garrick, D. A class of Bayesian methods to combine large numbers of genotyped and non-genotyped animals for whole-genome analyses. Genet. Sel. Evol. 2014. [Google Scholar] [CrossRef]
- Meuwissen, T.; Hayes, B.; Goddard, M. Prediction of total genetic value using genome-wide dense marker maps. Genetics 2001, 157, 1819–1829. [Google Scholar]
- Habier, D.; Fernando, R.; Kizilkaya, K.; Garrick, D. Extension of the bayesian alphabet for genomic selection. BMC Bioinform. 2011. [Google Scholar] [CrossRef]
- Lopez-Villalobos, N.; Garrick, D. Estimation of genetic parameters for lactation yields of milk, fat and protein of New Zealand dairy goats. J. Anim. Sci. 2001, 79, 70. [Google Scholar]
- Scholtens, M.; Smith, R.; Lopez-Lozano, S.; Lopez-Villalobos, N.; Burt, D.; Harper, L.; Tuohy, M.; Thomas, D.; Carr, A.; Gray, D.; et al. Brief communication: The current state of the New Zealand goat industry. Proc. N. Z. Soc. Anim. Prod. 2017, 77, 164–168. [Google Scholar]
- Sargent, F.; Lytton, V.; Wall, O. Test interval method of calculating dairy herd improvement association records. J. Dairy Sci. 1968, 51, 170–179. [Google Scholar] [CrossRef]
- Gilmour, A.; Gogel, B.; Cullis, B.; Thompson, R.; Butler, D. ASReml User Guide Release 3.0.; VSN International Ltd.: Hemel Hempstead, UK, 2009. [Google Scholar]
- Gregory, K.; Cundiff, L. Crossbreeding in beef cattle: Evaluation of systems. J. Anim. Sci. 1980, 51, 1224–1242. [Google Scholar] [CrossRef]
- Scholtens, M.; Jiang, A.; Smith, A.; Littlejohn, M.; Lehnert, K.; Snell, R.; Lopez-Villalobos, N.; Garrick, D.; Blair, H. Genome-wide association studies of lactation yields of milk, fat, protein and somatic cell score in New Zealand dairy goats. J. Anim. Sci. Biotechnol. 2020. [Google Scholar] [CrossRef]
- Cheng, H.; Garrick, D.; Fernando, R. JWAS: Julia Implementation of Whole-Genome Analyses Software Using Univariate and Multivariate Bayesian Mixed Effects Model. Available online: http://qtl.rocks/ (accessed on 13 March 2020).
- Hsu, W.; Garrick, D.; Fernando, R. The accuracy and bias of single-step genomic prediction for populations under selection. G3 Genes Genomes Genet. 2017, 7, 2685–2694. [Google Scholar] [CrossRef]
- Scholtens, M.; Lopez-Villalobos, N.; Garrick, D.; Blair, H.; Lehnert, K.; Snell, R. Genetic parameters for total lactation yields of milk, fat, protein, and somatic cell score in New Zealand dairy goats. Anim. Sci. J. 2019. [Google Scholar] [CrossRef]
- Geweke, J. Evaluating the Accuracy of Sampling-Based Approaches to the Calculation of Posterior Moments; Federal Reserve Bank of Minneapolis; Research Department Minneapolis: Minneapolis, MN, USA, 1991; Volume 196. [Google Scholar]
- Henderson, C. Best linear unbiased estimation and prediction under a selection model. Biometrics 1975, 31, 423–447. [Google Scholar] [CrossRef]
- Mucha, S.; Mrode, R.; MacLaren-Lee, I.; Coffey, M.; Conington, J. Estimation of genomic breeding values for milk yield in UK dairy goats. J. Dairy Sci. 2015, 98, 8201–8208. [Google Scholar] [CrossRef]
- Molina, A.; Munoz, E.; Diaz, C.; Menendez-Buxadera, A.; Ramon, M.; Sanchez, M.; Carabano, M.J.; Serradilla, J.M. Goat genomic selection: Impact of the integration of genomic information in the genetic evaluations of the Spanish Florida goats. Small Rumin. Res. 2018, 163, 72–75. [Google Scholar] [CrossRef]
- Carillier, C.; Larroque, H.; Robert-Granié, C. Comparison of joint versus purebred genomic evaluation in the French multi-breed dairy goat population. Genet. Sel. Evol. 2014. [Google Scholar] [CrossRef]
- Teissier, M.; Larroque, H.; Robert-Granie, C. Weighted single-step genomic BLUP improves accuracy of genomic breeding values for protein content in French dairy goats: A quantitative trait influenced by a major gene. Genet. Sel. Evol. 2018. [Google Scholar] [CrossRef] [PubMed]
- Cooper, T.; Wiggans, G.; VanRaden, P. Including cow information in genomic prediction of Holstein dairy cattle in the US. In Proceedings of the 10th World Congress of Genetics Applied to Livestock Production, Vancouver, BC, Canada, 17–22 August 2014. [Google Scholar]
- Daetwyler, H.; Kemper, K.; Van der Werf, J.; Hayes, B. Components of the accuracy of genomic prediction in a multi-breed sheep population. J. Anim. Sci. 2012, 90, 3375–3384. [Google Scholar] [CrossRef] [PubMed]
- Clark, S.; Hickey, J.; Daetwyler, H.; van der Werf, J. The importance of information on relatives for the prediction of genomic breeding values and the implications for the makeup of reference data sets in livestock breeding schemes. Genet. Sel. Evol. 2012. [Google Scholar] [CrossRef] [PubMed]
- Hayes, B.; Bowman, P.; Chamberlain, A.; Goddard, M. Invited review: Genomic selection in dairy cattle: Progress and challenges. J. Dairy Sci. 2009, 92, 433–443. [Google Scholar] [CrossRef] [PubMed]
- Harris, B.; Johnson, D.; Spelman, R.; Sattler, J. Genomic selection in New Zealand and the implications for national genetic evaluation. In Proceedings of the Interbull Meeting, Niagara Falls, ON, Canada, 1 January 2008. [Google Scholar]
- Habier, D.; Fernando, R.; Dekkers, J. The impact of genetic relationship information on genome-assisted breeding values. Genetics 2007, 177, 2389–2397. [Google Scholar]

| Trait | Polygenic Variance | SNP Variance | Residual Variance | π | h2 1 |
|---|---|---|---|---|---|
| Milk yield | 9098.6 | 1011.0 | 30,345.0 | 0.98 | 0.25 |
| Fat yield | 8.66 | 1.33 | 35.20 | 0.98 | 0.24 |
| Protein yield | 5.69 | 0.88 | 23.10 | 0.98 | 0.24 |
| Somatic cell score | 0.51 | 0.08 | 2.50 | 0.98 | 0.21 |
| Trait | Mean | SD 1 | Min | Max | CV 2 |
|---|---|---|---|---|---|
| Lactation length (days) | 288 | 23 | 185 | 305 | 8 |
| Yields (up to 305 days) | |||||
| Milk yield (kg) | 1002.0 | 268.3 | 292.4 | 1811.3 | 27 |
| Fat yield (kg) | 31.8 | 8.9 | 8.2 | 67.1 | 28 |
| Protein yield (kg) | 31.8 | 8.4 | 8.9 | 58.5 | 26 |
| SCS 3 (units) | 9.5 | 1.2 | 6.5 | 12.6 | 12 |
| EBV | GBV | Gain (%) | |||
|---|---|---|---|---|---|
| Trait | r | SE 3 | r | SE 3 | |
| Milk yield | 0.22 | 0.01 | 0.38 | 0.01 | +73% |
| Fat yield | 0.21 | 0.01 | 0.43 | 0.01 | +103% |
| Protein yield | 0.21 | 0.01 | 0.34 | 0.01 | +64% |
| Somatic cell score | 0.20 | 0.01 | 0.39 | 0.01 | +95% |
| Breed Coefficient of Traits | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Model | Breed | Milk Yield | SE | Fat Yield | SE | Protein Yield | SE | SCS 3 | SE |
| PBLUP | |||||||||
| Saanen | 0 | 0 | 0 | 0 | |||||
| ANTO | −100.60 | 138.70 | −0.52 | 4.69 | −1.07 | 3.80 | −0.02 | 1.22 | |
| Unknown | −33.12 | 45.10 | −1.18 | 1.52 | −1.50 | 1.24 | 0.11 | 0.40 | |
| ssBC | |||||||||
| Saanen | −112.54 | 2.52 | −0.59 | 0.08 | −2.11 | 0.07 | 0.47 | 0.02 | |
| ANTO | −364.56 | 5.82 | −5.28 | 0.19 | −8.87 | 0.16 | 0.01 | 0.05 | |
| Unknown | −64.04 | 2.34 | −0.92 | 0.08 | −1.42 | 0.07 | 0.46 | 0.02 | |
| Breed and J Coefficients of Traits | ||||||||
| Breed | Milk yield | SE | Fat Yield | SE | Protein yield | SE | SCS 2 | SE |
| Saanen | 0 | 0 | 0 | 0 | ||||
| ANTO | 94.04 | 247.57 | 4.99 | 8.54 | 7.03 | 6.73 | 1.03 | 2.25 |
| Unknown | −164.89 | 83.04 | −2.29 | 2.67 | −4.53 | 2.37 | 0.58 | 0.72 |
| Breed-Specific J Covariate Coefficient of Traits | ||||||||
| JSaanen | −112.54 | 78.95 | −0.59 | 2.56 | −2.11 | 2.25 | 0.47 | 0.71 |
| JANTO | −458.60 | 300.00 | −10.28 | 10.23 | −15.90 | 8.19 | −1.02 | 2.76 |
| JUnknown | 100.85 | 34.88 | 1.38 | 1.14 | 3.11 | 1.00 | −0.12 | 0.31 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Scholtens, M.; Lopez-Villalobos, N.; Lehnert, K.; Snell, R.; Garrick, D.; Blair, H.T. Advantage of including Genomic Information to Predict Breeding Values for Lactation Yields of Milk, Fat, and Protein or Somatic Cell Score in a New Zealand Dairy Goat Herd. Animals 2021, 11, 24. https://doi.org/10.3390/ani11010024
Scholtens M, Lopez-Villalobos N, Lehnert K, Snell R, Garrick D, Blair HT. Advantage of including Genomic Information to Predict Breeding Values for Lactation Yields of Milk, Fat, and Protein or Somatic Cell Score in a New Zealand Dairy Goat Herd. Animals. 2021; 11(1):24. https://doi.org/10.3390/ani11010024
Chicago/Turabian StyleScholtens, Megan, Nicolas Lopez-Villalobos, Klaus Lehnert, Russell Snell, Dorian Garrick, and Hugh T. Blair. 2021. "Advantage of including Genomic Information to Predict Breeding Values for Lactation Yields of Milk, Fat, and Protein or Somatic Cell Score in a New Zealand Dairy Goat Herd" Animals 11, no. 1: 24. https://doi.org/10.3390/ani11010024
APA StyleScholtens, M., Lopez-Villalobos, N., Lehnert, K., Snell, R., Garrick, D., & Blair, H. T. (2021). Advantage of including Genomic Information to Predict Breeding Values for Lactation Yields of Milk, Fat, and Protein or Somatic Cell Score in a New Zealand Dairy Goat Herd. Animals, 11(1), 24. https://doi.org/10.3390/ani11010024






