Fish Pathology Research and Diagnosis in Aquaculture of Farmed Fish; a Proteomics Perspective
Abstract
Simple Summary
Abstract
1. Introduction
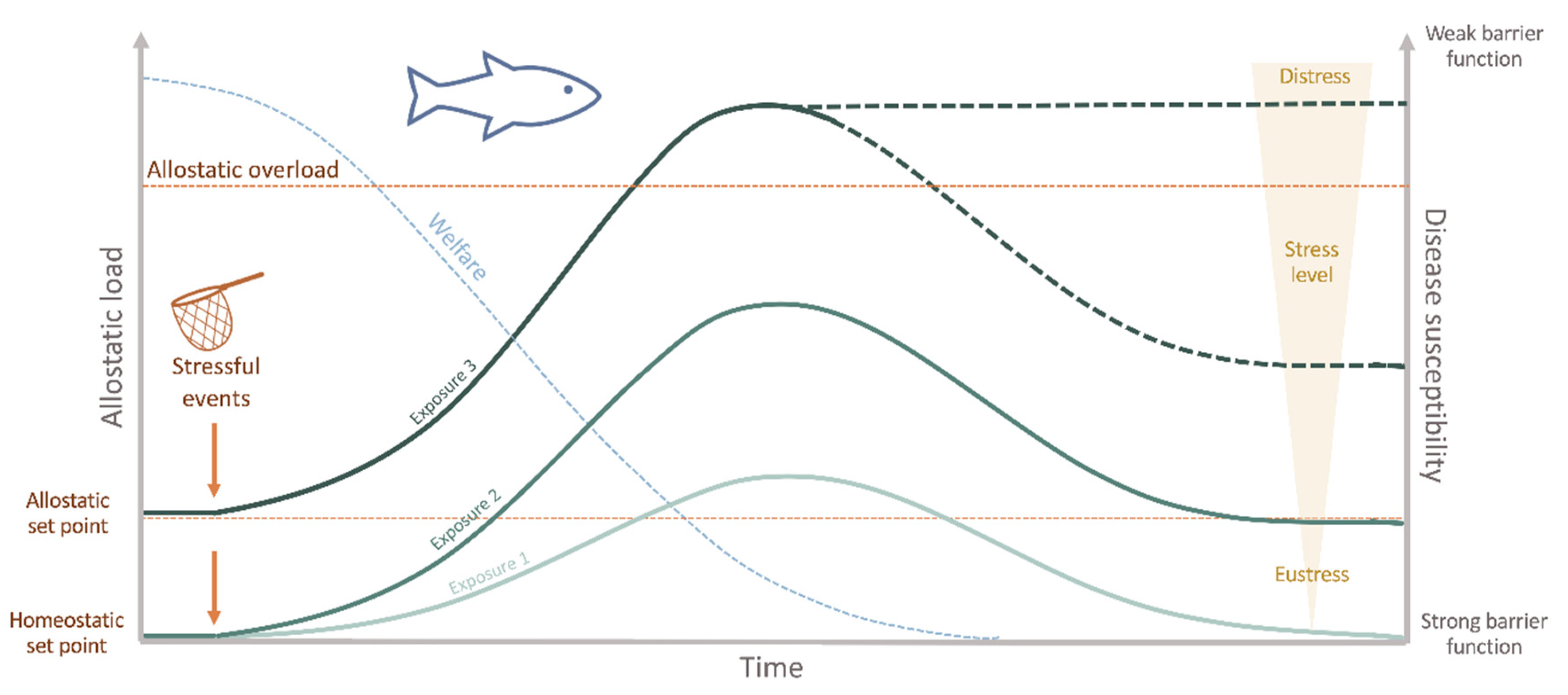
2. Fish Health, Stress and Welfare
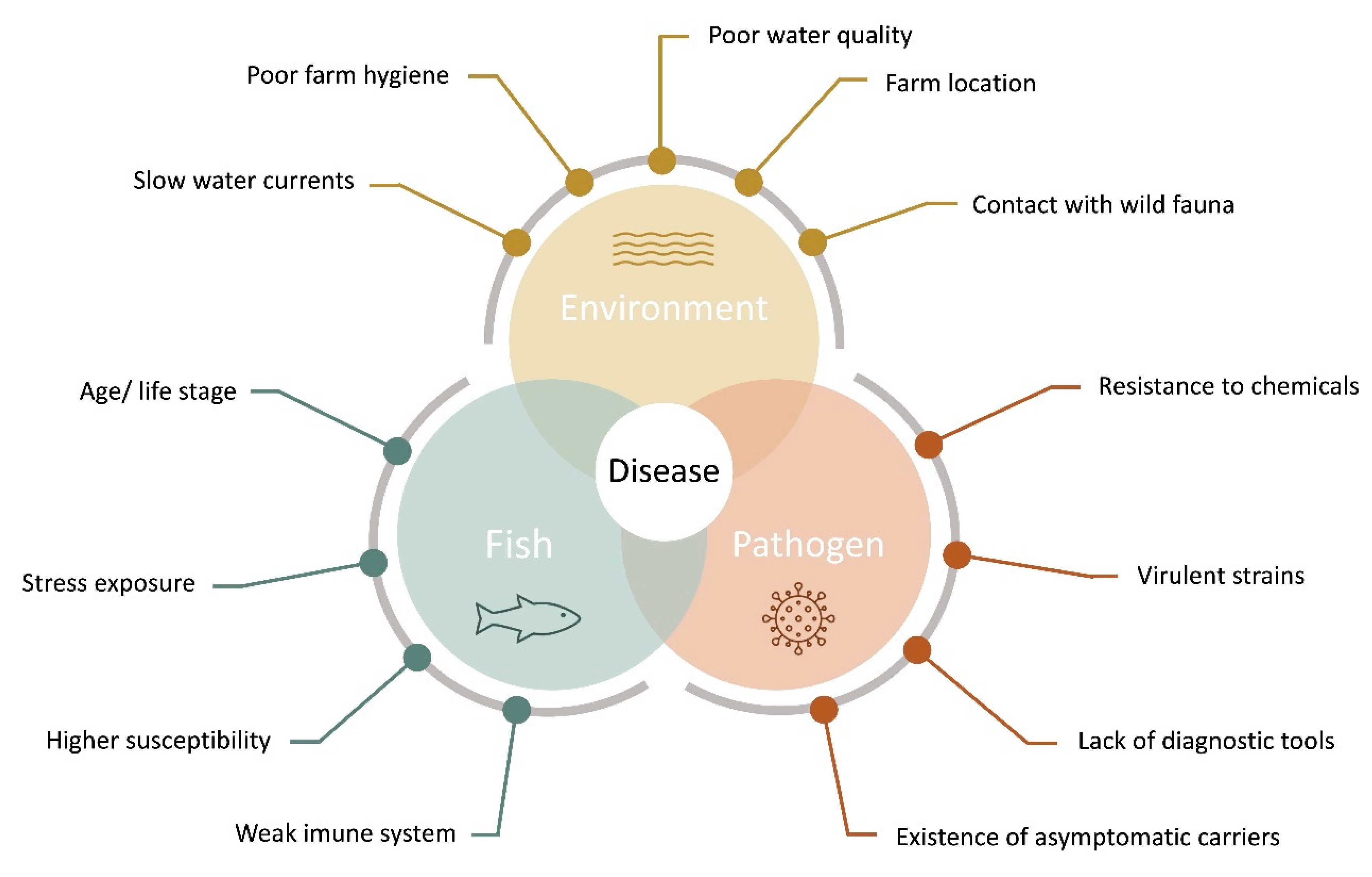
3. Disease Diagnostics
3.1. Pathogen Identification
3.2. Symptomatology
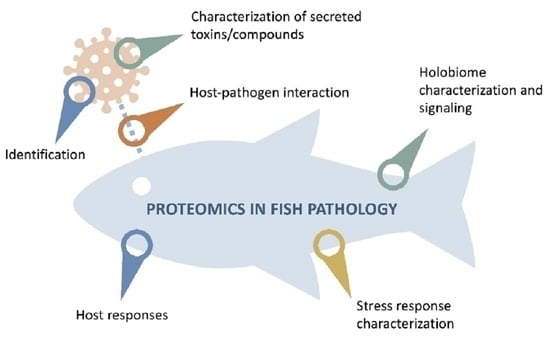
4. Tools for the Study of Host-Pathogen Interactions
4.1. The “Holobiome” Approach: Metagenomics and Metaproteomics
4.2. Omics-Based Strategies and Protein-Protein Interaction (PPI) Networks
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bank, T.W. Fish to 2030—Prospects for Fisheries and Aquaculture; International Bank for Reconstruction and Development/International Development Association or The World Bank: Washington, DC, USA, 2013; p. 77. [Google Scholar]
- FAO. The State of World Fisheries and Aquaculture 2020. Sustainability in Action; FAO: Rome, Italy, 2020. [Google Scholar]
- Rodrigues, P.M.; Schrama, D.; Campos, A.; Osório, H.; Freitas, M. Applications of Proteomics in Aquaculture. In Agricultural Proteomics Volume 1: Crops, Horticulture, Farm Animals, Food, Insect and Microorganisms; Salekdeh, G.H., Ed.; Springer International Publishing: Cham, Switzerland, 2016; pp. 165–199. [Google Scholar]
- Murray, A.G.; Peeler, E.J. A framework for understanding the potential for emerging diseases in aquaculture. Prev. Vet. Med. 2005, 67, 223–235. [Google Scholar] [CrossRef] [PubMed]
- Waagbø, R. Chapter 13 Feeding and disease resistance in fish. In Biology of Growing Animals; Mosenthin, R., Zentek, J., Żebrowska, T., Eds.; Elsevier: Amsterdam, The Netherlands, 2006; Volume 4, pp. 387–415. [Google Scholar]
- Barber, I. Parasites, behaviour and welfare in fish. Appl. Anim. Behav. Sci. 2007, 104, 251–264. [Google Scholar] [CrossRef]
- Brugere, C.; Onuigbo, D.M.; Morgan, K.L. People matter in animal disease surveillance: Challenges and opportunities for the aquaculture sector. Aquaculture 2017, 467, 158–169. [Google Scholar] [CrossRef]
- Chintagari, S.; Hazard, N.; Edwards, G.; Jadeja, R.; Janes, M. Risks associated with fish and seafood. Preharvest Food Saf. 2018, 123–142. [Google Scholar] [CrossRef]
- Hill, B.J. The need for effective disease control in international aquaculture. Dev. Biol. 2005, 121, 3–12. [Google Scholar]
- Shinn, A.P.; Pratoomyot, J.; Bron, J.E.; Paladini, G.; Brooker, E.E.; Brooker, A.J. Economic costs of protistan and metazoan parasites to global mariculture. Parasitology 2015, 142, 196–270. [Google Scholar] [CrossRef]
- Iwama, G.K. The welfare of fish. Dis. Aquat. Org. 2007, 75, 155–158. [Google Scholar] [CrossRef]
- Adams, A.; Thompson, K.D. Biotechnology offers revolution to fish health management. Trends Biotechnol. 2006, 24, 201–205. [Google Scholar] [CrossRef]
- Aung, M.M.; Chang, Y.S. Traceability in a food supply chain: Safety and quality perspectives. Food Control 2014, 39, 172–184. [Google Scholar] [CrossRef]
- Trienekens, J.; Zuurbier, P. Quality and safety standards in the food industry, developments and challenges. Int. J. Prod. Econ. 2008, 113, 107–122. [Google Scholar] [CrossRef]
- Peeler, E.J.; Taylor, N.G.H. The application of epidemiology in aquatic animal health -opportunities and challenges. Vet. Res. 2011, 42, 94. [Google Scholar] [CrossRef] [PubMed]
- Hedrick, R.P. Movement of pathogens with the international trade of live fish: Problems and solutions. Rev. Sci. Tech. 1996, 15, 523–531. [Google Scholar] [CrossRef] [PubMed]
- Cameron, A. Survey Toolbox for Aquatic Animal Diseases: A Practical Manual and Software Package; ACIAR Monograph No. 94; Australian Centre for International Agricultural Research: Canberra, Australia, 2002; 375p.
- Oidtmann, B.C.; Peeler, E.J.; Thrush, M.A.; Cameron, A.R.; Reese, R.A.; Pearce, F.M.; Dunn, P.; Lyngstad, T.M.; Tavornpanich, S.; Brun, E.; et al. Expert consultation on risk factors for introduction of infectious pathogens into fish farms. Prev. Vet. Med. 2014, 115, 238–254. [Google Scholar] [CrossRef] [PubMed]
- Sitjà-Bobadilla, A.; Oidtmann, B. Chapter 5—Integrated Pathogen Management Strategies in Fish Farming. In Fish Diseases; Jeney, G., Ed.; Academic Press: Cambridge, MA, USA, 2017; pp. 119–144. [Google Scholar]
- Freitas, J.; Vaz-Pires, P.; Câmara, J.S. From aquaculture production to consumption: Freshness, safety, traceability and authentication, the four pillars of quality. Aquaculture 2020, 518, 734857. [Google Scholar] [CrossRef]
- Scarfe, A.D.; Palić, D. Aquaculture biosecurity: Practical approach to prevent, control, and eradicate diseases. In Aquaculture Health Management; Elsevier: Amsterdam, The Netherlands, 2020; pp. 75–116. [Google Scholar]
- Rodrigues, P.M.; Martin, S.A.M.; Silva, T.S.; Boonanuntanasarn, S.; Schrama, D.; Moreira, M.; Raposo, C. Proteomics in Fish and Aquaculture Research. In Proteomics in Domestic Animals: From Farm to Systems Biology; de Almeida, A.M., Eckersall, D., Miller, I., Eds.; Springer International Publishing: Cham, Switzerland, 2018; pp. 311–338. [Google Scholar]
- Cash, P. Proteomics in the study of the molecular taxonomy and epidemiology of bacterial pathogens. Electrophoresis 2009, 30, S133–S141. [Google Scholar] [CrossRef]
- Lafferty, K.D. The ecology of climate change and infectious diseases. Ecology 2009, 90, 888–900. [Google Scholar] [CrossRef]
- Parrington, J.; Coward, K. Use of emerging genomic and proteomic technologies in fish physiology. Aquat. Living Resour. 2002, 15, 193–196. [Google Scholar] [CrossRef]
- Burge, C.A.; Friedman, C.S.; Getchell, R.; House, M.; Lafferty, K.D.; Mydlarz, L.D.; Prager, K.C.; Sutherland, K.P.; Renault, T.; Kiryu, I.; et al. Complementary approaches to diagnosing marine diseases: A union of the modern and the classic. Philos. Trans. R. Soc. B Biol. Sci. 2016, 371. [Google Scholar] [CrossRef]
- Gotesman, M.; Menanteau-Ledouble, S.; Saleh, M.; Bergmann, S.M.; El-Matbouli, M. A new age in AquaMedicine: Unconventional approach in studying aquatic diseases. BMC Vet. Res. 2018, 14, 178. [Google Scholar] [CrossRef]
- Oskoueian, E.; Eckersall, P.D.; Bencurova, E.; Dandekar, T. Application of Proteomic Biomarkers in Livestock Disease Management. In Agricultural Proteomics Volume 2: Environmental Stresses; Salekdeh, G.H., Ed.; Springer International Publishing: Cham, Switzerland, 2016; pp. 299–310. [Google Scholar]
- Alves, R.N.; Cordeiro, O.; Silva, T.S.; Richard, N.; de Vareilles, M.; Marino, G.; Di Marco, P.; Rodrigues, P.M.; Conceição, L.E.C. Metabolic molecular indicators of chronic stress in gilthead seabream (Sparus aurata) using comparative proteomics. Aquaculture 2010, 299, 57–66. [Google Scholar] [CrossRef]
- Rodrigues, P.M.; Silva, T.S.; Dias, J.; Jessen, F. PROTEOMICS in aquaculture: Applications and trends. J. Proteom. 2012, 75, 4325–4345. [Google Scholar] [CrossRef] [PubMed]
- Wilkins, M.R.; Pasquali, C.; Appel, R.D.; Ou, K.; Golaz, O.; Sanchez, J.-C.; Yan, J.X.; Gooley, A.A.; Hughes, G.; Humphery-Smith, I.; et al. From Proteins to Proteomes: Large Scale Protein Identification by Two-Dimensional Electrophoresis and Arnino Acid Analysis. Bio/Technology 1996, 14, 61–65. [Google Scholar] [CrossRef] [PubMed]
- Cox, J.; Mann, M. Quantitative, high-resolution proteomics for data-driven systems biology. Annu. Rev. Biochem. 2011, 80, 273–299. [Google Scholar] [CrossRef]
- Aebersold, R.; Mann, M. Mass-spectrometric exploration of proteome structure and function. Nature 2016, 537, 347–355. [Google Scholar] [CrossRef] [PubMed]
- Parker, C.G.; Pratt, M.R. Click Chemistry in Proteomic Investigations. Cell 2020, 180, 605–632. [Google Scholar] [CrossRef]
- Huntingford, F.A.; Adams, C.; Braithwaite, V.A.; Kadri, S.; Pottinger, T.G.; Sandøe, P.; Turnbull, J.F. Current issues in fish welfare. J. Fish Biol. 2006, 68, 332–372. [Google Scholar] [CrossRef]
- Brown, C. Fish intelligence, sentience and ethics. Anim. Cogn. 2015, 18, 1–17. [Google Scholar] [CrossRef]
- Ashley, P.J. Fish welfare: Current issues in aquaculture. Appl. Anim. Behav. Sci. 2007, 104, 199–235. [Google Scholar] [CrossRef]
- Martins, C.I.M.; Galhardo, L.; Noble, C.; Damsgård, B.; Spedicato, M.T.; Zupa, W.; Beauchaud, M.; Kulczykowska, E.; Massabuau, J.-C.; Carter, T.; et al. Behavioural indicators of welfare in farmed fish. Fish Physiol. Biochem. 2012, 38, 17–41. [Google Scholar] [CrossRef]
- Dawkins, M.S. Evolution and Animal Welfare. Q. Rev. Biol. 1998, 73, 305–328. [Google Scholar] [CrossRef]
- Saraiva, J.L.; Castanheira, M.F.; Arechavala-López, P.; Volstorf, J.; Studer, B.H. Domestication and welfare in farmed fish. In Animal Domestication; IntechOpen: London, UK, 2018. [Google Scholar]
- Wall, T. Disease and Medicines—The Welfare Implications. In Fish Welfare; Blackwell Publishing Ltd.: Oxford, Uk, 2008; pp. 195–201. [Google Scholar]
- Sørum, U.; Damsgård, B. Effects of anaesthetisation and vaccination on feed intake and growth in Atlantic salmon (Salmo salar L.). Aquaculture 2004, 232, 333–341. [Google Scholar] [CrossRef]
- Huntingford, F.A.; Kadri, S. Defining, assessing and promoting the welfare of farmed fish. Rev. Sci. Tech. 2014, 33, 233–244. [Google Scholar] [CrossRef] [PubMed]
- Segner, H.; Sundh, H.; Buchmann, K.; Douxfils, J.; Sundell, K.S.; Mathieu, C.; Ruane, N.; Jutfelt, F.; Toften, H.; Vaughan, L. Health of farmed fish: Its relation to fish welfare and its utility as welfare indicator. Fish Physiol. Biochem. 2012, 38, 85–105. [Google Scholar] [CrossRef] [PubMed]
- Conte, F.S. Stress and the welfare of cultured fish. Appl. Anim. Behav. Sci. 2004, 86, 205–223. [Google Scholar] [CrossRef]
- Ndong, D.; Chen, Y.-Y.; Lin, Y.-H.; Vaseeharan, B.; Chen, J.-C. The immune response of tilapia Oreochromis mossambicus and its susceptibility to Streptococcus iniae under stress in low and high temperatures. Fish Shellfish Immunol. 2007, 22, 686–694. [Google Scholar] [CrossRef] [PubMed]
- Ellison, A.R.; Uren Webster, T.M.; Rey, O.; Garcia de Leaniz, C.; Consuegra, S.; Orozco-terWengel, P.; Cable, J. Transcriptomic response to parasite infection in Nile tilapia (Oreochromis niloticus) depends on rearing density. BMC Genom. 2018, 19, 723. [Google Scholar] [CrossRef]
- Jiang, I.-F.; Bharath Kumar, V.; Lee, D.-N.; Weng, C.-F. Acute osmotic stress affects Tilapia (Oreochromis mossambicus) innate immune responses. Fish Shellfish Immunol. 2008, 25, 841–846. [Google Scholar] [CrossRef]
- Caipang, C.M.A.; Brinchmann, M.F.; Berg, I.; Iversen, M.; Eliassen, R.; Kiron, V. Changes in selected stress and immune-related genes in Atlantic cod, Gadus morhua, following overcrowding. Aquac. Res. 2008, 39, 1533–1540. [Google Scholar] [CrossRef]
- Korytář, T.; Nipkow, M.; Altmann, S.; Goldammer, T.; Köllner, B.; Rebl, A. Adverse Husbandry of Maraena Whitefish Directs the Immune System to Increase Mobilization of Myeloid Cells and Proinflammatory Responses. Front. Immunol. 2016, 7, 631. [Google Scholar] [CrossRef]
- Caipang, C.M.A.; Berg, I.; Brinchmann, M.F.; Kiron, V. Short-term crowding stress in Atlantic cod, Gadus morhua L. modulates the humoral immune response. Aquaculture 2009, 295, 110–115. [Google Scholar] [CrossRef]
- Costas, B.; Conceição, L.E.C.; Aragão, C.; Martos, J.A.; Ruiz-Jarabo, I.; Mancera, J.M.; Afonso, A. Physiological responses of Senegalese sole (Solea senegalensis Kaup, 1858) after stress challenge: Effects on non-specific immune parameters, plasma free amino acids and energy metabolism. Aquaculture 2011, 316, 68–76. [Google Scholar] [CrossRef]
- Fast, M.D.; Hosoya, S.; Johnson, S.C.; Afonso, L.O.B. Cortisol response and immune-related effects of Atlantic salmon (Salmo salar Linnaeus) subjected to short- and long-term stress. Fish Shellfish Immunol. 2008, 24, 194–204. [Google Scholar] [CrossRef] [PubMed]
- Vazzana, M.; Cammarata, M.; Cooper, E.L.; Parrinello, N. Confinement stress in sea bass (Dicentrarchus labrax) depresses peritoneal leukocyte cytotoxicity. Aquaculture 2002, 210, 231–243. [Google Scholar] [CrossRef]
- Mauri, I.; Romero, A.; Acerete, L.; MacKenzie, S.; Roher, N.; Callol, A.; Cano, I.; Alvarez, M.C.; Tort, L. Changes in complement responses in Gilthead seabream (Sparus aurata) and European seabass (Dicentrarchus labrax) under crowding stress, plus viral and bacterial challenges. Fish Shellfish Immunol. 2011, 30, 182–188. [Google Scholar] [CrossRef]
- MacKenzie, S.; Iliev, D.; Liarte, C.; Koskinen, H.; Planas, J.V.; Goetz, F.W.; Mölsä, H.; Krasnov, A.; Tort, L. Transcriptional analysis of LPS-stimulated activation of trout (Oncorhynchus mykiss) monocyte/macrophage cells in primary culture treated with cortisol. Mol. Immunol. 2006, 43, 1340–1348. [Google Scholar] [CrossRef] [PubMed]
- Douxfils, J.; Lambert, S.; Mathieu, C.; Milla, S.; Mandiki, S.N.M.; Henrotte, E.; Wang, N.; Dieu, M.; Raes, M.; Rougeot, C.; et al. Influence of domestication process on immune response to repeated emersion stressors in Eurasian perch (Perca fluviatilis L.). Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2014, 173, 52–60. [Google Scholar] [CrossRef]
- de Magalhães, C.R.; Schrama, D.; Farinha, A.P.; Revets, D.; Kuehn, A.; Planchon, S.; Rodrigues, P.M.; Cerqueira, M. Protein changes as robust signatures of fish chronic stress: A proteomics approach to fish welfare research. BMC Genom. 2020, 21, 309. [Google Scholar] [CrossRef]
- Moberg, G.P. Biological response to stress: Implications for animal welfare. Biol. Anim. Stress Basic Princ. Implic. Anim. Welf. 2000, 1, 21. [Google Scholar]
- McEwen, B.S.; Wingfield, J.C. What is in a name? Integrating homeostasis, allostasis and stress. Horm. Behav. 2010, 57, 105–111. [Google Scholar] [CrossRef]
- Wendelaar Bonga, S.E. The stress response in fish. Physiol. Rev. 1997, 77, 591–625. [Google Scholar] [CrossRef]
- Pankhurst, N.W. The endocrinology of stress in fish: An environmental perspective. Gen. Comp. Endocrinol. 2011, 170, 265–275. [Google Scholar] [CrossRef] [PubMed]
- Korte, S.M.; Olivier, B.; Koolhaas, J.M. A new animal welfare concept based on allostasis. Physiol. Behav. 2007, 92, 422–428. [Google Scholar] [CrossRef] [PubMed]
- Boonstra, R. Reality as the leading cause of stress: Rethinking the impact of chronic stress in nature. Funct. Ecol. 2013, 27, 11–23. [Google Scholar] [CrossRef]
- Holden, C. Researchers Pained by Effort to Define Distress Precisely. Science 2000, 290, 1474–1475. [Google Scholar] [CrossRef] [PubMed]
- Yada, T.; Tort, L. Interactions. In Fish Physiology; Schreck, C.B., Tort, L., Farrell, A.P., Brauner, C.J., Eds.; Academic Press: Cambridge, MA, USA, 2016; Volume 35, pp. 365–403. [Google Scholar]
- Gomez, D.; Sunyer, J.O.; Salinas, I. The mucosal immune system of fish: The evolution of tolerating commensals while fighting pathogens. Fish Shellfish Immunol. 2013, 35, 1729–1739. [Google Scholar] [CrossRef]
- Olsen, R.E.; Sundell, K.; Hansen, T.; Hemre, G.-I.; Myklebust, R.; Mayhew, T.M.; Ringø, E. Acute stress alters the intestinal lining of Atlantic salmon, Salmo salar L.: An electron microscopical study. Fish Physiol. Biochem. 2002, 26, 211–221. [Google Scholar] [CrossRef]
- Olsen, R.E.; Sundell, K.; Mayhew, T.M.; Myklebust, R.; Ringø, E. Acute stress alters intestinal function of rainbow trout, Oncorhynchus mykiss (Walbaum). Aquaculture 2005, 250, 480–495. [Google Scholar] [CrossRef]
- Sundh, H.; Kvamme, B.O.; Fridell, F.; Olsen, R.E.; Ellis, T.; Taranger, G.L.; Sundell, K. Intestinal barrier function of Atlantic salmon (Salmo salar L.) post smolts is reduced by common sea cage environments and suggested as a possible physiological welfare indicator. BMC Physiol. 2010, 10, 22. [Google Scholar] [CrossRef]
- Douxfils, J.; Mathieu, C.; Mandiki, S.N.M.; Milla, S.; Henrotte, E.; Wang, N.; Vandecan, M.; Dieu, M.; Dauchot, N.; Pigneur, L.M.; et al. Physiological and proteomic evidences that domestication process differentially modulates the immune status of juvenile Eurasian perch (Perca fluviatilis) under chronic confinement stress. Fish Shellfish Immunol. 2011, 31, 1113–1121. [Google Scholar] [CrossRef]
- Saeij, J.P.; Verburg-van Kemenade, L.B.; van Muiswinkel, W.B.; Wiegertjes, G.F. Daily handling stress reduces resistance of carp to Trypanoplasma borreli: In vitro modulatory effects of cortisol on leukocyte function and apoptosis. Dev. Comp. Immunol. 2003, 27, 233–245. [Google Scholar] [CrossRef]
- Pruett, S.B. Stress and the immune system. Pathophysiology 2003, 9, 133–153. [Google Scholar] [CrossRef]
- Esteban, M.A.; Rodríguez, A.; Ayala, A.G.; Meseguer, J. Effects of high doses of cortisol on innate cellular immune response of seabream (Sparus aurata L.). Gen. Comp. Endocrinol. 2004, 137, 89–98. [Google Scholar] [CrossRef] [PubMed]
- Baschant, U.; Tuckermann, J. The role of the glucocorticoid receptor in inflammation and immunity. J. Steroid Biochem. Mol. Biol. 2010, 120, 69–75. [Google Scholar] [CrossRef] [PubMed]
- Castanheira, M.F.; Conceição, L.E.C.; Millot, S.; Rey, S.; Bégout, M.-L.; Damsgård, B.; Kristiansen, T.; Höglund, E.; Øverli, Ø.; Martins, C.I.M. Coping styles in farmed fish: Consequences for aquaculture. Rev. Aquac. 2017, 9, 23–41. [Google Scholar] [CrossRef]
- Koakoski, G.; Oliveira, T.A.; da Rosa, J.G.; Fagundes, M.; Kreutz, L.C.; Barcellos, L.J. Divergent time course of cortisol response to stress in fish of different ages. Physiol. Behav. 2012, 106, 129–132. [Google Scholar] [CrossRef]
- Madaro, A.; Fernö, A.; Kristiansen, T.S.; Olsen, R.E.; Gorissen, M.; Flik, G.; Nilsson, J. Effect of predictability on the stress response to chasing in Atlantic salmon (Salmo salar L.) parr. Physiol. Behav. 2016, 153, 1–6. [Google Scholar] [CrossRef]
- Martinez-Porchas, M.; Martinez-Cordova, L.R.; Ramos-Enriquez, R. Cortisol and glucose: Reliable indicators of fish stress. Pan-Am. J. Aquat. Sci. 2009, 4, 158–178. [Google Scholar]
- Tort, L. Stress and immune modulation in fish. Dev. Comp. Immunol. 2011, 35, 1366–1375. [Google Scholar] [CrossRef]
- Peng, X.-X. Proteomics and its applications to aquaculture in China: Infection, immunity, and interaction of aquaculture hosts with pathogens. Dev. Comp. Immunol. 2013, 39, 63–71. [Google Scholar] [CrossRef]
- Provan, F.; Jensen, L.B.; Uleberg, K.E.; Larssen, E.; Rajalahti, T.; Mullins, J.; Obach, A. Proteomic analysis of epidermal mucus from sea lice–infected Atlantic salmon, Salmo salar L. J. Fish Dis. 2013, 36, 311–321. [Google Scholar] [CrossRef]
- Rajan, B.; Lokesh, J.; Kiron, V.; Brinchmann, M.F. Differentially expressed proteins in the skin mucus of Atlantic cod (Gadus morhua) upon natural infection with Vibrio anguillarum. BMC Vet. Res. 2013, 9, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Easy, R.H.; Ross, N.W. Changes in Atlantic salmon Salmo salar mucus components following short- and long-term handling stress. J. Fish Biol. 2010, 77, 1616–1631. [Google Scholar] [CrossRef] [PubMed]
- Cordero, H.; Morcillo, P.; Cuesta, A.; Brinchmann, M.F.; Esteban, M.A. Differential proteome profile of skin mucus of gilthead seabream (Sparus aurata) after probiotic intake and/or overcrowding stress. J. Proteom. 2016, 132, 41–50. [Google Scholar] [CrossRef]
- Guardiola, F.A.; Cuartero, M.; Del Mar Collado-González, M.; Díaz Baños, F.G.; Cuesta, A.; Moriñigo, M.; Esteban, M. Terminal carbohydrates abundance, immune related enzymes, bactericidal activity and physico-chemical parameters of the Senegalese sole (Solea senegalensis, Kaup) skin mucus. Fish Shellfish Immunol. 2017, 60, 483–491. [Google Scholar] [CrossRef] [PubMed]
- Isani, G.; Andreani, G.; Carpenè, E.; Di Molfetta, S.; Eletto, D.; Spisni, E. Effects of waterborne Cu exposure in gilthead sea bream (Sparus aurata): A proteomic approach. Fish Shellfish Immunol. 2011, 31, 1051–1058. [Google Scholar] [CrossRef]
- Moreira, M.; Schrama, D.; Soares, F.; Wulff, T.; Pousão-Ferreira, P.; Rodrigues, P. Physiological responses of reared sea bream (Sparus aurata Linnaeus, 1758) to an Amyloodinium ocellatum outbreak. J. Fish Dis. 2017, 40, 1545–1560. [Google Scholar] [CrossRef]
- Xiong, X.-P.; Dong, C.-F.; Xu, X.; Weng, S.-P.; Liu, Z.-Y.; He, J.-G. Proteomic analysis of zebrafish (Danio rerio) infected with infectious spleen and kidney necrosis virus. Dev. Comp. Immunol. 2011, 35, 431–440. [Google Scholar] [CrossRef]
- Ji, C.; Wu, H.; Wei, L.; Zhao, J.; Wang, Q.; Lu, H. Responses of Mytilus galloprovincialis to bacterial challenges by metabolomics and proteomics. Fish Shellfish Immunol. 2013, 35, 489–498. [Google Scholar] [CrossRef]
- Lü, A.; Hu, X.; Wang, Y.; Shen, X.; Li, X.; Zhu, A.; Tian, J.; Ming, Q.; Feng, Z. iTRAQ analysis of gill proteins from the zebrafish (Danio rerio) infected with Aeromonas hydrophila. Fish Shellfish Immunol. 2014, 36, 229–239. [Google Scholar] [CrossRef]
- Buján, N.; Hernández-Haro, C.; Monteoliva, L.; Gil, C.; Magariños, B. Comparative proteomic study of Edwardsiella tarda strains with different degrees of virulence. J. Proteom. 2015, 127 Part B, 310–320. [Google Scholar] [CrossRef]
- Xu, D.; Song, L.; Wang, H.; Xu, X.; Wang, T.; Lu, L. Proteomic analysis of cellular protein expression profiles in response to grass carp reovirus infection. Fish Shellfish Immunol. 2015, 44, 515–524. [Google Scholar] [CrossRef] [PubMed]
- Braceland, M.; Bickerdike, R.; Tinsley, J.; Cockerill, D.; McLoughlin, M.F.; Graham, D.A.; Burchmore, R.J.; Weir, W.; Wallace, C.; Eckersall, P.D. The serum proteome of Atlantic salmon, Salmo salar, during pancreas disease (PD) following infection with salmonid alphavirus subtype 3 (SAV3). J. Proteom. 2013, 94, 423–436. [Google Scholar] [CrossRef]
- Kumar, G.; Hummel, K.; Noebauer, K.; Welch, T.J.; Razzazi-Fazeli, E.; El-Matbouli, M. Proteome analysis reveals a role of rainbow trout lymphoid organs during Yersinia ruckeri infection process. Sci. Rep. 2018, 8, 13998. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.D.; Hsu, L.S.; Chien, C.C.; Chen, S.C. Proteomic analysis of ametryn toxicity in zebrafish embryos. Environ. Toxicol. 2018, 33, 579–586. [Google Scholar] [CrossRef] [PubMed]
- Veiseth-Kent, E.; Grove, H.; Færgestad, E.M.; Fjæra, S.O. Changes in muscle and blood plasma proteomes of Atlantic salmon (Salmo salar) induced by crowding. Aquaculture 2010, 309, 272–279. [Google Scholar] [CrossRef]
- Singhal, N.; Kumar, M.; Kanaujia, P.; Virdi, J. MALDI-TOF mass spectrometry: An emerging technology for microbial identification and diagnosis. Front Microbiol. 2015, 6, 791. [Google Scholar] [CrossRef] [PubMed]
- Song, W.J.; Qin, Q.W.; Qiu, J.; Huang, C.H.; Wang, F.; Hew, C.L. Functional Genomics Analysis of Singapore Grouper Iridovirus: Complete Sequence Determination and Proteomic Analysis. J. Virol. 2004, 78, 12576–12590. [Google Scholar] [CrossRef]
- Zhou, S.; Wan, Q.; Huang, Y.; Huang, X.; Cao, J.; Ye, L.; Lim, T.K.; Lin, Q.; Qin, Q. Proteomic analysis of Singapore grouper iridovirus envelope proteins and characterization of a novel envelope protein VP088. Proteomics 2011, 11, 2236–2248. [Google Scholar] [CrossRef]
- Vancsok, C.; Peñaranda, M.M.D.; Raj, V.S.; Leroy, B.; Jazowiecka-Rakus, J.; Boutier, M.; Gao, Y.; Wilkie, G.S.; Suárez, N.M.; Wattiez, R.; et al. Proteomic and Functional Analyses of the Virion Transmembrane Proteome of Cyprinid Herpesvirus 3. J. Virol. 2017, 91, e01209–e01217. [Google Scholar] [CrossRef]
- Chuang, M.-Y.; Tsai, W.-C.; Kuo, T.-Y.; Chen, H.-M.; Chen, W.-J. Comparative proteome analysis reveals proteins involved in salt adaptation in Photobacterium damselae subsp. piscicida. J. Basic Microbiol. 2016, 56, 1234–1243. [Google Scholar] [CrossRef]
- Puentes, B.; Balado, M.; Bermúdez-Crespo, J.; Osorio, C.R.; Lemos, M.L. A proteomic analysis of the iron response of Photobacterium damselae subsp. damselae reveals metabolic adaptations to iron levels changes and novel potential virulence factors. Vet. Microbiol. 2017, 201, 257–264. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Sancho, M.; Vela Ana, I.; Awad, M.; Kostrzewa, M.; Domínguez, L.; Fernández-Garayzábal, J.F. Differentiation of Photobacterium damselae subspecies using Matrix-Assisted Laser-Desorption/Ionization Time-of-Flight Mass Spectrometry (MALDI-TOF MS) in fish isolates. Aquaculture 2016, 464, 159–164. [Google Scholar] [CrossRef]
- Tan, Y.P.; Lin, Q.; Wang, X.H.; Joshi, S.; Hew, C.L.; Leung, K.Y. Comparative Proteomic Analysis of Extracellular Proteins of Edwardsiella tarda. Infect. Immun. 2002, 70, 6475–6480. [Google Scholar] [CrossRef]
- Srinivasa Rao, P.S.; Yamada, Y.; Tan, Y.P.; Leung, K.Y. Use of proteomics to identify novel virulence determinants that are required for Edwardsiella tarda pathogenesis. Mol. Microbiol. 2004, 53, 573–586. [Google Scholar] [CrossRef] [PubMed]
- Kumar, G.; Hummel, K.; Welch, T.J.; Razzazi-Fazeli, E.; El-Matbouli, M. Global proteomic profiling of Yersinia ruckeri strains. Vet. Res. 2017, 48, 55. [Google Scholar] [CrossRef] [PubMed]
- Kumar, G.; Hummel, K.; Ahrens, M.; Menanteau-Ledouble, S.; Welch, T.J.; Eisenacher, M.; Razzazi-Fazeli, E.; El-Matbouli, M. Shotgun proteomic analysis of Yersinia ruckeri strains under normal and iron-limited conditions. Vet. Res. 2016, 47, 100. [Google Scholar] [CrossRef]
- Ormsby, M.J.; Grahame, E.; Burchmore, R.; Davies, R.L. Comparative bioinformatic and proteomic approaches to evaluate the outer membrane proteome of the fish pathogen Yersinia ruckeri. J. Proteom. 2019, 199, 135–147. [Google Scholar] [CrossRef]
- Dumpala, P.R.; Gülsoy, N.; Lawrence, M.L.; Karsi, A. Proteomic analysis of the fish pathogen Flavobacterium columnare. Proteome Sci. 2010, 8, 26. [Google Scholar] [CrossRef]
- LaFrentz, B.R.; LaPatra, S.E.; Call, D.R.; Wiens, G.D.; Cain, K.D. Proteomic analysis of Flavobacterium psychrophilum cultured in vivo and in iron-limited media. Dis. Aquat. Org. 2009, 87, 171–182. [Google Scholar] [CrossRef]
- Pérez-Sancho, M.; Vela, A.I.; Wiklund, T.; Kostrzewa, M.; Domínguez, L.; Fernández-Garayzábal, J.F. Differentiation of Flavobacterium psychrophilum from Flavobacterium psychrophilum-like species by MALDI-TOF mass spectrometry. Res. Vet. Sci. 2017, 115, 345–352. [Google Scholar] [CrossRef]
- Shen, C.-J.; Kuo, T.-Y.; Lin, C.-C.; Chow, L.-P.; Chen, W.-J. Proteomic identification of membrane proteins regulating antimicrobial peptide resistance in Vibrio parahaemolyticus. J. Appl. Microbiol. 2010, 108, 1398–1407. [Google Scholar] [CrossRef] [PubMed]
- Tavares, G.C.; Carvalho, A.F.; Pereira, F.L.; Rezende, C.P.; Azevedo, V.A.C.; Leal, C.A.G.; Figueiredo, H.C.P. Transcriptome and Proteome of Fish-Pathogenic Streptococcus agalactiae Are Modulated by Temperature. Front. Microbiol. 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Yolanda, T.-C.; Clara, F.-Á.; Ysabel, S. Proteomic and molecular fingerprinting for identification and tracking of fish pathogenic Streptococcus. Aquaculture 2019, 498, 322–334. [Google Scholar] [CrossRef]
- Kim, S.W.; Jang, H.B.; Lee, J.S.; Im, S.P.; Lazarte, J.M.S.; Seo, J.P.; Lee, W.J.; Kim, J.S.; Jung, T.S. Comparison of proteome typing and serotyping of Streptococcus parauberis isolates from olive flounder (Paralichthys olivaceus). J. Microbiol. Methods 2015, 118, 168–172. [Google Scholar] [CrossRef] [PubMed]
- Torres-Corral, Y.; Santos, Y. Identification and typing of Vagococcus salmoninarum using genomic and proteomic techniques. J. Fish Dis. 2019, 42, 597–612. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.-C.; Zhang, J.; Sun, L. Streptococcus iniae SF1: Complete Genome Sequence, Proteomic Profile, and Immunoprotective Antigens. PLoS ONE 2014, 9, e91324. [Google Scholar] [CrossRef] [PubMed]
- López-Cortés, X.A.; Nachtigall, F.M.; Olate, V.R.; Araya, M.; Oyanedel, S.; Diaz, V.; Jakob, E.; Ríos-Momberg, M.; Santos, L.S. Fast detection of pathogens in salmon farming industry. Aquaculture 2017, 470, 17–24. [Google Scholar] [CrossRef]
- Fernández-Álvarez, C.; Torres-Corral, Y.; Saltos-Rosero, N.; Santos, Y. MALDI-TOF mass spectrometry for rapid differentiation of Tenacibaculum species pathogenic for fish. Appl. Microbiol. Biotechnol. 2017, 101, 5377–5390. [Google Scholar] [CrossRef]
- Fernández-Álvarez, C.; Torres-Corral, Y.; Santos, Y. Use of ribosomal proteins as biomarkers for identification of Flavobacterium psychrophilum by MALDI-TOF mass spectrometry. J. Proteom. 2018, 170, 59–69. [Google Scholar] [CrossRef]
- Kurokawa, S.; Kabayama, J.; Fukuyasu, T.; Hwang, S.D.; Park, C.-I.; Park, S.-B.; del Castillo, C.S.; Hikima, J.-i.; Jung, T.-S.; Kondo, H.; et al. Bacterial Classification of Fish-Pathogenic Mycobacterium Species by Multigene Phylogenetic Analyses and MALDI Biotyper Identification System. Mar. Biotechnol. 2013, 15, 340–348. [Google Scholar] [CrossRef]
- Böhme, K.; Fernández-No, I.C.; Barros-Velázquez, J.; Gallardo, J.M.; Calo-Mata, P.; Cañas, B. Species Differentiation of Seafood Spoilage and Pathogenic Gram-Negative Bacteria by MALDI-TOF Mass Fingerprinting. J. Proteome Res. 2010, 9, 3169–3183. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, V.; Rezinciuc, S.; Bulone, V. Quantitative Proteomic Analysis of Four Developmental Stages of Saprolegnia parasitica. Front. Microbiol. 2018, 8. [Google Scholar] [CrossRef] [PubMed]
- Tan, Y.P.; Rao, P.S.S.; Zhang, Y.L.; Hew, C.L.; Leung, K.Y. Identification of virulence genes in bacterial fish pathogens: A genomic and proteomic approach. In Aquatic Genomics: Steps Toward a Great Future; Shimizu, N., Aoki, T., Hirono, I., Takashima, F., Eds.; Springer: Tokyo, Japan, 2003; pp. 340–351. [Google Scholar]
- Ahmed, F.; Kumar, G.; Soliman, F.M.; Adly, M.A.; Soliman, H.A.M.; El-Matbouli, M.; Saleh, M. Proteomics for understanding pathogenesis, immune modulation and host pathogen interactions in aquaculture. Comp. Biochem. Physiol. Part D Genom. Proteom. 2019, 32, 100625. [Google Scholar] [CrossRef] [PubMed]
- Toranzo, A.E.; Magariños, B.; Romalde, J.L. A review of the main bacterial fish diseases in mariculture systems. Aquaculture 2005, 246, 37–61. [Google Scholar] [CrossRef]
- Dickerson, H.W.; Dawe, D. Ichthyophthirius multifiliis and Cryptocaryon irritans (phylum Ciliophora). Fish Dis. Disord. 2006, 1, 116–153. [Google Scholar]
- Rimstad, E.; Dale, O.B.; Dannevig, B.H.; Falk, K. Infectious salmon anaemia. Fish Dis. Disord. 2011, 3, 143–165. [Google Scholar]
- Noga, E.J. Fish Disease: Diagnosis and Treatment; John Wiley & Sons: Hoboken, NJ, USA, 2010. [Google Scholar]
- Reddacliff, L.A.; Whittington, R.J. Pathology of epizootic haematopoietic necrosis virus (EHNV) infection in rainbow trout (Oncorhynchus mykiss Walbaum) and redfin perch (Perca fluviatilis L). J. Comp. Pathol. 1996, 115, 103–115. [Google Scholar] [CrossRef]
- Bootland, L.M.; Leong, J. Infectious haematopoietic necrosis virus. Fish Dis. Disord. 2011, 3, 66–109. [Google Scholar]
- Ahmadivand, S.; Soltani, M.; Mardani, K.; Shokrpoor, S.; Hassanzadeh, R.; Ahmadpoor, M.; Rahmati-Holasoo, H.; Meshkini, S. Infectious hematopoietic necrosis virus (IHNV) outbreak in farmed rainbow trout in Iran: Viral isolation, pathological findings, molecular confirmation, and genetic analysis. Virus Res. 2017, 229, 17–23. [Google Scholar] [CrossRef]
- Wolf, K. Fish Viruses and Fish Viral Diseases; Cornell University Press: Ithaca, NY, USA, 2019. [Google Scholar]
- Munro, E.S.; Midtlyng, P.J. Infectious pancreatic necrosis and associated aquatic birnaviruses. Fish Dis. Disord. 2006, 3, 1–65. [Google Scholar]
- Roberts, R.J.; Pearson, M.D. Infectious pancreatic necrosis in Atlantic salmon, Salmo salar L. J. Fish Dis. 2005, 28, 383–390. [Google Scholar] [CrossRef] [PubMed]
- Smail, D.A.; Munro, E.S. The Virology of Teleosts. In Fish Pathology; Wiley-Blackwell: Oxford, UK, 2012; pp. 186–291. [Google Scholar]
- Thorud, K.; Djupvik, H. Infectious anaemia in Atlantic salmon (Salmo salar L.). Bull. Eur. Assoc. Fish Pathol. 1988, 8, 109–111. [Google Scholar]
- McLoughlin, M.F.; Nelson, R.N.; McCormick, J.I.; Rowley, H.M.; Bryson, D.B. Clinical and histopathological features of naturally occurring pancreas disease in farmed Atlantic salmon, Salmo salar L. J. Fish Dis. 2002, 25, 33–43. [Google Scholar] [CrossRef]
- Taksdal, T.; Olsen, A.B.; Bjerkås, I.; Hjortaas, M.J.; Dannevig, B.H.; Graham, D.A.; McLoughlin, M.F. Pancreas disease in farmed Atlantic salmon, Salmo salar L., and rainbow trout, Oncorhynchus mykiss (Walbaum), in Norway. J. Fish Dis. 2007, 30, 545–558. [Google Scholar] [CrossRef] [PubMed]
- Smail, D.A.; Snow, M. Viral haemorrhagic septicaemia. Fish Dis. Disord. 2011, 3, 110–142. [Google Scholar]
- Volpe, E.; Gustinelli, A.; Caffara, M.; Errani, F.; Quaglio, F.; Fioravanti, M.L.; Ciulli, S. Viral nervous necrosis outbreaks caused by the RGNNV/SJNNV reassortant betanodavirus in gilthead sea bream (Sparus aurata) and European sea bass (Dicentrarchus labrax). Aquaculture 2020, 523, 735155. [Google Scholar] [CrossRef]
- Qin, Y.; Liu, J.; Liu, W.; Shi, H.; Jia, A.; Lu, Y.; Liu, X. First isolation and identification of red-grouper nervous necrosis virus (RGNNV) from adult hybrid Hulong grouper (Epinephelus fuscoguttatus × Epinephelus lanceolatus) in China. Aquaculture 2020, 529, 735662. [Google Scholar] [CrossRef]
- Austin, B.; Austin, D.A. Bacterial Fish Pathogens: Disease of Farmed and Wild Fish; Springer: Berlin/Heidelberg, Germany, 2016. [Google Scholar]
- Actis, L.; Tolmasky, M.; Crosa, J. Vibriosis. In Fish Diseases and Disorders; CABI: Wallingford, UK, 2011; Volume 3, pp. 570–605. [Google Scholar]
- Nagano, I.; Inoue, S.; Kawai, K.; Oshima, S.-I. Repeatable immersion infection with Photobacterium damselae subsp. piscicida reproducing clinical signs and moderate mortality. Fish. Sci. 2009, 75, 707–714. [Google Scholar] [CrossRef]
- Toranzo, A.E.; Barreiro, S.N.; Casal, J.F.; Figueras, A.; Magarinos, B.; Barja, J.L. Pasteurellosis in cultured gilthead seabream (Sparus aurata): First report in Spain. Aquaculture 1991, 99, 1–15. [Google Scholar] [CrossRef]
- Gudmundsdottir, B.K.; Bjornsdottir, B. Aeromonas salmonicida and A. hydrophila. In Fish Viruses and Bacteria: Pathobiology and Protection; CABI: Boston, MA, USA, 2017; pp. 173–189. [Google Scholar]
- Bricknell, I.R.; Bowden, T.J.; Bruno, D.W.; MacLachlan, P.; Johnstone, R.; Ellis, A.E. Susceptibility of Atlantic halibut, Hippoglossus hippoglossus (L.) to infection with typical and atypical Aeromonas salmonicida. Aquaculture 1999, 175, 1–13. [Google Scholar] [CrossRef]
- Handlinger, J.; Soltani, M.; Percival, S. The pathology of Flexibacter maritimus in aquaculture species in Tasmania, Australia. J. Fish Dis. 1997, 20, 159–168. [Google Scholar] [CrossRef]
- Ferguson, H.W.; Ostland, V.E.; Byrne, P.; Lumsdsen, J.S. Experimental Production of Bacterial Gill Disease in Trout by Horizontal Transmission and by Bath Challenge. J. Aquat. Anim. Health 1991, 3, 118–123. [Google Scholar] [CrossRef]
- Nilsen, H.; Olsen, A.B.; Vaagnes, Ø.; Hellberg, H.; Bottolfsen, K.; Skjelstad, H.; Colquhoun, D.J. Systemic Flavobacterium psychrophilum infection in rainbow trout, Oncorhynchus mykiss (Walbaum), farmed in fresh and brackish water in Norway. J. Fish Dis. 2011, 34, 403–408. [Google Scholar] [CrossRef] [PubMed]
- Doménech, A.; Fernández-Garayzábal, J.F.; García, J.A.; Cutuli, M.T.; Blanco, M.; Gibello, A.; Moreno, M.A.; Domínguez, L. Association of Pseudomonas anguilliseptica infection with ‘winter disease’ in sea bream, Sparus aurata L. J. Fish Dis. 1999, 22, 69–71. [Google Scholar] [CrossRef]
- Salati, F. Enterococcus seriolicida and streptococcus spp. (S. iniae, S. agalactiae and S. dysgalactiae). Fish Dis. Disord. 2011, 3, 375–396. [Google Scholar]
- Munday, B.; Foster, C.; Roubal, F.; Lester, R. Paramoebic gill infection and associated pathology of Atlantic salmon, Salmo salar and rainbow trout, Salmo gairdneri in Tasmania. In Pathology in Marine Science, Proceedings of the Third International Colloquium on Pathology in Marine Aquaculture, Gloucester Point, VA, USA, 2–6 October 1988; Academic Press: Cambridge, MA, USA, 1990; pp. 215–222. [Google Scholar]
- Rodger, H.; McArdle, J. An outbreak of amoebic gill disease in Ireland. Vet. Rec. 1996, 139, 348. [Google Scholar] [CrossRef] [PubMed]
- Hines, R.S.; Spira, D.T. Ichthyophthirius multifiliis (Fouquet) in the mirror carp, Cyprinus carpio L. I. Course of infection. J. Fish Biol. 1973, 5, 385–392. [Google Scholar]
- Hines, R.S.; Spira, D.T. Ichthyophthiriasis in the mirror carp Cyprinus carpio (L.) V. Acquired immunity. J. Fish Biol. 1974, 6, 373–378. [Google Scholar] [CrossRef]
- Nigrelli, R.; Ruggieri, G. Enzootics in the New York Aquarium caused by Cryptocaryon irritans Brown, 1951, a histophagous ciliate in the skin, eyes and gills of marine fishes. Zoologica 1966, 51, 97–102. [Google Scholar]
- Kuperman, B.I.; Matey, V.E. Massive infestation by Amyloodinium ocellatum (Dinoflagellida) of fish in a highly saline lake, Salton Sea, California, USA. Dis. Aquat. Org. 1999, 39, 65–73. [Google Scholar] [CrossRef]
- Soares, F.; Quental Ferreira, H.; Cunha, E.; Pousão-Ferreira, P. Occurrence of Amyloodinium ocellatum in aquaculture fish production: A serious problem in semi-intensive earthen ponds. Aquacult. Eur. 2011, 36, 13–16. [Google Scholar]
- Marques, C.L.; Medeiros, A.; Moreira, M.; Quental-Ferreira, H.; Mendes, A.C.; Pousão-Ferreira, P.; Soares, F. Report and genetic identification of Amyloodinium ocellatum in a sea bass (Dicentrarchus labrax) broodstock in Portugal. Aquac. Rep. 2019, 14, 100191. [Google Scholar] [CrossRef]
- Basson, L.; Van As, J. Trichodinidae and other ciliophorans (Phylum Ciliophora). Fish Dis. Disord. 2006, 1, 154–182. [Google Scholar]
- Van As, J. An experimental elvaluation of the use of formalin to control trichodiniasis and other ectoparasitic protozoans on fry of Cyprinus carpio L. and Oreochromis mossambicus (Peters). S. Afr. J. Wildl. Res. 24 Mon. Delayed Open Access 1984, 14, 42–48. [Google Scholar]
- Buchmann, K.; Bresciani, J. Monogenea (Phylum Platyhelminthes). Fish Dis. Disord. 2006, 1, 297–344. [Google Scholar]
- Buchmann, K. 11 Gyrodactylus salaris and Gyrodactylus derjavinoides. In Fish Parasites: Pathobiology and Protection; CABI International: London, UK, 2012; p. 193. [Google Scholar]
- Jonsdottir, H.; Bron, J.; Wootten, R.; Turnbull, J. The histopathology associated with the pre-adult and adult stages of Lepeophtheirus salmonis on the Atlantic salmon, Salmo salar L. J. Fish Dis. 1992, 15, 521–527. [Google Scholar] [CrossRef]
- Carrera, M.; Piñeiro, C.; Martinez, I. Proteomic Strategies to Evaluate the Impact of Farming Conditions on Food Quality and Safety in Aquaculture Products. Foods 2020, 9, 1050. [Google Scholar] [CrossRef]
- Näpflin, K.; O’Connor, E.A.; Becks, L.; Bensch, S.; Ellis, V.A.; Hafer-Hahmann, N.; Harding, K.C.; Lindén, S.K.; Olsen, M.T.; Roved, J.; et al. Genomics of host-pathogen interactions: Challenges and opportunities across ecological and spatiotemporal scales. PeerJ 2019, 7, e8013. [Google Scholar] [CrossRef]
- Gallardo-Escárate, C.; Valenzuela-Muñoz, V.; Núñez-Acuña, G.; Carrera, C.; Gonçalves, A.T.; Valenzuela-Miranda, D.; Benavente, B.P.; Roberts, S. Catching the complexity of salmon-louse interactions. Fish Shellfish Immunol. 2019, 90, 199–209. [Google Scholar] [CrossRef]
- Sen, R.; Nayak, L.; De, R.K. A review on host-pathogen interactions: Classification and prediction. Eur. J. Clin. Microbiol. Infect. Dis. 2016, 35, 1581–1599. [Google Scholar] [CrossRef]
- Tello, M.; Valdes, N.; Vargas, R.; Rojas, J.; Parra, M.; Gajardo, G.; Gonzalez, A. Application of Metagenomics to Chilean Aquaculture. In Metagenomics—Basics, Methods and Applications; Hozzein, W.N., Ed.; IntechOpen: London, UK, 2019. [Google Scholar]
- Guerrero, R.; Margulis, L.; Berlanga, M. Symbiogenesis: The holobiont as a unit of evolution. Int. Microbiol. 2013, 16, 133–143. [Google Scholar] [CrossRef]
- Thaiss, C.A.; Zmora, N.; Levy, M.; Elinav, E. The microbiome and innate immunity. Nature 2016, 535, 65–74. [Google Scholar] [CrossRef] [PubMed]
- Saito, M.A.; Bertrand, E.M.; Duffy, M.E.; Gaylord, D.A.; Held, N.A.; Hervey, W.J.; Hettich, R.L.; Jagtap, P.D.; Janech, M.G.; Kinkade, D.B.; et al. Progress and Challenges in Ocean Metaproteomics and Proposed Best Practices for Data Sharing. J. Proteome Res. 2019, 18, 1461–1476. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Q.; Wang, Y.; Lu, J.; Lin, W.; Chen, F.; Jiao, N. Metagenomic and Metaproteomic Insights into Photoautotrophic and Heterotrophic Interactions in a. mBio 2020, 11. [Google Scholar] [CrossRef] [PubMed]
- Géron, A.; Werner, J.; Wattiez, R.; Lebaron, P.; Matallana-Surget, S. Deciphering the Functioning of Microbial Communities: Shedding Light on the Critical Steps in Metaproteomics. Front. Microbiol. 2019, 10, 2395. [Google Scholar] [CrossRef]
- Sudhagar, A.; Kumar, G.; El-Matbouli, M. Transcriptome Analysis Based on RNA-Seq in Understanding Pathogenic Mechanisms of Diseases and the Immune System of Fish: A Comprehensive Review. Int. J. Mol. Sci. 2018, 19, 245. [Google Scholar] [CrossRef]
- Low, C.F.; Rozaini, M.Z.H.; Musa, N.; Syarul Nataqain, B. Current knowledge of metabolomic approach in infectious fish disease studies. J. Fish Dis. 2017, 40, 1267–1277. [Google Scholar] [CrossRef]
- Díaz-Pascual, F.; Ortíz-Severín, J.; Varas, M.A.; Allende, M.L.; Chávez, F.P. Host-Pathogen Interaction as Revealed by Global Proteomic Profiling of Zebrafish Larvae. Front. Cell Infect. Microbiol. 2017, 7, 334. [Google Scholar] [CrossRef]
- Torraca, V.; Mostowy, S. Zebrafish Infection: From Pathogenesis to Cell Biology. Trends Cell Biol. 2018, 28, 143–156. [Google Scholar] [CrossRef]
- Li, H.; Zhu, Q.F.; Peng, X.X.; Peng, B. Interactome of E. piscicida and grouper liver proteins reveals strategies of bacterial infection and host immune response. Sci. Rep. 2017, 7, 39824. [Google Scholar] [CrossRef]
- Waiho, K.; Afiqah-Aleng, N.; Iryani, M.T.M.; Fazhan, H. Protein–protein interaction network: An emerging tool for understanding fish disease in aquaculture. Rev. Aquac. 2020, 1–22. [Google Scholar] [CrossRef]
- Forné, I.; Abián, J.; Cerdà, J. Fish proteome analysis: Model organisms and non-sequenced species. Proteomics 2010, 10, 858–872. [Google Scholar] [CrossRef]
- Li, Y.; Xie, W.; Li, Q. Understanding the lipopolysaccharide induced liver proteome changes and identification of immune genes in Lampetra morii. Aquac. Fish. 2016, 1, 9–14. [Google Scholar] [CrossRef][Green Version]
- Ye, H.; Lin, Q.; Luo, H. Applications of transcriptomics and proteomics in understanding fish immunity. Fish Shellfish Immunol. 2018, 77, 319–327. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Hu, Y.-H.; Xiao, Z.-Z.; Sun, L. Megalocytivirus-induced proteins of turbot (Scophthalmus maximus): Identification and antiviral potential. J. Proteom. 2013, 91, 430–443. [Google Scholar] [CrossRef] [PubMed]
- Encinas, P.; Rodriguez-Milla, M.A.; Novoa, B.; Estepa, A.; Figueras, A.; Coll, J. Zebrafish fin immune responses during high mortality infections with viral haemorrhagic septicemia rhabdovirus. A proteomic and transcriptomic approach. BMC Genom. 2010, 11, 518. [Google Scholar] [CrossRef]
- Medina-Gali, R.; Belló-Pérez, M.; Ciordia, S.; Mena, M.C.; Coll, J.; Novoa, B.; del Mar Ortega-Villaizán, M.; Perez, L. Plasma proteomic analysis of zebrafish following spring viremia of carp virus infection. Fish Shellfish Immunol. 2019, 86, 892–899. [Google Scholar] [CrossRef]
- Nombela, I.; Lopez-Lorigados, M.; Salvador-Mira, M.E.; Puente-Marin, S.; Chico, V.; Ciordia, S.; Mena, M.C.; Mercado, L.; Coll, J.; Perez, L.; et al. Integrated Transcriptomic and Proteomic Analysis of Red Blood Cells from Rainbow Trout Challenged with VHSV Point Towards Novel Immunomodulant Targets. Vaccines 2019, 7, 63. [Google Scholar] [CrossRef]
- Liu, M.; Wu, T.; Li, S.; Wei, P.; Yan, Y.; Gu, W.; Wang, W.; Meng, Q. Combined transcriptomic/proteomic analysis of crucian carp Carassius auratus gibelio in cyprinid herpesvirus 2 infection. Fish Shellfish Immunol. 2018, 82, 386–399. [Google Scholar] [CrossRef]
- Causey, D.R.; Pohl, M.A.N.; Stead, D.A.; Martin, S.A.M.; Secombes, C.J.; Macqueen, D.J. High-throughput proteomic profiling of the fish liver following bacterial infection. BMC Genom. 2018, 19, 719. [Google Scholar] [CrossRef]
- Long, M.; Zhao, J.; Li, T.; Tafalla, C.; Zhang, Q.; Wang, X.; Gong, X.; Shen, Z.; Li, A. Transcriptomic and proteomic analyses of splenic immune mechanisms of rainbow trout (Oncorhynchus mykiss) infected by Aeromonas salmonicida subsp. salmonicida. J. Proteom. 2015, 122, 41–54. [Google Scholar] [CrossRef]
- Di, G.; Li, H.; Zhang, C.; Zhao, Y.; Zhou, C.; Naeem, S.; Li, L.; Kong, X. Label-free proteomic analysis of intestinal mucosa proteins in common carp (Cyprinus carpio) infected with Aeromonas hydrophila. Fish Shellfish Immunol. 2017, 66, 11–25. [Google Scholar] [CrossRef]
- Booth, N.J.; Bilodeau-Bourgeois, A.L. Proteomic analysis of head kidney tissue from high and low susceptibility families of channel catfish following challenge with Edwardsiella ictaluri. Fish Shellfish Immunol. 2009, 26, 193–196. [Google Scholar] [CrossRef] [PubMed]
- Kumar, G.; Hummel, K.; Razzazi-Fazeli, E.; El-Matbouli, M. Modulation of posterior intestinal mucosal proteome in rainbow trout (Oncorhynchus mykiss) after Yersinia ruckeri infection. Vet. Res. 2019, 50, 54. [Google Scholar] [CrossRef] [PubMed]
- Cha, I.S.; Kwon, J.; Park, S.H.; Nho, S.W.; Jang, H.B.; Park, S.B.; del Castillo, C.S.; Hikima, J.; Aoki, T.; Jung, T.S. Kidney proteome responses in the teleost fish Paralichthys olivaceus indicate a putative immune response against Streptococcus parauberis. J. Proteom. 2012, 75, 5166–5175. [Google Scholar] [CrossRef] [PubMed]
- Addis, M.; Cappuccinelli, R.; Tedde, V.; Pagnozzi, D.; Viale, I.; Meloni, M.; Salati, F.; Roggio, T.; Uzzau, S. Influence of Moraxella sp. colonization on the kidney proteome of farmed gilthead sea breams (Sparus aurata, L.). Proteome Sci. 2010, 8, 50. [Google Scholar] [CrossRef] [PubMed]
- Saleh, M.; Kumar, G.; Abdel-Baki, A.-A.S.; Dkhil, M.A.; El-Matbouli, M.; Al-Quraishy, S. Quantitative proteomic profiling of immune responses to Ichthyophthirius multifiliis in common carp skin mucus. Fish Shellfish Immunol. 2019, 84, 834–842. [Google Scholar] [CrossRef] [PubMed]
- Valdenegro-Vega, V.A.; Crosbie, P.; Bridle, A.; Leef, M.; Wilson, R.; Nowak, B.F. Differentially expressed proteins in gill and skin mucus of Atlantic salmon (Salmo salar) affected by amoebic gill disease. Fish Shellfish Immunol. 2014, 40, 69–77. [Google Scholar] [CrossRef] [PubMed]
- Marcos-López, M.; Rodger, H.D.; O’Connor, I.; Braceland, M.; Burchmore, R.J.S.; Eckersall, P.D.; MacCarthy, E. A proteomic approach to assess the host response in gills of farmed Atlantic salmon Salmo salar L. affected by amoebic gill disease. Aquaculture 2017, 470, 1–10. [Google Scholar] [CrossRef]
- Dong, M.; Liang, Y.; Ramalingam, R.; Tang, S.W.; Shen, W.; Ye, R.; Gopalakrishnan, S.; Au, D.W.T.; Lam, Y.W. Proteomic characterization of the interactions between fish serum proteins and waterborne bacteria reveals the suppression of anti-oxidative defense as a serum-mediated antimicrobial mechanism. Fish Shellfish Immunol. 2017, 62, 96–106. [Google Scholar] [CrossRef]
- Sotton, B.; Paris, A.; Le Manach, S.; Blond, A.; Lacroix, G.; Millot, A.; Duval, C.; Huet, H.; Qiao, Q.; Labrut, S.; et al. Metabolic changes in Medaka fish induced by cyanobacterial exposures in mesocosms: An integrative approach combining proteomic and metabolomic analyses. Sci. Rep. 2017, 7, 4051. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Ali, F.; Cai, Q.; Yao, Z.; Sun, L.; Lin, W.; Lin, X. Quantitative proteomic analysis reveals that chemotaxis is involved in chlortetracycline resistance of Aeromonas hydrophila. J. Proteom. 2018, 172, 143–151. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Liu, W.; Jiang, Q.; Zuo, Y.; Su, Y.; Zhao, L.; Qin, Y.; Yan, Q. Integration of Transcriptomic and Proteomic Approaches Reveals the Temperature-Dependent Virulence of Pseudomonas plecoglossicida. Front. Cell. Infect. Microbiol. 2018, 8, 207. [Google Scholar] [CrossRef] [PubMed]
- Menanteau-Ledouble, S.; Kattlun, J.; Nöbauer, K.; El-Matbouli, M. Protein expression and transcription profiles of three strains of Aeromonas salmonicida ssp. salmonicida under normal and iron-limited culture conditions. Proteome Sci. 2014, 12, 29. [Google Scholar] [CrossRef] [PubMed]
- Yao, J.Y.; Xu, Y.; Yuan, X.M.; Yin, W.L.; Yang, G.L.; Lin, L.Y.; Pan, X.Y.; Wang, C.F.; Shen, J.Y. Proteomic analysis of differentially expressed proteins in the two developmental stages of Ichthyophthirius multifiliis. Parasitol. Res. 2017, 116, 637–646. [Google Scholar] [CrossRef]
- Stryiński, R.; Mateos, J.; Pascual, S.; González, Á.F.; Gallardo, J.M.; Łopieńska-Biernat, E.; Medina, I.; Carrera, M. Proteome profiling of L3 and L4 Anisakis simplex development stages by TMT-based quantitative proteomics. J. Proteom. 2019, 201, 1–11. [Google Scholar] [CrossRef]
- Lin, J.; Huang, S.; Zhang, Q. Outer membrane proteins: Key players for bacterial adaptation in host niches. Microbes. Infect. 2002, 4, 325–331. [Google Scholar] [CrossRef]
- Koebnik, R.; Locher, K.P.; Van Gelder, P. Structure and function of bacterial outer membrane proteins: Barrels in a nutshell. Mol. Microbiol. 2000, 37, 239–253. [Google Scholar] [CrossRef]
- Shahin, K.; Thompson, K.D.; Inglis, N.F.; McLean, K.; Ramirez-Paredes, J.G.; Monaghan, S.J.; Hoare, R.; Fontaine, M.; Metselaar, M.; Adams, A. Characterization of the outer membrane proteome of Francisella noatunensis subsp. orientalis. J. Appl. Microbiol. 2018, 125, 686–699. [Google Scholar] [CrossRef]
- Kumar, G.; Rathore, G.; El-Matbouli, M. Outer membrane protein assembly factor YaeT (omp85) and GroEL proteins of Edwardsiella tarda are immunogenic antigens for Labeo rohita (Hamilton). J. Fish Dis. 2014, 37, 1055–1059. [Google Scholar] [CrossRef]
- Luo, Z.; Fu, J.; Li, N.; Liu, Z.; Qin, T.; Zhang, X.; Nie, P. Immunogenic proteins and their vaccine development potential evaluation in outer membrane proteins (OMPs) of Flavobacterium columnare. Aquac. Fish. 2016, 1, 1–8. [Google Scholar] [CrossRef]

| Pathogen | Disease | Pathogen ID | Metabolic Process | Proteomic Method | Reference |
|---|---|---|---|---|---|
| Vírus | |||||
| Singapore grouper iridovirus (SGIV) | Sistemic diseases | Vírus proteín characterization | - | SDS-PAGE MALDI-TOF-MS | [99] |
| Singapore grouper iridovirus (SGIV) | Sistemic diseases | Vírus envelope proteín analysis and characterization | - | 1-DE MALDI-TOF/TOF-MS/MS and LC-MALDI-TOF/TOF-MS/MS | [100] |
| Cyprinid herpesvirus 3 (CyHV-3) | Koi herpesvirus (KHV) disease | Functional analyses of the virion transmembrane proteome | - | SDS-PAGE LC-MS/MS | [101] |
| Bacteria | |||||
| Photobacterium damselaesubsp. piscicida | Pasteurellosis | - | Importance of outer membrane proteins in osmotic adaptations and bacterial virulence | 2-DE LC-nano ESI-Q-TOF MS/MS | [102] |
| Photobacterium damselaesubsp. damselae | Vibriosis | - | Importance of iron in virulence | 2-DE MALDI-TOF/TOF-MS | [103] |
| Photobacterium damselae | Pasteurellosis | Differentiation of Photobacterium damselae subspecies | - | MALDI-TOF-MS | [104] |
| Edwardsiella tarda | Haemorrhagic septicaemia | Identification of proteins associated with virulent and avirulent strains | - | 2-DE MALDI-TOF MS | [105,106] |
| Edwardsiella tarda | Haemorrhagic septicaemia | - | Virulence determinants | 2-DE ESI MS/MS | [92] |
| Yersinia ruckeri | Enteric redmouth disease | Identification and characterization of biotype 1 and biotype 2 strains | - | Nano LC-MS/MS | [107] |
| Yersinia ruckeri | Enteric redmouth disease | - | Importance of iron in virulence | Nano LC-MS/MS | [108] |
| Yersinia ruckeri | Enteric redmouth disease | - | Importance of outer membrane proteins in bacterial virulence | Gell free and 1-D nLC-ESI-MS/MS | [109] |
| Flavobacterium columnare | Columnaris disease | - | Virulence determinants | 2-D LC ESI MS/MS and 2-DE MALDI TOF/TOF MS | [110] |
| Flavobacterium psychrophilum | Bacterial coldwater disease | Proteomic profiling of strains, the importance of iron in virulence | - | 2-DE LC-MS/MS | [111] |
| Flavobacterium psychrophilum | Bacterial coldwater disease | Differentiation of Flavobacterium psychrophilum from Flavobacterium psychrophilum-like species | - | MALDI-TOF MS | [112] |
| Vibrio parahaemolyticus | Vibriosis | - | Identify proteins regulating antimicrobial peptide resistance | 2-DE LC-ESI-Q-TOF MS/MS | [113] |
| Streptococcus agalactiae | Streptococcosis | - | Temperature effects on bacterial virulence | NanoUPLC-HDMSE | [114] |
| Streptococcus iniaeand Streptococcus parauberis | Streptococcosis | Identification and molecular fingerprinting | - | MALDI-TOF-MS | [115] |
| Streptococcus parauberis | Streptococcosis | Identification of bacterial strains | - | MALDI-TOF-MS | [116] |
| Vagococcus salmoninarum | Cold-water streptococcosis | Typing and characterization of bacterial strains | - | MALDI-TOF-MS | [117] |
| Streptococcus iniae | Streptococcosis | Proteomic profile of a pathogenic strain | - | 2-DE MALDI-TOF-MS | [118] |
| Piscirickettsia salmonis | Salmonid rickettsial syndrome | Detection and identification | Virulence determinants | MALDI-TOF-MS | [119] |
| Tenacibaculum sp. | Tenacibaculosis | Identification of Tenacibaculum species | - | MALDI-TOF-MS | [120,121] |
| Mycobacterium marinum | Mycobacteriosis | Identification of Mycobacterium marinum subspecies | - | MALDI-TOF-MS | [122] |
| Several bacterial species | - | Differentiate several gram-negative fish pathogenic bacteria | - | MALDI-TOF-MS | [123] |
| Fungus | |||||
| Saprolegnia parasitica | Saprolegniasis | Identification and characterization of developmental stages | - | iTRAQ SDS-PAGE Nano-LC-MS/MS | [124] |
| Parasites | |||||
| Caligus rogercresseyi | Sea lice | Detection and identification | Virulence determinants | MALDI-TOF-MS | [119] |
| Disease | Pathogen | Host | Clinical Signs/Pathology | References |
|---|---|---|---|---|
| Virus | ||||
| Epizootic haematopoietic necrosis (EHN) | Epizootic haematopoietic necrosis virus | Redfin perch (Perca fluviatilis), Rainbow trout (Oncorhynchus mykiss) | Erratic swimming, darkened skin, skin ulcers, exophthalmia, swollen spleen and kidney, petechial haemorrhages on fins, ascites, abdominal distension | [131] |
| Infectious haematopoietic necrosis (IHN) | Infectious haematopoietic necrosis virus | Salmonids | Lethargy, darkened skin, exophthalmia, eye haemorrhage, pale gills, swollen abdomen, opaque faecal casts, petechial haemorrhage on fins, visceral pallor | [132,133] |
| Infectious Pancreatic Necrosis (IPN) | Infectious pancreatic necrosis virus | Salmonids | Irregular swimming, loss of appetite, darkening of the skin, distended abdomen, exophthalmia, pale gills petechial haemorrhages, visceral ascites, intestine with catarrhal exudate and pale liver | [134,135,136] |
| Infectious salmon Anaemia (ISA) | Infectious Salmon Anaemia virus | Atlantic salmon (Salmo salar) | Lethargy, anaemia, exophthalmia, pale gills and internal organs, ascites, oedemas, petechiae in visceral fat, liver and spleen congestion | [137,138] |
| Lymphocystis disease | Lymphocystis disease virus | Broadly infectious | Nodular lesions on the skin, fins and internally | [137] |
| Pancreatic disease (PD) | Salmonid alphaviruses (SAV) | Rainbow trout (Oncorhynchus mykiss), Atlantic salmon (Salmo salar) | Lethargy, hang in the corners of the cage or rest in the bottom, loss of appetite, yellow faecal casts, ascites, petechial haemorrhages in pyloric caecal fat, lesions in pancreas and skeletal and cardiac muscle | [139,140] |
| Viral haemorrhagic septicaemia (VHS) | Viral haemorrhagic septicaemia virus | Broadly infectious | Aberrant swimming (spiral, leaping, flashing), exophthalmia, darkened skin, anaemia, pale gills and liver, internal haemorrhages, ascites leading to a swollen abdomen, swollen and hyperaemic kidney | [137,141] |
| Viral nervous necrosis (VNN) | Betanodaviruses | Broadly infectious | Lethargy, abnormal swimming; anorexia, skin darkening, abdominal distension, hyperinflation of the swim bladder | [142,143] |
| Bacteria | ||||
| Vibriosis | Vibrio anguillarum | Broadly infectious | Lethargy, cease feeding, darkened skin, exophthalmia and corneal opacity, pale gills, petechiae at fin bases and skin, ulcers, generalized septicaemia | [127,144,145] |
| Pasteurellosis | Photobacterium damselae subsp. piscicida | White perch (Morone americanus) yellowtail (Seriola quinqueradiata) gilthead seabream (Sparus aurata) | Darkened skin, swollen spleen, white-spotted lesions in spleen and kidney, bacterial accumulations on the tissues of internal organs | [146,147] |
| Furunculosis | Aeromonas salmonicida subsp. salmonicida | Broadly infectious | Lethargy, cease feeding, darkened skin, exophthalmia, haemorrhages at the base of the fins, enlarged spleen, lesions on the skin (furuncles), ulcers, pale liver, general septicaemia | [148,149] |
| Tenacibaculosis | Tenacibaculum maritimum | Broadly infectious | Flashing swimming behaviour, anorexic, erosion on the skin, fins (tail rot), head and gills, petechial haemorrhage on the abdominal peritoneum, ulcers | [150] |
| Bacterial gill disease (BGD) | Flavobacterium branchiophilum | Coldwater fish (mainly salmonids) | Loss of appetite, gill infestation, increased opercular movements, gasping at the water surface, respiratory distress | [151] |
| Rainbow Trout Fry Syndrom (RFTS) | Flavobacterium psychrophilum | Salmonids | Lethargy, anorexia, distended abdomen, darkened skin in caudal peduncle area, skin ulceration, swollen spleen, pale organs | [152] |
| Red spot disease (Winter disease) | Pseudomonas anguilliseptica | Eels, salmonids, gilthead seabream (Sparus aurata), European seabass (Dicentrarchus labrax), European cod (Gadus morhua) | Erratic swimming, petechial haemorrhages in the skin and liver, distended abdomen, ascitic fluid in the peritoneal cavity, pale liver, haemorrhaged kidney, intestine with yellowish exudate | [144,153] |
| Disease | Pathogen | Host | Clinical Signs/Pathology | References |
| Lactococcosis | Lactococcus garvieae | Broadly infectious | Lethargy, anorexia, exophthalmia, distended abdomen, ascitic fluid in the peritoneal cavity, congestion and haemorrhages of liver, intestine, spleen and kidney, haemorrhagic septicaemia | [154] |
| Parasites | ||||
| Amoebic gill disease (AGD) | Neoparamoeba perurans | Atlantic salmon (Salmo salar), rainbow trout (Oncorhynchus mykiss) | Lethargy, respiratory distress, increased opercular movements, whitish patches on gills and excessive mucus | [155,156] |
| White spot disease/Ichthyophthiriasis | Ichthyophthirius multifiliis | Freshwater fish | Hyperactive (initially), lethargic; pale gills, darkened skin, white spots in the skin, increase of mucus production, skin ulcers, frayed fins, pale liver, enlarged spleen and kidney | [128,157,158] |
| White spot disease/Cryptocaryoniasis | Cryptocaryon irritans | Saltwater fish | Hyperactive (initially), lethargic, numerous small whitish spots on the skin surface, petechial haemorrhages on the skin, excessive mucus production, skin ulcers, corneal clouding and blindness | [128,159] |
| Amyloodiniosis | Amyloodinium ocellatum | Broadly infectious | Cease feeding, scratch against the bottom, infested gills and skin, excessive mucus production, epithelial erosion on attachment sites | [160,161,162] |
| Trichodinosis | Trichodina sp. | Broadly infectious | Lethargy, cease feeding, infested gills, skin and fins, Greyish colour due to excessive mucus production, skin lesions in attachment sites, frayed fins | [163,164] |
| Gyrodactilosis | Gyrodactylus salaris | Salmonids | Infest mainly fins and skin, lethargy, anorexia, emaciated fins, darkened skin, epithelium lesions | [165,166] |
| Sea lice | Lepeophtheirus salmonis | Salmonids | Skin lesions, especially on the head, haemorrhages, scale loss, oedema, hyperplasia and cellular inflammations | [167] |
| Aetiological Agent | Species | Tissue | Modified Protein Groups | Reference |
|---|---|---|---|---|
| Vírus | ||||
| Infectious spleen and kidney necrosis virus (ISKNV) (Megalocytivirus) | Zebrafish (Danio rerio) | Spleen | Cytoskeletal proteins, stress response, lipoprotein and carbohydrate metabolism, signal transduction, proteolysis, metabolic and catabolic processes | [89] |
| Megalocytivirus | Turbot (Scophtalmus maximus) | Spleen | Cytoskeleton proteins, molecular biosynthesis, cellular signal transduction and chaperone proteins | [187] |
| Salmonid alphavirus subtype 3 (SAV3) | Atlantic salmon (Salmo salar) | Serum | Humoral components of immunity | [94] |
| Rhabdovirus | Zebrafish (Danio rerio) | Fins | Proteins of the glycolytic pathway and cytoskeleton components | [188] |
| Spring viremia of carp virus | Zebrafish (Danio rerio) | Plasma | Vitellogenin and Gig2 | [189] |
| Viral septicemia hemorrhagic virus | Rainbow trout (Oncorhynchus mykiss) | Red blood cells from blood and head kidney | Proteins related to viral transcription | [190] |
| Cyprinid herpesvirus-2 | Crucian carp (Carassius carassius) | Head kidney | Cytoskeleton, transport, immunologic, intracellular and physiologic proteins | [191] |
| Bacteria | ||||
| Aeromonas salmonicida | Rainbow trout (Oncorhynchus mykiss) | Liver | Complement system and acute phase response proteins | [192] |
| Aeromonas salmonicida | Rainbow trout (Oncorhynchus mykiss) | Spleen | Immune system, signaling molecules and interaction | [193] |
| Aeromonas hydrophila | Common carp (Cyprinus carpio) | Intestinal mucosa | Proteins involved in stress and immune response | [194] |
| Aeromonas hydrophila | Zebrafish (Danio rerio) | Gills | Stress and immune response | [91] |
| Edwardsiella ictaluri | Channel catfish (Ictalurus punctatus) | Head kidney | Macrophage function, cellular stress response, cellular energy production and metabolism | [195] |
| Yersinia ruckeri | Rainbow trout (Oncorhynchus mykiss) | Head kidney and spleen | Immune system, cellular, metabolic, developmental, multicellular, adhesion, regulation and response to stimulus | [95] |
| Yersinia ruckeri | Rainbow trout (Oncorhynchus mykiss) | Intestine | Metabolic process biological regulation, cellular processes and component organization | [196] |
| Vibrio anguillarum | Atlantic cod (Gadus morhua) | Mucus | Proteins involved in the immune system | [83] |
| Streptococcus parauberis | Olive flounder (Paralichthys olivaceus) | Kidney | Proteins involved in immune response, cellular recovery and glycoprotein synthesis | [197] |
| Moraxella sp. | Gilthead seabream (Sparus aurata) | Kidney | Mitochondrial proteins, cellular response to oxidative stress, infection and inflammation | [198] |
| Parasites | ||||
| Ichthyophthirius multifiliis | Common carp (Cyprinus carpio) | Mucus | Proteins involved in the immune and inflammatory response | [199] |
| Lepeophtheirus salmonis | Atlantic salmon (Salmo salar) | Mucus | Proteins involved in glycolysis, peptide synthesis, immune and defence response | [82] |
| Amyloodinium ocellatum | Gilthead seabream (Sparus aurata) | Plasma | Proteins involved in the acute-phase response, inflammation, lipid transport, homeostasis and wound healing | [88] |
| Neoparamoeba perurans | Atlantic salmon (Salmo salar) | Gill and skin mucus | Proteins involved in cell to cell signalling and inflammation pathway | [189] |
| Neoparamoeba perurans | Atlantic salmon (Salmo salar) | Gill and skin mucus | Proteins involved in cell cycle regulation, cytoskeletal regulation, oxidative metabolism and immunity | [201] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Moreira, M.; Schrama, D.; Farinha, A.P.; Cerqueira, M.; Raposo de Magalhães, C.; Carrilho, R.; Rodrigues, P. Fish Pathology Research and Diagnosis in Aquaculture of Farmed Fish; a Proteomics Perspective. Animals 2021, 11, 125. https://doi.org/10.3390/ani11010125
Moreira M, Schrama D, Farinha AP, Cerqueira M, Raposo de Magalhães C, Carrilho R, Rodrigues P. Fish Pathology Research and Diagnosis in Aquaculture of Farmed Fish; a Proteomics Perspective. Animals. 2021; 11(1):125. https://doi.org/10.3390/ani11010125
Chicago/Turabian StyleMoreira, Márcio, Denise Schrama, Ana Paula Farinha, Marco Cerqueira, Cláudia Raposo de Magalhães, Raquel Carrilho, and Pedro Rodrigues. 2021. "Fish Pathology Research and Diagnosis in Aquaculture of Farmed Fish; a Proteomics Perspective" Animals 11, no. 1: 125. https://doi.org/10.3390/ani11010125
APA StyleMoreira, M., Schrama, D., Farinha, A. P., Cerqueira, M., Raposo de Magalhães, C., Carrilho, R., & Rodrigues, P. (2021). Fish Pathology Research and Diagnosis in Aquaculture of Farmed Fish; a Proteomics Perspective. Animals, 11(1), 125. https://doi.org/10.3390/ani11010125