Selection for Favorable Health Traits: A Potential Approach to Cope with Diseases in Farm Animals
Simple Summary
Abstract
1. Introduction
2. Farm Animal Diseases: Influence, Prevalence, and Controlling Issues
2.1. The Influence, Prevalence, and Controlling Issues of Common Diseases in Farm Animals
2.2. Current Methods to Control Diseases in Farm Animals
2.2.1. Vaccination
2.2.2. Medical Treatments
2.2.3. Culling
2.2.4. Genome Editing
2.2.5. Biosensor
2.2.6. Probiotics
3. Selection for Animals with Favorable Health Traits
3.1. Health Traits in Farm Animals: Definition, Classification, and Components
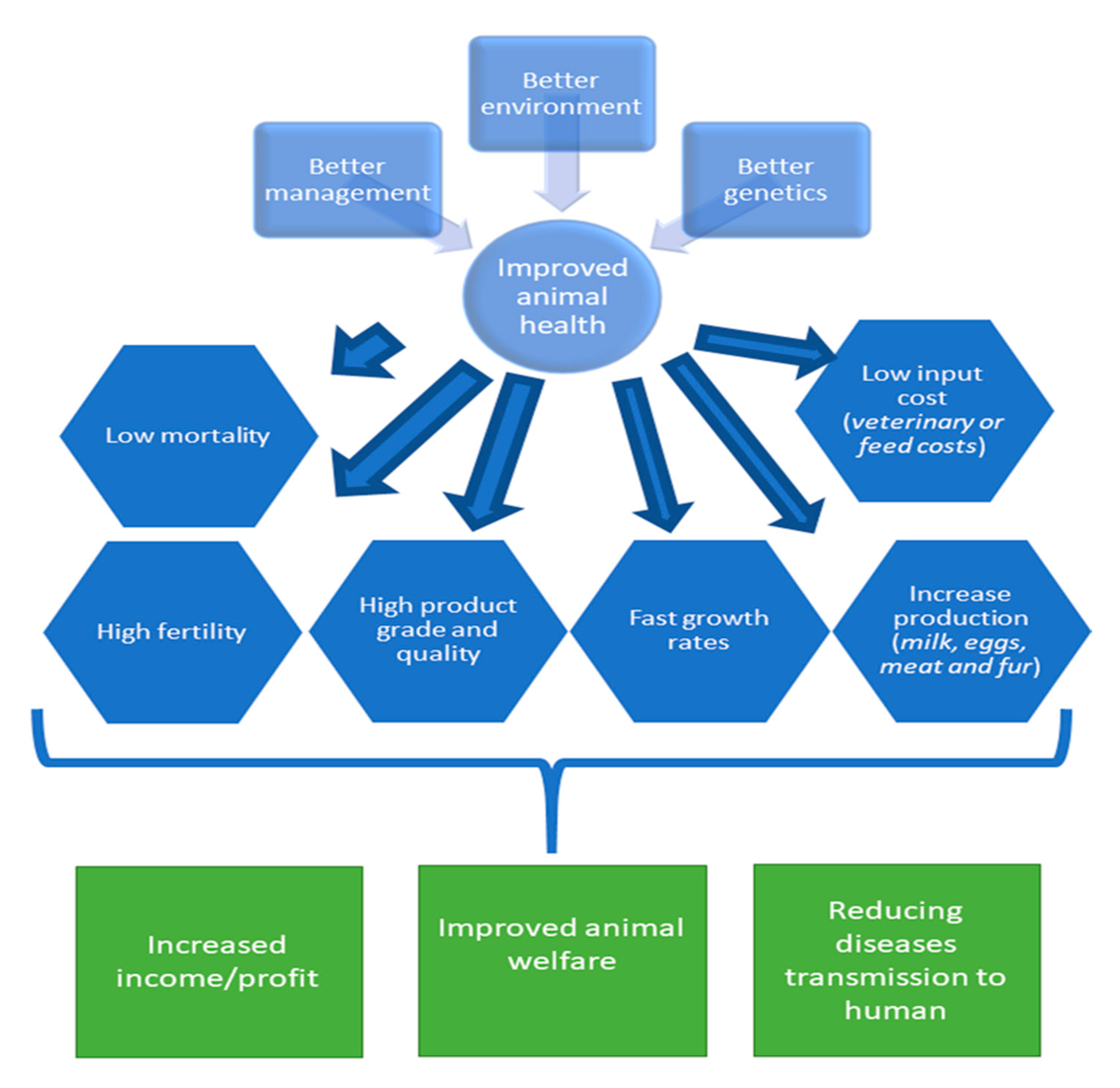
3.2. The Benefits of Selecting Farm Animals with Favorable Health Traits
3.3. Methods of Selection for Health Traits
3.3.1. Traditional Genetic Selection
3.3.2. Marker-Assisted Selection
3.3.3. Genomic Selection
3.4. Selection for Different Types of Health Traits
3.4.1. Selection for Disease Response Traits (Resistance, Tolerance, and Resilience)
3.4.2. Selections for Immune Response Traits
3.5. Challenges in the Selection of Health Traits
3.5.1. Desirability
3.5.2. Feasibility
3.5.3. Sustainability
3.6. Promise of Selection for Health Traits
3.6.1. High-Throughput Phenotyping and Sequencing, and Generation of Big Data
3.6.2. Data Sharing and International Corporations
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bishop, S.C.; Woolliams, J.A. Genomics and disease resistance studies in livestock. Livest. Sci. 2014, 166, 190–198. [Google Scholar] [CrossRef] [PubMed]
- Rashid, A.; Rasheed, K.; Asim, M.; Hussain, A. Risks of vaccination: A review. J. Venom. Anim. Toxins Incl. Trop. Dis. 2009, 15. [Google Scholar] [CrossRef]
- Yeruham, I.; Yadin, H.; Haymovich, M.; Perl, S. Adverse reactions to FMD vaccine. Vet. Dermatol. 2001, 12, 197–201. [Google Scholar] [CrossRef] [PubMed]
- Beyene, T. Veterinary drug residues in food-animal products: Its risk factors and potential effects on public health. J. Vet. Sci. Technol. 2016, 7, 1–7. [Google Scholar] [CrossRef]
- de Jong, A.; Thomas, V.; Simjee, S.; Moyaert, H.; El Garch, F.; Maher, K.; Morrissey, I.; Butty, P.; Klein, U.; Marion, H.; et al. Antimicrobial susceptibility monitoring of respiratory tract pathogens isolated from diseased cattle and pigs across Europe: The VetPath study. Vet. Microbiol. 2014, 172, 202–215. [Google Scholar] [CrossRef]
- Ibrahim, A.; Junaidu, A.; Garba, M. Multiple antibiotic residues in meat from slaughtered cattle in Nigeria. Internet J. Vet. Med. 2010, 8, 1–7. [Google Scholar]
- Kehinde, O.G.; Junaidu, K.; Mohammed, M.; AbdulRahman, A.M. Detection of antimicrobial drug residues in commercial eggs using Premi® Test. Int. J. Poult. Sci. 2012, 11, 50–54. [Google Scholar] [CrossRef]
- Rokka, M.; Eerola, S.; Perttila, U.; Rossow, L.; Venalainen, E.; Valkonen, E.; Valaja, J.; Peltonen, K. The residue levels of narasin in eggs of laying hens fed with unmedicated and medicated feed. Mol. Nutr. Food Res. 2005, 49, 38–42. [Google Scholar] [CrossRef]
- Wilson, M.E. The afterlife of antibiotics. J. Travel Med. 2020, 27. [Google Scholar] [CrossRef]
- Pritchard, T.C.; Coffey, M.P.; Bond, K.S.; Hutchings, M.R.; Wall, E. Phenotypic effects of subclinical paratuberculosis (Johne’s disease) in dairy cattle. J. Dairy Sci. 2017, 100, 679–690. [Google Scholar] [CrossRef]
- Themudo, G.E.; Østergaard, J.; Ersbøll, A.K. Persistent spatial clusters of plasmacytosis among Danish mink farms. Prev. Vet. Med. 2011, 102, 75–82. [Google Scholar] [CrossRef] [PubMed]
- Holmberg, M.; Andersson-Eklund, L. Quantitative trait loci affecting health traits in Swedish dairy cattle. J. Dairy Sci. 2004, 87, 2653–2659. [Google Scholar] [CrossRef]
- Doeschl-Wilson, A.B.; Villanueva, B.; Kyriazakis, I. The first step toward genetic selection for host tolerance to infectious pathogens: Obtaining the tolerance phenotype through group estimates. Front. Genet. 2012, 3, 265. [Google Scholar] [CrossRef] [PubMed]
- Mallard, B.A.; Emam, M.; Paibomesai, M.; Thompson-Crispi, K.; Wagter-Lesperance, L. Genetic selection of cattle for improved immunity and health. Jpn. J. Vet. Res. 2015, 63, 37–44. [Google Scholar] [CrossRef]
- Cowley, D.B.; Graham, D.A.; Guelbenzu, M.; Doherty, M.L.; More, S.J. Aspects of bovine herpesvirus 1 and bovine viral diarrhoea virus herd-level seroprevalence and vaccination in dairy and beef herds in Northern Ireland. Ir. Vet. J. 2014, 67, 18. [Google Scholar] [CrossRef]
- Carman, S.; van Dreumel, T.; Ridpath, J.; Hazlett, M.; Alves, D.; Dubovi, E.; Tremblay, R.; Bolin, S.; Godkin, A.; Anderson, N. Severe acute bovine viral diarrhea in Ontario, 1993-1995. J. Vet. Diagn. Invest. 1998, 10, 27–35. [Google Scholar] [CrossRef]
- Houe, H. Economic impact of BVDV infection in dairies. Biologicals 2003, 31, 137–143. [Google Scholar] [CrossRef]
- Brownlie, J. BVD—Why vaccination alone is not the complete answer to eradication. Livestock 2014, 19, 221–224. [Google Scholar] [CrossRef]
- Pinior, B.; Firth, C.L.; Richter, V.; Lebl, K.; Trauffler, M.; Dzieciol, M.; Hutter, S.E.; Burgstaller, J.; Obritzhauser, W.; Winter, P.; et al. A systematic review of financial and economic assessments of bovine viral diarrhea virus (BVDV) prevention and mitigation activities worldwide. Prev. Vet. Med. 2017, 137, 77–92. [Google Scholar] [CrossRef]
- Attalla, S.A.; Seykora, A.J.; Cole, J.B.; Heins, B.J. Genetic parameters of milk ELISA scores for Johne’s disease. J. Dairy Sci. 2010, 93, 1729–1735. [Google Scholar] [CrossRef]
- Cho, J.; Tauer, L.W.; Schukken, Y.H.; Smith, R.L.; Lu, Z.; Grohn, Y.T. Cost-Effective Control Strategies for Johne’s Disease in Dairy Herds. Can. J. Agric. Econ. 2013, 61, 583–608. [Google Scholar] [CrossRef]
- Gershwin, L.J.; Van Eenennaam, A.L.; Anderson, M.L.; McEligot, H.A.; Shao, M.X.; Toaff-Rosenstein, R.; Taylor, J.F.; Neibergs, H.L.; Womack, J.; Bovine Respiratory Disease Complex Coordinated Agricultural Project Research Team. Single Pathogen Challenge with Agents of the Bovine Respiratory Disease Complex. PLoS ONE 2015, 10, e0142479. [Google Scholar] [CrossRef] [PubMed]
- Miles, D.G. Overview of the North American beef cattle industry and the incidence of bovine respiratory disease (BRD). Anim. Health Res. Rev. 2009, 10, 101–103. [Google Scholar] [CrossRef]
- Neibergs, H.; Neibergs, J.; Wojtowicz, A.; Taylor, J.; Seabury, C.; Womack, J. Economic benefits of using genetic selection to reduce the prevalence of bovine respiratory disease complex in beef feedlot cattle. In Proceedings of the 2014 Beef Improvement Federation Annual Meeting and Convention, Lincoln, NE, USA, 18–21 June 2014; pp. 82–87. [Google Scholar]
- DeDonder, K.D.; Apley, M.D. A literature review of antimicrobial resistance in Pathogens associated with bovine respiratory disease. Anim. Health Res. Rev. 2015, 16, 125–134. [Google Scholar] [CrossRef] [PubMed]
- Carvelli, A.; Sala, M.; Autorino, G.L.; Scicluna, M.T.; Iacoponi, F.; Rombola, P.; Scaramozzino, P. A cross-sectional serosurvey in a sheep population in central Italy following a bluetongue epidemic. PLoS ONE 2019, 14, e0208074. [Google Scholar] [CrossRef] [PubMed]
- Elhassan, A.M.; Fadol, M.A.; El Hussein, A.R.M. Seroprevalence of bluetongue virus in dairy herds with reproductive problems in Sudan. Int. Schol. Res. Notices (ISRN) Vet. Sci. 2014, 2014, 595724. [Google Scholar] [CrossRef]
- Velthuis, A.G.; Saatkamp, H.W.; Mourits, M.C.; de Koeijer, A.A.; Elbers, A.R. Financial consequences of the Dutch bluetongue serotype 8 epidemics of 2006 and 2007. Prev. Vet. Med. 2010, 93, 294–304. [Google Scholar] [CrossRef]
- Kyriakis, C.S.; Billinis, C.; Papadopoulos, E.; Vasileiou, N.G.; Athanasiou, L.V.; Fthenakis, G.C. Bluetongue in small ruminants: An opinionated review, with a brief appraisal of the 2014 outbreak of the disease in Greece and the south-east Europe. Vet. Microbiol. 2015, 181, 66–74. [Google Scholar] [CrossRef]
- Boumart, Z.; Daouam, S.; Belkourati, I.; Rafi, L.; Tuppurainen, E.; Tadlaoui, K.O.; El Harrak, M. Comparative innocuity and efficacy of live and inactivated sheeppox vaccines. BMC Vet. Res. 2016, 12, 133. [Google Scholar] [CrossRef]
- Hota, A.; Biswal, S.; Sahoo, N.; Venkatesan, G.; Arya, S.; Kumar, A.; Ramakrishnan, M.A.; Pandey, A.B.; Rout, M. Seroprevalence of Capripoxvirus infection in sheep and goats among different agro-climatic zones of Odisha, India. Vet. World 2018, 11, 66. [Google Scholar] [CrossRef]
- Hurisa, T.; Jing, Z.; Jia, H.; Chen, G.; He, X. A Review on Sheeppox and Goatpox: Insight of Epidemiology, Diagnosis, Treatment and Control Measures in Ethiopia. J. Infect. Dis. Epidemiol. 2018, 4, 2474–3658. [Google Scholar] [CrossRef]
- Garner, M.; Sawarkar, S.; Brett, E.; Edwards, J.; Kulkarni, V.; Boyle, D.; Singh, S. The extent and impact of sheep pox and goat pox in the state of Maharashtra, India. Trop. Anim. Health Prod. 2000, 32, 205–223. [Google Scholar] [CrossRef] [PubMed]
- Lunney, J.K.; Fang, Y.; Ladinig, A.; Chen, N.; Li, Y.; Rowland, B.; Renukaradhya, G.J. Porcine reproductive and respiratory syndrome virus (PRRSV): Pathogenesis and interaction with the immune system. Annu. Rev. Anim. Biosci. 2016, 4, 129–154. [Google Scholar] [CrossRef] [PubMed]
- Arruda, A.G.; Poljak, Z.; Friendship, R.; Carpenter, J.; Hand, K. Descriptive analysis and spatial epidemiology of porcine reproductive and respiratory syndrome (PRRS) for swine sites participating in area regional control and elimination programs from 3 regions of Ontario. Can. J. Vet. Res. 2015, 79, 268–278. [Google Scholar] [PubMed]
- Holtkamp, D.J.; Kliebenstein, J.B.; Neumann, E.; Zimmerman, J.J.; Rotto, H.; Yoder, T.K.; Wang, C.; Yeske, P.; Mowrer, C.L.; Haley, C.A. Assessment of the economic impact of porcine reproductive and respiratory syndrome virus on United States pork producers. J. Swine Health Prod. 2013, 21, 72. [Google Scholar]
- Mussell, A.; Oginskyy, A.; Grier, K.; Morin, M.; Lachance, M.; Whittington, L.; Friendship, R. A Risk, Benefit, Strength, Weakness, Opportunity and Threat Analysis for the Control. and Possible Eradication of Porcine Reproductive and Respiratory Syndrome (PRRS) Virus Within the Canadian Swine Herd; George Morris Centre: Guelph, ON, Canada, 2011. [Google Scholar]
- Hess, A.S.; Islam, Z.; Hess, M.K.; Rowland, R.R.; Lunney, J.K.; Doeschl-Wilson, A.; Plastow, G.S.; Dekkers, J.C. Comparison of host genetic factors influencing pig response to infection with two North American isolates of porcine reproductive and respiratory syndrome virus. Genet. Sel. Evol. 2016, 48, 43. [Google Scholar] [CrossRef]
- Sun, N.; Sun, P.; Lv, H.; Sun, Y.; Guo, J.; Wang, Z.; Luo, T.; Wang, S.; Li, H. Matrine displayed antiviral activity in porcine alveolar macrophages co-infected by porcine reproductive and respiratory syndrome virus and porcine circovirus type 2. Sci. Rep. 2016, 6, 1–13. [Google Scholar] [CrossRef]
- Brown, V.R.; Bevins, S.N. A Review of African Swine Fever and the Potential for Introduction into the United States and the Possibility of Subsequent Establishment in Feral Swine and Native Ticks. Front. Vet. Sci. 2018, 5, 11. [Google Scholar] [CrossRef]
- Liu, J.; Liu, B.; Shan, B.; Wei, S.; An, T.; Shen, G.; Chen, Z. Prevalence of African Swine Fever in China, 2018–2019. J. Med. Virol. 2019, 92, 1023–1034. [Google Scholar] [CrossRef]
- USDA. GAIN Report: Russia 2017 Livestock and Products Annual RS1757; United States Department of Agriculture: Washington, DC, USA, 2017. [Google Scholar]
- Bello, M.B.; Yusoff, K.; Ideris, A.; Hair-Bejo, M.; Peeters, B.P.H.; Omar, A.R. Diagnostic and Vaccination Approaches for Newcastle Disease Virus in Poultry: The Current and Emerging Perspectives. Biomed. Res. Int. 2018, 2018, 7278459. [Google Scholar] [CrossRef]
- Cross, T.A.; Arsnoe, D.; Minnis, R.; King, D.; Swafford, S.; Pedersen, K.; Owen, J. Prevalence of avian paramyxovirus 1 and avian influenza virus in double-crested cormorants (Phalacrocorax auritus) in eastern North America. J. Wildl. Dis. 2013, 49, 965–977. [Google Scholar] [CrossRef] [PubMed]
- Wise, M.G.; Suarez, D.L.; Seal, B.S.; Pedersen, J.C.; Senne, D.A.; King, D.J.; Kapczynski, D.R.; Spackman, E. Development of a real-time reverse-transcription PCR for detection of Newcastle disease virus RNA in clinical samples. J. Clin. Microbiol. 2004, 42, 329–338. [Google Scholar] [CrossRef] [PubMed]
- Dimitrov, K.M.; Afonso, C.L.; Yu, Q.; Miller, P.J. Newcastle disease vaccines—A solved problem or a continuous challenge? Vet. Microbiol. 2017, 206, 126–136. [Google Scholar] [CrossRef] [PubMed]
- Mayers, J.; Mansfield, K.L.; Brown, I.H. The role of vaccination in risk mitigation and control of Newcastle disease in poultry. Vaccine 2017, 35, 5974–5980. [Google Scholar] [CrossRef]
- Morrow, C.; Fehler, F. Marek’s disease: A worldwide problem. In Marek’s Disease; Davison, F., Nair, V., Eds.; Elsevier Academic Press: London, UK, 2004; pp. 49–61. [Google Scholar]
- Wajid, S.J.; Katz, M.E.; Renz, K.G.; Walkden-Brown, S.W. Prevalence of Marek’s disease virus in different chicken populations in Iraq and indicative virulence based on sequence variation in the EcoRI-Q (meq) gene. Avian Dis. 2013, 57, 562–568. [Google Scholar] [CrossRef]
- Boodhoo, N.; Gurung, A.; Sharif, S.; Behboudi, S. Marek’s disease in chickens: A review with focus on immunology. Vet. Res. 2016, 47, 119. [Google Scholar] [CrossRef]
- Reddy, S.M.; Izumiya, Y.; Lupiani, B. Marek’s disease vaccines: Current status, and strategies for improvement and development of vector vaccines. Vet. Microbiol. 2017, 206, 113–120. [Google Scholar] [CrossRef]
- Dunn, J.R.; Gimeno, I.M. Current status of Marek’s disease in the United States and worldwide based on a questionnaire survey. Avian Dis. 2013, 57, 483–490. [Google Scholar] [CrossRef]
- Farid, A.; Ferns, L. Aleutian mink disease virus infection may cause hair depigmentation. Scientifur 2011, 35, 55–59. [Google Scholar]
- Hansen, M.; Lund, E. Pregnancy rate and foetal mortality in Aleutian disease virus infected mink. Acta Vet. Scand. 1988, 29, 271. [Google Scholar]
- McDonald, R.A.; Lariviere, S. Diseases and pathogens of Mustela spp, with special reference to the biological control of introduced stoat Mustela erminea populations in New Zealand. J. R. Soc. N. Z. 2001, 31, 721–744. [Google Scholar] [CrossRef]
- Reichert, M.; Kostro, K. Effect of persistent infection of mink with Aleutian mink disease virus on reproductive failure. Bull. Vet. Inst. Pulawy 2014, 58, 369–373. [Google Scholar] [CrossRef]
- Farid, A.; Zillig, M.; Finley, G.; Smith, G. Prevalence of the Aleutian mink disease virus infection in Nova Scotia, Canada. Prev. Vet. Med. 2012, 106, 332–338. [Google Scholar] [CrossRef] [PubMed]
- Farid, A.; Daftarian, P.; Fatehi, J. Transmission Dynamics of Aleutian Mink Disease Virus on a Farm Under Test and Removal Scheme. J. Vet. Sci. Med. Diagn. 2018, 7, 2. [Google Scholar] [CrossRef]
- Christensen, L.S.; Gram-Hansen, L.; Chriel, M.; Jensen, T.H. Diversity and stability of Aleutian mink disease virus during bottleneck transitions resulting from eradication in domestic mink in Denmark. Vet. Microbiol. 2011, 149, 64–71. [Google Scholar] [CrossRef]
- Aasted, B.; Alexandersen, S.; Christensen, J. Vaccination with Aleutian mink disease parvovirus (AMDV) capsid proteins enhances disease, while vaccination with the major non-structural AMDV protein causes partial protection from disease. Vaccine 1998, 16, 1158–1165. [Google Scholar] [CrossRef]
- Heuer, C.; Healy, A.; Zerbini, C. Economic Effects of Exposure to Bovine Viral Diarrhea Virus on Dairy Herds in New Zealand. J. Dairy Sci. 2007, 90, 5428–5438. [Google Scholar] [CrossRef]
- Roeder, P.; Jeffrey, M.; Cranwell, M. Pestivirus fetopathogenicity in cattle: Changing sequelae with fetal maturation. Vet. Rec. 1986, 118, 44–48. [Google Scholar] [CrossRef]
- Khodakaram-Tafti, A.; Farjanikish, G. Persistent bovine viral diarrhea virus (BVDV) infection in cattle herds. Iran. J. Vet. Res. 2017, 18, 154. [Google Scholar]
- Losinger, W.C. Economic impact of reduced milk production associated with Johne’s disease on dairy operations in the USA. J. Dairy. Res. 2005, 72, 425–432. [Google Scholar] [CrossRef]
- VanLeeuwen, J.; Haddad, J.; Dohoo, I.; Keefe, G.; Tiwari, A.; Tremblay, R. Associations between reproductive performance and seropositivity for bovine leukemia virus, bovine viral-diarrhea virus, Mycobacterium avium subspecies paratuberculosis, and Neospora caninum in Canadian dairy cows. Prev. Vet. Med. 2010, 94, 54–64. [Google Scholar] [CrossRef] [PubMed]
- Urban-Chmiel, R.; Wernicki, A.; Puchalski, A.; Dec, M.; Stęgierska, D.; Grooms, D.L.; Barbu, N.I. Detection of bovine respiratory syncytial virus infections in young dairy and beef cattle in Poland. Vet. Quart. 2015, 35, 33–36. [Google Scholar] [CrossRef] [PubMed]
- Barnard, B.; Gerdes, G.H.; Meiswinkel, R. Some epidemiological and economic aspects of a bluetongue-like disease in cattle in South Africa-1995/96 and 1997. Onderstepoort J. Vet. Res. 1998, 65, 145–151. [Google Scholar] [PubMed]
- Toussaint, J.-F.; Sailleau, C.; Mast, J.; Houdart, P.; Czaplicki, G.; Demeestere, L.; VandenBussche, F.; Van Dessel, W.; Goris, N.; Bréard, E. Bluetongue in Belgium, 2006. Emerg. Infect. Dis. 2007, 13, 614. [Google Scholar] [CrossRef]
- Conraths, F.J.; Gethmann, J.M.; Staubach, C.; Mettenleiter, T.C.; Beer, M.; Hoffmann, B. Epidemiology of bluetongue virus serotype 8, Germany. Emerg. Infect. Dis. 2009, 15, 433. [Google Scholar] [CrossRef]
- Limon, G.; Gamawa, A.A.; Ahmed, A.I.; Lyons, N.A.; Beard, P.M. Epidemiological Characteristics and Economic Impact of Lumpy Skin Disease, Sheeppox and Goatpox Among Subsistence Farmers in Northeast Nigeria. Front. Vet. Sci. 2020, 7, 8. [Google Scholar] [CrossRef]
- Pena, R.N.; Fernández, C.; Blasco-Felip, M.; Fraile, L.J.; Estany, J. Genetic Markers Associated with Field PRRSV-Induced Abortion Rates. Viruses 2019, 11, 706. [Google Scholar] [CrossRef]
- Pils, M.C.; Dreckmann, K.; Jansson, K.; Glage, S.; Held, N.; Sommer, W.; Länger, F.; Avsar, M.; Warnecke, G.; Bleich, A. Mortality Due to Porcine Reproductive and Respiratory Syndrome Virus in Immunocompromised Göttingen Minipigs (Sus scrofa domestica). Comp. Med. 2016, 66, 392–398. [Google Scholar]
- Schlafer, D.; Mebus, C. Abortion in sows experimentally infected with African swine fever virus: Clinical features. Am. J. Vet. Res. 1984, 45, 1353–1360. [Google Scholar]
- Sánchez-Cordón, P.; Montoya, M.; Reis, A.; Dixon, L. African swine fever: A re-emerging viral disease threatening the global pig industry. Vet. J. 2018, 233, 41–48. [Google Scholar] [CrossRef]
- Van Eck, J.; Davelaar, F.; Van Den Heuvel-Plesman, T.A.; Van Kol, N.; Kouwenhoven, B.; Guldie, F. Dropped egg production, soft shelled and shell-less eggs associated with appearance of precipitins to adenovirus in flocks of laying fowls. Avian Dis. 1976, 5, 261–272. [Google Scholar] [CrossRef] [PubMed]
- Sedeik, M.; Elbestawy, A.; El-Shall, N.; Abd El-Hack, M.; Saadeldin, I.; Swelum, A. Comparative efficacy of commercial inactivated Newcastle disease virus vaccines against Newcastle disease virus genotype VII in broiler chickens. Poult. Sci. 2019, 98, 2000–2007. [Google Scholar] [CrossRef] [PubMed]
- Purchase, H. Clinical disease and its economic impact. In Marek’s Disease; Springer: Berlin, Germany, 1985; pp. 17–42. [Google Scholar]
- Biggs, P.M.; Nair, V. The long view: 40 years of Marek’s disease research and Avian Pathology. Avian. Pathol. 2012, 41, 3–9. [Google Scholar] [CrossRef] [PubMed]
- Bastida, F.; Juste, R.A. Paratuberculosis control: A review with a focus on vaccination. J. Immune Based Ther. Vaccines 2011, 9, 8. [Google Scholar] [CrossRef]
- Buchy, P.; Ascioglu, S.; Buisson, Y.; Datta, S.; Nissen, M.; Tambyah, P.A.; Vong, S. Impact of vaccines on antimicrobial resistance. Int. J. Infect. Dis. 2020, 90, 188–196. [Google Scholar] [CrossRef]
- Grubman, M.J.; Baxt, B. Foot-and-mouth disease. Clin. Microbiol. Rev. 2004, 17, 465–493. [Google Scholar] [CrossRef]
- Zientara, S.; MacLachlan, N.J.; Calistri, P.; Sanchez-Vizcaino, J.M.; Savini, G. Bluetongue vaccination in Europe. Expert Rev. Vaccines 2010, 9, 989–991. [Google Scholar] [CrossRef]
- Chen, R.T. Vaccine risks: Real, perceived and unknown. Vaccine 1999, 17, S41–S46. [Google Scholar] [CrossRef]
- Tago, D.; Sall, B.; Lancelot, R.; Pradel, J. VacciCost—A tool to estimate the resource requirements for implementing livestock vaccination campaigns. Application to peste des petits ruminants (PPR) vaccination in Senegal. Prev. Vet. Med. 2017, 144, 13–19. [Google Scholar] [CrossRef]
- Page, S.; Gautier, P. Use of antimicrobial agents in livestock. Rev. Sci. Tech. 2012, 31, 145–188. [Google Scholar] [CrossRef]
- Wang, J.; Deng, Y.; Diao, H. Perceived Risk, Expected Benefits and Pig Farmers’ Behaviors of Veterinary Drug Usage. Int. J. Environ. Res. Public Health 2018, 15, 1716. [Google Scholar] [CrossRef] [PubMed]
- Council, N.R. The Use of Drugs in Food Animals: Benefits and Risks; National Academies Press: Washington, DC, USA, 1999. [Google Scholar]
- Shao, Y.; Li, M.; Zhang, W.; Ji, Y.; Hayes, D.J. World’s Largest Pork Producer in Crisis: China’s African Swine Fever Outbreak. Agric. Policy Rev. 2018, 2018, 1. [Google Scholar]
- Zhang, H.; Kono, H.; Kubota, S. An integrated epidemiological and economic analysis of vaccination against highly pathogenic porcine reproductive and respiratory syndrome (PRRS) in Thua Thien Hue Province, Vietnam. Asian Austral. J. Amin. 2014, 27, 1499. [Google Scholar] [CrossRef] [PubMed]
- Canuti, M.; O’Leary, K.E.; Hunter, B.D.; Spearman, G.; Ojkic, D.; Whitney, H.G.; Lang, A.S. Driving forces behind the evolution of the Aleutian mink disease parvovirus in the context of intensive farming. Virus Evol. 2016, 2, vew004. [Google Scholar] [CrossRef]
- Kashtanov, S.; Salnikova, L. Aleutian mink disease: Epidemiological and genetic aspects. Biol. Bull. Rev. 2018, 8, 104–113. [Google Scholar] [CrossRef]
- Kalds, P.; Zhou, S.; Cai, B.; Liu, J.; Wang, Y.; Petersen, B.; Sonstegard, T.; Wang, X.; Chen, Y. Sheep and Goat Genome Engineering: From Random Transgenesis to the CRISPR Era. Front. Genet. 2019, 10. [Google Scholar] [CrossRef]
- Lillico, S.G.; Proudfoot, C.; King, T.J.; Tan, W.; Zhang, L.; Mardjuki, R.; Paschon, D.E.; Rebar, E.J.; Urnov, F.D.; Mileham, A.J.; et al. Mammalian interspecies substitution of immune modulatory alleles by genome editing. Sci. Rep. 2016, 6, 21645. [Google Scholar] [CrossRef] [PubMed]
- Proudfoot, C.; Lillico, S.; Tait-Burkard, C. Genome editing for disease resistance in pigs and chickens. Anim. Front. 2019, 9, 6–12. [Google Scholar] [CrossRef]
- Ruan, J.; Xu, J.; Chen-Tsai, R.Y.; Li, K. Genome editing in livestock: Are we ready for a revolution in animal breeding industry? Transgenic Res. 2017, 26, 715–726. [Google Scholar] [CrossRef] [PubMed]
- Tait-Burkard, C.; Doeschl-Wilson, A.; McGrew, M.J.; Archibald, A.L.; Sang, H.M.; Houston, R.D.; Whitelaw, C.B.; Watson, M. Livestock 2.0–genome editing for fitter, healthier, and more productive farmed animals. Genome Biol. 2018, 19, 204. [Google Scholar] [CrossRef]
- Van Eenennaam, A.L. Application of genome editing in farm animals: Cattle. Transgenic Res. 2019, 28, 93–100. [Google Scholar] [CrossRef] [PubMed]
- Whitworth, K.M.; Rowland, R.R.; Ewen, C.L.; Trible, B.R.; Kerrigan, M.A.; Cino-Ozuna, A.G.; Samuel, M.S.; Lightner, J.E.; McLaren, D.G.; Mileham, A.J.; et al. Gene-edited pigs are protected from porcine reproductive and respiratory syndrome virus. Nat. Biotechnol. 2016, 34, 20–22. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Wang, Y.; Zhang, Y.; Yang, M.; Lv, J.; Liu, J.; Zhang, Y. TALE nickase-mediated SP110 knockin endows cattle with increased resistance to tuberculosis. Proc. Natl. Acad. Sci. USA 2015, 112, E1530–E1539. [Google Scholar] [CrossRef]
- Van Gorp, H.; Van Breedam, W.; Van Doorsselaere, J.; Delputte, P.L.; Nauwynck, H.J. Identification of the CD163 protein domains involved in infection of the porcine reproductive and respiratory syndrome virus. J. Virol. 2010, 84, 3101–3105. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Zhang, J.; Zhang, X.; Shi, J.; Pan, Y.; Zhou, R.; Li, G.; Li, Z.; Cai, G.; Wu, Z. CD163 knockout pigs are fully resistant to highly pathogenic porcine reproductive and respiratory syndrome virus. Antivir. Res. 2018, 151, 63–70. [Google Scholar] [CrossRef]
- Burkard, C.; Lillico, S.G.; Reid, E.; Jackson, B.; Mileham, A.J.; Ait-Ali, T.; Whitelaw, C.B.A.; Archibald, A.L. Precision engineering for PRRSV resistance in pigs: Macrophages from genome edited pigs lacking CD163 SRCR5 domain are fully resistant to both PRRSV genotypes while maintaining biological function. PLoS Pathog. 2017, 13, e1006206. [Google Scholar] [CrossRef] [PubMed]
- Burkard, C.; Opriessnig, T.; Mileham, A.J.; Stadejek, T.; Ait-Ali, T.; Lillico, S.G.; Whitelaw, C.B.A.; Archibald, A.L. Pigs lacking the scavenger receptor cysteine-rich domain 5 of CD163 are resistant to porcine reproductive and respiratory syndrome virus 1 infection. J. Virol. 2018, 92. [Google Scholar] [CrossRef]
- Murtaugh, M.P.; Stadejek, T.; Abrahante, J.E.; Lam, T.T.; Leung, F.C.-C. The ever-expanding diversity of porcine reproductive and respiratory syndrome virus. Virus Res. 2010, 154, 18–30. [Google Scholar] [CrossRef]
- Velasco-Garcia, M.N.; Mottram, T. Biosensor technology addressing agricultural problems. Biosyst. Eng. 2003, 84, 1–12. [Google Scholar] [CrossRef]
- Luo, Y.; Nartker, S.; Miller, H.; Hochhalter, D.; Wiederoder, M.; Wiederoder, S.; Setterington, E.; Drzal, L.T.; Alocilja, E.C. Surface functionalization of electrospun nanofibers for detecting E. coli O157:H7 and BVDV cells in a direct-charge transfer biosensor. Biosens. Bioelectron. 2010, 26, 1612–1617. [Google Scholar] [CrossRef]
- Montrose, A.; Creedon, N.; Sayers, R.; Barry, S.; O’riordan, A. Novel single gold nanowire-based electrochemical immunosensor for rapid detection of bovine viral diarrhoea antibodies in serum. J. Biosens. Bioelectron. 2015, 6, 1–7. [Google Scholar] [CrossRef]
- Neethirajan, S.; Tuteja, S.K.; Huang, S.T.; Kelton, D. Recent advancement in biosensors technology for animal and livestock health management. Biosens. Bioelectron. 2017, 98, 398–407. [Google Scholar] [CrossRef]
- Neitzel, A.C.; Stamer, E.; Junge, W.; Thaller, G. Calibration of an automated California mastitis test with focus on the device-dependent variation. Springerplus 2014, 3, 760. [Google Scholar] [CrossRef] [PubMed]
- Schaefer, A.L.; Cook, N.J.; Bench, C.; Chabot, J.B.; Colyn, J.; Liu, T.; Okine, E.K.; Stewart, M.; Webster, J.R. The non-invasive and automated detection of bovine respiratory disease onset in receiver calves using infrared thermography. Res. Vet. Sci. 2012, 93, 928–935. [Google Scholar] [CrossRef] [PubMed]
- Tarasov, A.; Gray, D.W.; Tsai, M.Y.; Shields, N.; Montrose, A.; Creedon, N.; Lovera, P.; O’Riordan, A.; Mooney, M.H.; Vogel, E.M. A potentiometric biosensor for rapid on-site disease diagnostics. Biosens. Bioelectron. 2016, 79, 669–678. [Google Scholar] [CrossRef]
- Ye, W.W.; Tsang, M.K.; Liu, X.; Yang, M.; Hao, J. Upconversion luminescence resonance energy transfer (LRET)-based biosensor for rapid and ultrasensitive detection of avian influenza virus H7 subtype. Small 2014, 10, 2390–2397. [Google Scholar] [CrossRef]
- Galyean, M.L.; Eng, K.S. Application of research findings and summary of research needs: Bud Britton Memorial Symposium on Metabolic Disorders of Feedlot Cattle. J. Anim. Sci. 1998, 76, 323–327. [Google Scholar] [CrossRef]
- Reid, G.; Friendship, R. Alternatives to antibiotic use: Probiotics for the gut. Anim. Biotechnol. 2002, 13, 97–112. [Google Scholar] [CrossRef]
- Corcionivoschi, N.; Drinceanu, D.; Pop, I.M.; Stack, D.; Ştef, L.; Julean, C.; Bourke, B. The effect of probiotics on animal health. Anim. Sci. Biotechnol. 2010, 43, 35–41. [Google Scholar]
- Markowiak, P.; Śliżewska, K. The role of probiotics, prebiotics and synbiotics in animal nutrition. Gut Pathog. 2018, 10, 21. [Google Scholar] [CrossRef]
- Roy, A.; Panja, A.S.; Maitra, M.; Bandopyadhyay, B. Isolation and Characterizations of Probiotics from Bovine (Cow) Milk. Res. Rev. Biotechnol. Biosci. 2019, 6, 24–32. [Google Scholar] [CrossRef]
- Rautray, A.K.; Patra, R.; Sardar, K.; Sahoo, G. Potential of probiotics in livestock production. Explor. Anim. Medical Res. 2011, 1, 20–28. [Google Scholar]
- Sanchez, B.; Delgado, S.; Blanco-Miguez, A.; Lourenco, A.; Gueimonde, M.; Margolles, A. Probiotics, gut microbiota, and their influence on host health and disease. Mol. Nutr. Food Res. 2017, 61. [Google Scholar] [CrossRef] [PubMed]
- Haskell, M.J.; Simm, G.; Turner, S.P. Genetic selection for temperament traits in dairy and beef cattle. Front. Genet. 2014, 5, 368. [Google Scholar] [CrossRef]
- Golik, W.; Dameron, O.; Bugeon, J.; Fatet, A.; Hue, I.; Hurtaud, C.; Reichstadt, M.; Salaün, M.-C.; Vernet, J.; Joret, L. ATOL: The multi-species livestock trait ontology. In Proceedings of the 6th Metadata and Semantics Research Conference (MTSR 2012), Cádiz, Spain, 28–30 November 2012; pp. 289–300. [Google Scholar]
- Hughes, L.M.; Bao, J.; Hu, Z.-L.; Honavar, V.; Reecy, J.M. Animal trait ontology: The importance and usefulness of a unified trait vocabulary for animal species. J. Anim. Sci. 2008, 86, 1485–1491. [Google Scholar] [CrossRef]
- Mulder, H. Is GxE a burden or a blessing? Opportunities for genomic selection and big data. J. Anim. Breed. Genet. 2017, 134, 435–436. [Google Scholar] [CrossRef]
- Gonda, M.; Chang, Y.; Shook, G.; Collins, M.; Kirkpatrick, B. Genetic variation of Mycobacterium avium ssp. paratuberculosis infection in US Holsteins. J. Dairy Sci. 2006, 89, 1804–1812. [Google Scholar] [CrossRef]
- Koets, A.; Adugna, G.; Janss, L.; Van Weering, H.; Kalis, C.; Wentink, G.; Rutten, V.; Schukken, Y. Genetic variation of susceptibility to Mycobacterium avium subsp. paratuberculosis infection in dairy cattle. J. Dairy Sci. 2000, 83, 2702–2708. [Google Scholar] [CrossRef]
- Mortensen, H.; Nielsen, S.S.; Berg, P. Genetic variation and heritability of the antibody response to Mycobacterium avium subspecies paratuberculosis in Danish Holstein cows. J. Dairy Sci. 2004, 87, 2108–2113. [Google Scholar] [CrossRef]
- Morris, C.A. A review of genetic resistance to disease in Bos taurus cattle. Vet. J. 2007, 174, 481–491. [Google Scholar] [CrossRef]
- Nicholas, F.W. Veterinary Genetics, 1st ed.; Oxford Science Publications: New York City, NY, USA; Clarendon Press: Oxford, UK, 1987. [Google Scholar]
- McManus, C.; do Prado Paim, T.; de Melo, C.B.; Brasil, B.S.; Paiva, S.R. Selection methods for resistance to and tolerance of helminths in livestock. Parasite 2014, 21, 56. [Google Scholar] [CrossRef] [PubMed]
- Kaplan, R.M. Drug resistance in nematodes of veterinary importance: A status report. Trends Parasitol. 2004, 20, 477–481. [Google Scholar] [CrossRef] [PubMed]
- Sayers, G.; Sweeney, T. Gastrointestinal nematode infection in sheep—A review of the alternatives to anthelmintics in parasite control. Anim. Health Res. Rev. 2005, 6, 159–171. [Google Scholar] [CrossRef] [PubMed]
- Guterbock, W.M.; Van Eenennaam, A.L.; Anderson, R.J.; Gardner, I.A.; Cullor, J.S.; Holmberg, C.A. Efficacy of intramammary antibiotic therapy for treatment of clinical mastitis caused by environmental pathogens. J. Dairy Sci. 1993, 76, 3437–3444. [Google Scholar] [CrossRef]
- Myllys, V.; Asplund, K.; Brofeldt, E.; Hirvelä-Koski, V.; Honkanen-Buzalski, T.; Junttila, J.; Kulkas, L.; Myllykangas, O.; Niskanen, M.; Saloniemi, H. Bovine mastitis in Finland in 1988 and 1995—Changes in prevalence and antimicrobial resistance. Acta Vet. Scand. 1998, 39, 119–126. [Google Scholar] [PubMed]
- Boireau, C.; Cazeau, G.; Jarrige, N.; Calavas, D.; Madec, J.-Y.; Leblond, A.; Haenni, M.; Gay, É. Antimicrobial resistance in bacteria isolated from mastitis in dairy cattle in France, 2006–2016. J. Dairy Sci. 2018, 101, 9451–9462. [Google Scholar] [CrossRef] [PubMed]
- Heringstad, B.; Klemetsdal, G.; Ruane, J. Selection for mastitis resistance in dairy cattle: A review with focus on the situation in the Nordic countries. Livest. Prod. Sci. 2000, 64, 95–106. [Google Scholar] [CrossRef]
- Gibson, J.P.; Bishop, S.C. Use of molecular markers to enhance resistance of livestock to disease: A global approach. Rev. Sci. Tech. 2005, 24, 343–353. [Google Scholar] [CrossRef]
- Abernethy, D.A.; Upton, P.; Higgins, I.M.; McGrath, G.; Goodchild, A.V.; Rolfe, S.J.; Broughan, J.M.; Downs, S.H.; Clifton-Hadley, R.; Menzies, F.D.; et al. Bovine tuberculosis trends in the UK and the Republic of Ireland, 1995-2010. Vet. Rec. 2013, 172, 312. [Google Scholar] [CrossRef]
- Råberg, L.; Graham, A.L.; Read, A.F. Decomposing health: Tolerance and resistance to parasites in animals. Philos. Trans. R. Soc. B Biol. Sci. 2009, 364, 37–49. [Google Scholar] [CrossRef]
- Bourdon, R.M. Understanding Animal Breeding, 2nd ed.; Bourdon, R.M., Ed.; Prentice Hall: Upper Saddler River, NJ, USA, 2014. [Google Scholar]
- Hart, B.L. Behavioural defences in animals against pathogens and parasites: Parallels with the pillars of medicine in humans. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2011, 366, 3406–3417. [Google Scholar] [CrossRef] [PubMed]
- Weigel, K.A.; VanRaden, P.M.; Norman, H.D.; Grosu, H. A 100-Year Review: Methods and impact of genetic selection in dairy cattle-From daughter-dam comparisons to deep learning algorithms. J. Dairy Sci. 2017, 100, 10234–10250. [Google Scholar] [CrossRef] [PubMed]
- Rauw, W.M.; Kanis, E.; Noordhuizen-Stassen, E.N.; Grommers, F.J. Undesirable side effects of selection for high production efficiency in farm animals: A review. Livest. Prod. Sci. 1998, 56, 15–33. [Google Scholar] [CrossRef]
- Emmans, G.; Kyriazakis, I. Consequences of genetic change in farm animals on food intake and feeding behaviour. Proc. Nutr. Soc. 2001, 60, 115–125. [Google Scholar] [CrossRef] [PubMed]
- Heringstad, B.; Østerås, O. More than 30 years of health recording in Norway. ICAR Tech. Ser. 2013, 17, 39–46. [Google Scholar]
- Beavers, L.; Van Doormal, B. Improving Existing Traits and Adding Exciting New Ones. Available online: https://www.cdn.ca/Articles/GEBAPR2016/10_Vision%20of%20%20GE%20Services%20-%20ENGLISH.pdf (accessed on 7 July 2020).
- Miglior, F.; Koeck, A.; Kistemaker, G.; Van Doormaal, B. A New Index for Mastitis Resistance. Available online: https://www.cdn.ca/Articles/GEBMAR2014/DCBGC%20Report_mastitis%20-%20FINAL.pdf (accessed on 7 July 2020).
- Durmaz, A.A.; Karaca, E.; Demkow, U.; Toruner, G.; Schoumans, J.; Cogulu, O. Evolution of genetic techniques: Past, present, and beyond. Biomed. Res. Int. 2015, 2015. [Google Scholar] [CrossRef]
- Gogolin-Ewens, K.; Meeusen, E.; Scott, P.; Adams, T.; Brandon, M. Genetic selection for disease resistance and traits of economic importance in animal production. Rev. Sci. Tech. 1990, 9, 865–896. [Google Scholar] [CrossRef]
- Andersson, L.; Haley, C.S.; Ellegren, H.; Knott, S.A.; Johansson, M.; Andersson, K.; Andersson-Eklund, L.; Edfors-Lilja, I.; Fredholm, M.; Hansson, I.; et al. Genetic mapping of quantitative trait loci for growth and fatness in pigs. Science 1994, 263, 1771–1774. [Google Scholar] [CrossRef]
- Näslund, J.; Fikse, W.; Pielberg, G.; Lundén, A. Frequency and effect of the bovine acyl-CoA: Diacylglycerol acyltransferase 1 (DGAT1) K232A polymorphism in Swedish dairy cattle. J. Dairy. Sci. 2008, 91, 2127–2134. [Google Scholar] [CrossRef]
- Thaller, G.; Kramer, W.; Winter, A.; Kaupe, B.; Erhardt, G.; Fries, R. Effects of DGAT1 variants on milk production traits in German cattle breeds. J. Anim. Sci. 2003, 81, 1911–1918. [Google Scholar] [CrossRef]
- Do, D.; Schenkel, F.; Miglior, F.; Zhao, X.; Ibeagha-Awemu, E. Targeted genotyping to identify potential functional variants associated with cholesterol content in bovine milk. Anim. Genet. 2020, 51, 200–209. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Liu, J.; Sun, D.; Ma, P.; Ding, X.; Yu, Y.; Zhang, Q. Genome wide association studies for milk production traits in Chinese Holstein population. PLoS ONE 2010, 5, e13661. [Google Scholar] [CrossRef] [PubMed]
- Williams, J.L. The use of marker-assisted selection in animal breeding and biotechnology. Rev. Sci. Tech. 2005, 24, 379–391. [Google Scholar] [CrossRef] [PubMed]
- Wakchaure, R.; Ganguly, S.; Praveen, P.; Kumar, A.; Sharma, S.; Mahajan, T. Marker assisted selection (MAS) in animal breeding: A review. Drug. Metab. Toxicol. 2015, 6, e127. [Google Scholar] [CrossRef]
- Ruane, J.; Colleau, J.J. Marker-assisted selection for a sex-limited character in a nucleus breeding population. J. Dairy Sci. 1996, 79, 1666–1678. [Google Scholar] [CrossRef]
- Wolc, A.; Arango, J.; Jankowski, T.; Settar, P.; Fulton, J.E.; O’Sullivan, N.P.; Fernando, R.; Garrick, D.J.; Dekkers, J.C.J.A.D. Genome-wide association study for Marek’s disease mortality in layer chickens. Avian. Dis. 2013, 57, 395–400. [Google Scholar] [CrossRef]
- Meuwissen, T.H.E.; Hayes, B.J.; Goddard, M.E. Prediction of Total Genetic Value Using Genome—Wide Dense Marker Maps. Genetics 2001, 157, 1819. [Google Scholar]
- Piccoli, M.L.; Brito, L.F.; Braccini, J.; Brito, F.V.; Cardoso, F.F.; Cobuci, J.A.; Sargolzaei, M.; Schenkel, F.S. A comprehensive comparison between single-and two-step GBLUP methods in a simulated beef cattle population. Can. J. Anim. Sci. 2018, 98, 565–575. [Google Scholar] [CrossRef]
- Goddard, M.E.; Hayes, B.J.; Meuwissen, T.H. Genomic selection in livestock populations. Genet. Res. 2010, 92, 413–421. [Google Scholar] [CrossRef]
- Miar, Y.; Plastow, G.; Wang, Z. Genomic selection, a new era for pork quality Improvement. Springer Sci. Rev. 2015, 3, 27–37. [Google Scholar] [CrossRef]
- Meuwissen, T.; Hayes, B.; Goddard, M. Accelerating improvement of livestock with genomic selection. Annu. Rev. Anim. Biosci. 2013, 1, 221–237. [Google Scholar] [CrossRef]
- Bishop, S.; Morris, C. Genetics of disease resistance in sheep and goats. Small. Rumin. Res. 2007, 70, 48–59. [Google Scholar] [CrossRef]
- Martin, P.; Barkema, H.W.; Brito, L.F.; Narayana, S.G.; Miglior, F. Symposium review: Novel strategies to genetically improve mastitis resistance in dairy cattle. J. Dairy. Sci. 2018, 101, 2724–2736. [Google Scholar] [CrossRef] [PubMed]
- Weigel, K.A.; Shook, G.E. Genetic Selection for Mastitis Resistance. Vet. Clin. Food Anim. Pract. 2018, 34, 457–472. [Google Scholar] [CrossRef] [PubMed]
- Fang, L.; Sahana, G.; Ma, P.; Su, G.; Yu, Y.; Zhang, S.; Lund, M.S.; Sørensen, P. Exploring the genetic architecture and improving genomic prediction accuracy for mastitis and milk production traits in dairy cattle by mapping variants to hepatic transcriptomic regions responsive to intra-mammary infection. Genet. Sel. Evol. 2017, 49, 44. [Google Scholar] [CrossRef]
- Schneider, M.; Tait Jr, R.G.; Ruble, M.V.; Busby, W.D.; Reecy, J.M. Evaluation of fixed sources of variation and estimation of genetic parameters for incidence of bovine respiratory disease in preweaned calves and feedlot cattle. J. Anim. Sci. 2010, 88, 1220–1228. [Google Scholar] [CrossRef]
- Snowder, G.; Van Vleck, L.D.; Cundiff, L.; Bennett, G. Influence of breed, heterozygosity, and disease incidence on estimates of variance components of respiratory disease in preweaned beef calves. J. Anim. Sci. 2005, 83, 1247–1261. [Google Scholar] [CrossRef]
- Boddicker, N.; Waide, E.H.; Rowland, R.; Lunney, J.K.; Garrick, D.J.; Reecy, J.M.; Dekkers, J. Evidence for a major QTL associated with host response to porcine reproductive and respiratory syndrome virus challenge. J. Anim. Sci. 2012, 90, 1733–1746. [Google Scholar] [CrossRef]
- Cheng, H.; Niikura, M.; Kim, T.; Mao, W.; MacLea, K.S.; Hunt, H.; Dodgson, J.; Burnside, J.; Morgan, R.; Ouyang, M.; et al. Using integrative genomics to elucidate genetic resistance to Marek’s disease in chickens. Dev. Biol. 2008, 132, 365–372. [Google Scholar] [CrossRef]
- Heifetz, E.M.; Fulton, J.E.; O’Sullivan, N.P.; Arthur, J.A.; Cheng, H.; Wang, J.; Soller, M.; Dekkers, J.C. Mapping QTL affecting resistance to Marek’s disease in an F6 advanced intercross population of commercial layer chickens. BMC Genom. 2009, 10, 20. [Google Scholar] [CrossRef]
- McElroy, J.P.; Dekkers, J.; Fulton, J.; O’Sullivan, N.P.; Soller, M.; Lipkin, E.; Zhang, W.; Koehler, K.J.; Lamont, S.J.; Cheng, H. Microsatellite markers associated with resistance to Marek’s disease in commercial layer chickens. Poult. Sci. 2005, 84, 1678–1688. [Google Scholar] [CrossRef] [PubMed]
- Vallejo, R.L.; Bacon, L.D.; Liu, H.-C.; Witter, R.L.; Groenen, M.A.; Hillel, J.; Cheng, H.H. Genetic mapping of quantitative trait loci affecting susceptibility to Marek’s disease virus induced tumors in F2 intercross chickens. Genetics 1998, 148, 349–360. [Google Scholar] [PubMed]
- Yonash, N.; Bacon, L.; Witter, R.; Cheng, H. High resolution mapping and identification of new quantitative trait loci (QTL) affecting susceptibility to Marek’s disease. Anim. Genet. 1999, 30, 126–135. [Google Scholar] [CrossRef]
- Medzhitov, R.; Schneider, D.S.; Soares, M.P. Disease tolerance as a defense strategy. Science 2012, 335, 936–941. [Google Scholar] [CrossRef] [PubMed]
- Lough, G.; Rashidi, H.; Kyriazakis, I.; Dekkers, J.C.; Hess, A.; Hess, M.; Deeb, N.; Kause, A.; Lunney, J.K.; Rowland, R.R. Use of multi-trait and random regression models to identify genetic variation in tolerance to porcine reproductive and respiratory syndrome virus. Genet. Sel. Evol. 2017, 49, 37. [Google Scholar] [CrossRef]
- Zanella, R.; Settles, M.; McKay, S.; Schnabel, R.; Taylor, J.; Whitlock, R.; Schukken, Y.; Van Kessel, J.; Smith, J.; Neibergs, H. Identification of loci associated with tolerance to Johne’s disease in Holstein cattle. Anim. Genet. 2011, 42, 28–38. [Google Scholar] [CrossRef] [PubMed]
- Hanotte, O.; Ronin, Y.; Agaba, M.; Nilsson, P.; Gelhaus, A.; Horstmann, R.; Sugimoto, Y.; Kemp, S.; Gibson, J.; Korol, A.; et al. Mapping of quantitative trait loci controlling trypanotolerance in a cross of tolerant West African N’Dama and susceptible East African Boran cattle. Proc. Natl. Acad. Sci. USA 2003, 100, 7443–7448. [Google Scholar] [CrossRef]
- Phua, S.H.; Hyndman, D.L.; Baird, H.J.; Auvray, B.; McEwan, J.C.; Lee, M.A.; Dodds, K.G. Towards genomic selection for facial eczema disease tolerance in the New Zealand sheep industry. Anim. Genet. 2014, 45, 559–564. [Google Scholar] [CrossRef]
- Restif, O.; Koella, J.C. Concurrent evolution of resistance and tolerance to pathogens. Am. Nat. 2004, 164, E90–E102. [Google Scholar] [CrossRef]
- Roy, B.; Kirchner, J. Evolutionary dynamics of pathogen resistance and tolerance. Evolution 2000, 54, 51–63. [Google Scholar] [CrossRef]
- Drake, J.W.; Holland, J.J. Mutation rates among RNA viruses. Proc. Natl. Acad. Sci. USA 1999, 96, 13910–13913. [Google Scholar] [CrossRef]
- Ayres, J.S.; Schneider, D.S. Tolerance of infections. Annu. Rev. Immunol. 2012, 30, 271–294. [Google Scholar] [CrossRef] [PubMed]
- Raberg, L.; Sim, D.; Read, A.F. Disentangling genetic variation for resistance and tolerance to infectious diseases in animals. Science 2007, 318, 812–814. [Google Scholar] [CrossRef]
- Colditz, I.G.; Hine, B.C. Resilience in farm animals: Biology, management, breeding and implications for animal welfare. Anim. Prod. Sci. 2016, 56, 1961–1983. [Google Scholar] [CrossRef]
- Elgersma, G.G.; de Jong, G.; van der Linde, R.; Mulder, H.A. Fluctuations in milk yield are heritable and can be used as a resilience indicator to breed healthy cows. J. Dairy Sci. 2018, 101, 1240–1250. [Google Scholar] [CrossRef] [PubMed]
- Albers, G.A.; Gray, G.D.; Piper, L.R.; Barker, J.S.; Le Jambre, L.F.; Barger, I.A. The genetics of resistance and resilience to Haemonchus contortus infection in young merino sheep. Int. J. Parasitol. 1987, 17, 1355–1363. [Google Scholar] [CrossRef]
- Bisset, S.A.; Morris, C.A. Feasibility and implications of breeding sheep for resilience to nematode challenge. Int. J. Parasitol. 1996, 26, 857–868. [Google Scholar] [CrossRef]
- Mulder, H.A.; Rashidi, H. Selection on resilience improves disease resistance and tolerance to infections. J. Anim. Sci. 2017, 95, 3346–3358. [Google Scholar] [CrossRef]
- Berghof, T.V.L.; Poppe, M.; Mulder, H.A. Opportunities to Improve Resilience in Animal Breeding Programs. Front. Genet. 2018, 9, 692. [Google Scholar] [CrossRef]
- Abdel-Azim, G.A.; Freeman, A.E.; Kehrli, M.E., Jr.; Kelm, S.C.; Burton, J.L.; Kuck, A.L.; Schnell, S. Genetic basis and risk factors for infectious and noninfectious diseases in US Holsteins. I. Estimation of genetic parameters for single diseases and general health. J. Dairy Sci. 2005, 88, 1199–1207. [Google Scholar] [CrossRef]
- Thompson-Crispi, K.A.; Sewalem, A.; Miglior, F.; Mallard, B.A. Genetic parameters of adaptive immune response traits in Canadian Holsteins. J. Dairy Sci. 2012, 95, 401–409. [Google Scholar] [CrossRef] [PubMed]
- Henryon, M.; Heegaard, P.M.; Nielsen, J.; Berg, P.; Juul-Madsen, H.R. Immunological traits have the potential to improve selection of pigs for resistance to clinical and subclinical disease. Anim. Sci. 2006, 82, 597–606. [Google Scholar] [CrossRef]
- Psifidi, A.; Banos, G.; Matika, O.; Desta, T.T.; Bettridge, J.; Hume, D.A.; Dessie, T.; Christley, R.; Wigley, P.; Hanotte, O. Genome-wide association studies of immune, disease and production traits in indigenous chicken ecotypes. Genet. Sel. Evol. 2016, 48, 74. [Google Scholar] [CrossRef]
- Stear, M.J.; Bishop, S.C.; Mallard, B.A.; Raadsma, H. The sustainability, feasibility and desirability of breeding livestock for disease resistance. Res. Vet. Sci. 2001, 71, 1–7. [Google Scholar] [CrossRef]
- Jie, H.; Liu, Y.-P. Breeding for disease resistance in poultry: Opportunities with challenges. Worlds Poult. Sci. J. 2011, 67, 687–696. [Google Scholar] [CrossRef]
- Schulman, N.F.; Viitala, S.M.; de Koning, D.J.; Virta, J.; Maki-Tanila, A.; Vilkki, J.H. Quantitative trait Loci for health traits in Finnish Ayrshire cattle. J. Dairy Sci. 2004, 87, 443–449. [Google Scholar] [CrossRef]
- Simianer, H.; Solbu, H.; Schaeffer, L. Estimated genetic correlations between disease and yield traits in dairy cattle. J. Dairy Sci. 1991, 74, 4358–4365. [Google Scholar] [CrossRef]
- Van Dorp, T.; Dekkers, J.; Martin, S.; Noordhuizen, J. Genetic parameters of health disorders, and relationships with 305-day milk yield and conformation traits of registered Holstein cows. J. Dairy Sci. 1998, 81, 2264–2270. [Google Scholar] [CrossRef]
- Emanuelson, U. Recording of production diseases in cattle and possibilities for genetic improvements: A review. Livest. Prod. Sci. 1988, 20, 89–106. [Google Scholar] [CrossRef]
- Li, Z.; Nestor, K.E.; Saif, Y.M.; Anderson, J.W.; Patterson, R.A. Effect of selection for increased body weight in turkeys on lymphoid organ weights, phagocytosis, and antibody responses to fowl cholera and Newcastle disease-inactivated vaccines. Poult. Sci. 2001, 80, 689–694. [Google Scholar] [CrossRef]
- van der Most, P.J.; de Jong, B.; Parmentier, H.K.; Verhulst, S. Trade-off between growth and immune function: A meta-analysis of selection experiments. Funct. Ecol. 2011, 25, 74–80. [Google Scholar] [CrossRef]
- Hazel, L.N. The genetic basis for constructing selection indexes. Genetics 1943, 28, 476–490. [Google Scholar] [PubMed]
- Hirooka, H. Economic selection index in the genomic era. J. Anim. Breed. Genet. 2019, 136, 151–152. [Google Scholar] [CrossRef] [PubMed]
- Snowder, G. Genetic selection for disease resistance: Challenges and opportunities. In Proceedings of the Beef Improvement Federation Conference Proceedings, Omaha, NE, USA, 10–13 July 2002; pp. 52–60. [Google Scholar]
- Nardone, A.; Ronchi, B.; Lacetera, N.; Ranieri, M.S.; Bernabucci, U. Effects of climate changes on animal production and sustainability of livestock systems. Livest. Sci. 2010, 130, 57–69. [Google Scholar] [CrossRef]
- Meuwissen, T.; Hayes, B.; Goddard, M. Genomic selection: A paradigm shift in animal breeding. Anim. Front. 2016, 6, 6–14. [Google Scholar] [CrossRef]
- Asokan, G.V.; Asokan, V. Leveraging “big data” to enhance the effectiveness of “one health” in an era of health informatics. J. Epidemiol. Glob. Health 2015, 5, 311–314. [Google Scholar] [CrossRef]
- Normandeau, K. Beyond Volume, Variety and velocity Is the Issue of Big Data Veracity. Available online: http://insidebigdata.com/2013/09/12/beyond-volume-variety-velocity-issue-big-data-veracity/ (accessed on 7 July 2020).
- Berckmans, D. General introduction to precision livestock farming. Anim. Front. 2017, 7, 6–11. [Google Scholar] [CrossRef]
- Ip, R.H.; Ang, L.-M.; Seng, K.P.; Broster, J.; Pratley, J. Big data and machine learning for crop protection. Comput. Electron. Agric. 2018, 151, 376–383. [Google Scholar] [CrossRef]
- Bansal, S.; Chowell, G.; Simonsen, L.; Vespignani, A.; Viboud, C. Big data for infectious disease surveillance and modeling. J. Infect. Dis. 2016, 214, S375–S379. [Google Scholar] [CrossRef]
- Koltes, J.E.; Cole, J.B.; Clemmens, R.; Dilger, R.N.; Kramer, L.M.; Lunney, J.K.; McCue, M.E.; McKay, S.D.; Mateescu, R.G.; Murdoch, B.M. A vision for development and utilization of high-throughput phenotyping and big data analytics in livestock. Front. Genet. 2019, 10. [Google Scholar] [CrossRef]
- Rexroad, C.; Vallet, J.; Matukumalli, L.K.; Reecy, J.; Bickhart, D.; Blackburn, H.; Boggess, M.; Cheng, H.; Clutter, A.; Cockett, N. Genome to phenome: Improving animal health, production, and well-being–a new USDA blueprint for animal genome research 2018–2027. Front. Genet. 2019, 10, 327. [Google Scholar] [CrossRef] [PubMed]
- Halachmi, I.; Guarino, M.; Bewley, J.; Pastell, M. Smart animal agriculture: Application of real-time sensors to improve animal well-being and production. Annu. Rev. Anim. Biosci. 2019, 7, 403–425. [Google Scholar] [CrossRef] [PubMed]
- Khatun, M.; Thomson, P.C.; Clark, C.E.; García, S.C. Prediction of quarter level subclinical mastitis by combining in-line and on-animal sensor data. Anim. Prod. Sci. 2020, 60, 180–186. [Google Scholar] [CrossRef]
- Cole, J.B.; Eaglen, S.A.E.; Maltecca, C.; Mulder, H.A.; Pryce, J.E. The future of phenomics in dairy cattle breeding. Anim. Front. 2020, 10, 37–44. [Google Scholar] [CrossRef] [PubMed]
- Morota, G.; Jarquin, D.; Campbell, M.T.; Iwata, H. Statistical methods for the quantitative genetic analysis of high-throughput phenotyping data. arXiv 2019, arXiv:1904.12341. [Google Scholar]
- Morota, G.; Ventura, R.V.; Silva, F.F.; Koyama, M.; Fernando, S.C. Big Data Analytics and Precision Animal Agriculture Symposium: Machine learning and data mining advance predictive big data analysis in precision animal agriculture. J. Anim. Sci. 2018, 96, 1540–1550. [Google Scholar] [CrossRef] [PubMed]
- Ibeagha-Awemu, E.M.; Do, D.N.; Dudemaine, P.-L.; Fomenky, B.E.; Bissonnette, N. Integration of lncRNA and mRNA transcriptome analyses reveals genes and pathways potentially involved in calf intestinal growth and development during the early weeks of life. Genes 2018, 9, 142. [Google Scholar] [CrossRef]
- Suravajhala, P.; Kogelman, L.J.; Kadarmideen, H.N. Multi-omic data integration and analysis using systems genomics approaches: Methods and applications in animal production, health and welfare. Genet. Sel. Evol. 2016, 48, 38. [Google Scholar] [CrossRef]
- Ibeagha-Awemu, E.M.; Peters, S.O.; Bemji, M.N.; Adeleke, M.A.; Do, D.N. Leveraging available resources and stakeholder involvement for improved productivity of African livestock in the era of genomic breeding. Front. Genet. 2019, 10. [Google Scholar] [CrossRef]
- Giuffra, E.; Tuggle, C.K.; Consortium, F. Functional annotation of animal genomes (FAANG): Current achievements and roadmap. Annu. Rev. Anim. Biosci. 2019, 7, 65–88. [Google Scholar] [CrossRef]
- Hayes, B.J.; Daetwyler, H.D. 1000 Bull Genomes Project to Map Simple and Complex Genetic Traits in Cattle: Applications and Outcomes. Annu. Rev. Anim. Biosci. 2019, 7, 89–102. [Google Scholar] [CrossRef] [PubMed]
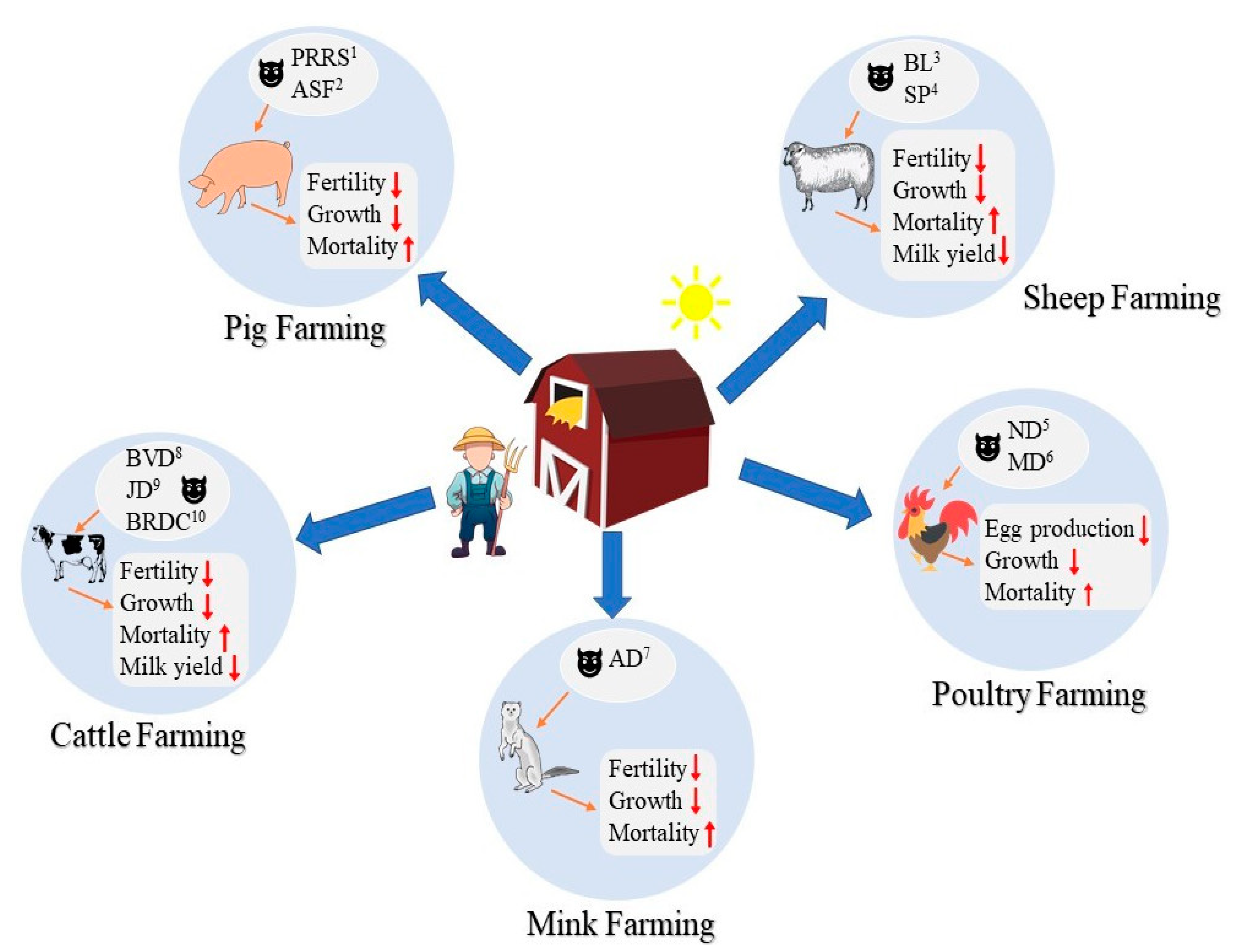
| Species | Disease | Prevalence | Economic Losses | Milk Yield | Fertility/Egg Production | Growth Rate | Mortality | Vaccine Available? | Specific Treatment? |
|---|---|---|---|---|---|---|---|---|---|
| Cattle | Bovine Viral Diarrhea | Up to 98.5% and 98.3% in non-vaccinated dairy and beef herds, respectively [15] | 40–100 thousand USD per herd in Canada [16] and 10–40 million USD per million calvings in Europe [17] | Reduced (~0.074 kg/day [61]) | Reduced (21% abortion rate [62]) | Reduced | High (~50% in calves [63]) | Yes | No |
| Johne’s Disease | 68.1% of US dairy operations were infected [20] | 15 million CAD annually in Canada and 200–250 million USD in US [21] | Reduced (up to 25% [64]) | Reduced (7% lower rate of conception [65]) | Reduced | Culling infected individuals | Yes | No | |
| Bovine Respiratory Disease Complex | 45.9% in UK dairy heifers | One billion USD annually in US [24] | N/A | N/A | Reduced | Moderate (~20% in calves [66]) | Yes | No | |
| Sheep | Bluetongue | 19% in Italy [26] and up to 94.3% in Sudan [27] | In 2007, 12.6 million euros in the Netherlands [28] | Reduced (up to 42% [67]) | Reduced (25% abortion rate and 50% decrease in fertility [68]) | Reduced | High (up to 41.5% [69]) | Yes | No |
| Sheeppox | Up to 22% [31] in India and 40% in Ethiopia [32] | 2.4 million USD annually in Maharashtra, India [33] | N/A | N/A | Reduced | High (up to 40% [70]) | Yes | No | |
| Swine | Porcine Reproductive and Respiratory Syndrome | Up to 48% of pig farms in Ontario, Canada [35] | 664 million USD annually in US [36] and 130 million CAD annually in Canada [37] | N/A | Reduced (up to 40% abortion rate [71]) | Reduced | High (up to 100% [72]) | Yes | No |
| African Swine Fever | 12.5% in China from August 2018 to July 2019 [41] | 1.25 billion USD from 2007 to 2017 in Russia [42] | N/A | Reduced (54% abortion rate [73]) | N/A | High (30–70% [74]) | No | No | |
| Poultry | Newcastle Disease | 85.2% in eastern North America between 2009 and 2011 [44] | 200 million USD from 2002 to 2003 in California, US [45] | N/A | Reduced (55% of egg production [75]) | Reduced | High (up to 100% [76]) | Yes | No |
| Marek’s Disease | 49.5% in Iraq [49] | 1–2 billion USD annually worldwide [52] | N/A | Reduced (decrease 5% egg production [77]) | Reduced | Moderate (10–30% [78]) | Yes | No | |
| Mink | Aleutian Disease | Up to 71% in Nova Scotia, Canada between 1998 and 2005 [57] | 10 million USD in Denmark during 1984 [57] | N/A | Reduced fertility (~2.5 kits per whelping [56]) | Reduced | High (30–100% [55]) | No | No |
| Controlling Method | Advantages | Disadvantages |
|---|---|---|
| Vaccination |
|
|
| Medical treatment |
|
|
| Culling |
|
|
| Genome editing |
|
|
| Biosensor |
|
|
| Probiotics |
|
|
| Species | Number of Publications | Number of Traits | Overall | Health | Disease Suppressibility | Immune Capacity | Pathogens and Parasites | Blood Parameters |
|---|---|---|---|---|---|---|---|---|
| Cattle | 1001 | 646 | 142,261 | 6380 | 2771 | 232 | 124 | 355 |
| Chicken | 328 | 430 | 12,246 | 820 | 739 | NA | NA | 294 |
| Horse | 94 | 56 | 2446 | 1128 | 1026 | 19 | NA | 1 |
| Swine | 698 | 691 | 30,580 | 6598 | 586 | 3230 | 81 | 2747 |
| Sheep | 173 | 262 | 3305 | 619 | 135 | 39 | 335 | 37 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hu, G.; Do, D.N.; Gray, J.; Miar, Y. Selection for Favorable Health Traits: A Potential Approach to Cope with Diseases in Farm Animals. Animals 2020, 10, 1717. https://doi.org/10.3390/ani10091717
Hu G, Do DN, Gray J, Miar Y. Selection for Favorable Health Traits: A Potential Approach to Cope with Diseases in Farm Animals. Animals. 2020; 10(9):1717. https://doi.org/10.3390/ani10091717
Chicago/Turabian StyleHu, Guoyu, Duy Ngoc Do, Janine Gray, and Younes Miar. 2020. "Selection for Favorable Health Traits: A Potential Approach to Cope with Diseases in Farm Animals" Animals 10, no. 9: 1717. https://doi.org/10.3390/ani10091717
APA StyleHu, G., Do, D. N., Gray, J., & Miar, Y. (2020). Selection for Favorable Health Traits: A Potential Approach to Cope with Diseases in Farm Animals. Animals, 10(9), 1717. https://doi.org/10.3390/ani10091717







